Abstract
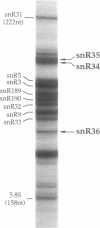
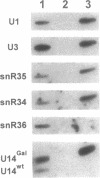
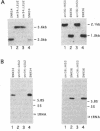
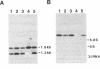
Genes for three novel snRNAs of Saccharomyces cerevisiae have been isolated, sequenced and tested for essentiality. The RNAs encoded by these genes are designated snR34, snR35 and snR36 respectively and contain 203, 204 and 182 nucleotides. Each RNA is derived from a single copy gene and all three RNAs are believed to be nucleolar, i.e. snoRNAs, based on extraction properties and association with fibrillarin. SnR34 and snR35 contain a trimethylguanosine cap, but this feature is absent from snR36. The novel RNAs lack elements conserved among several other snoRNAs, including box C, box D and long sequence complementarities with rRNA. Genetic disruption analyses showed each of the RNAs to be dispensable and a haploid strain lacking all three RNAs and a previously characterized fourth snoRNA (snR33) is also viable. No differences in the levels of precursors or mature rRNAs were apparent in the four gene knock-out strain. Possible roles for the new RNAs in ribosome biogenesis are discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balakin A. G., Lempicki R. A., Huang G. M., Fournier M. J. Saccharomyces cerevisiae U14 small nuclear RNA has little secondary structure and appears to be produced by post-transcriptional processing. J Biol Chem. 1994 Jan 7;269(1):739–746. [PubMed] [Google Scholar]
- Balakin A. G., Schneider G. S., Corbett M. S., Ni J., Fournier M. J. SnR31, snR32, and snR33: three novel, non-essential snRNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 1993 Nov 25;21(23):5391–5397. doi: 10.1093/nar/21.23.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berben G., Dumont J., Gilliquet V., Bolle P. A., Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991 Jul;7(5):475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- Clayton D. A. A nuclear function for RNase MRP. Proc Natl Acad Sci U S A. 1994 May 24;91(11):4615–4617. doi: 10.1073/pnas.91.11.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar T., Tollervey D. Identification and functional analysis of a novel yeast small nucleolar RNA. Nucleic Acids Res. 1993 Nov 25;21(23):5386–5390. doi: 10.1093/nar/21.23.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darr S. C., Brown J. W., Pace N. R. The varieties of ribonuclease P. Trends Biochem Sci. 1992 May;17(5):178–182. doi: 10.1016/0968-0004(92)90262-8. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Kiss T. Structure and function of nucleolar snRNPs. Mol Biol Rep. 1993 Aug;18(2):149–156. doi: 10.1007/BF00986770. [DOI] [PubMed] [Google Scholar]
- Fournier M. J., Maxwell E. S. The nucleolar snRNAs: catching up with the spliceosomal snRNAs. Trends Biochem Sci. 1993 Apr;18(4):131–135. doi: 10.1016/0968-0004(93)90020-n. [DOI] [PubMed] [Google Scholar]
- Goebl M. G., Petes T. D. Most of the yeast genomic sequences are not essential for cell growth and division. Cell. 1986 Sep 26;46(7):983–992. doi: 10.1016/0092-8674(86)90697-5. [DOI] [PubMed] [Google Scholar]
- Jones E. W. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977 Jan;85(1):23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Marshallsay C., Filipowicz W. 7-2/MRP RNAs in plant and mammalian cells: association with higher order structures in the nucleolus. EMBO J. 1992 Oct;11(10):3737–3746. doi: 10.1002/j.1460-2075.1992.tb05459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrer K., Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- Li H. D., Zagorski J., Fournier M. J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Mar;10(3):1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W., Tollervey D., Séraphin B. Small nuclear RNAs in messenger RNA and ribosomal RNA processing. FASEB J. 1993 Jan;7(1):47–53. doi: 10.1096/fasebj.7.1.8422974. [DOI] [PubMed] [Google Scholar]
- Maxwell E. S., Fournier M. J. The small nucleolar RNAs. Annu Rev Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- Newman A. Small nuclear RNAs and pre-mRNA splicing. Curr Opin Cell Biol. 1994 Jun;6(3):360–367. doi: 10.1016/0955-0674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Parker R., Simmons T., Shuster E. O., Siliciano P. G., Guthrie C. Genetic analysis of small nuclear RNAs in Saccharomyces cerevisiae: viable sextuple mutant. Mol Cell Biol. 1988 Aug;8(8):3150–3159. doi: 10.1128/mcb.8.8.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S., Durovic P., Dennis P. P. Ribosomal RNA precursor processing by a eukaryotic U3 small nucleolar RNA-like molecule in an archaeon. Science. 1995 May 19;268(5213):1056–1060. doi: 10.1126/science.7538698. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B. Novel intron-encoded small nucleolar RNAs. Cell. 1993 Nov 5;75(3):403–405. doi: 10.1016/0092-8674(93)90374-y. [DOI] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987 Dec 20;6(13):4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E., Keller W. The biochemistry of 3'-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- Zubenko G. S., Mitchell A. P., Jones E. W. Mapping of the proteinase b structural gene PRB1, in Saccharomyces cerevisiae and identification of nonsense alleles within the locus. Genetics. 1980 Sep;96(1):137–146. doi: 10.1093/genetics/96.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]