Abstract
Cellular prion protein (PRNP) is a glycoprotein involved in the pathogenesis of transmissible spongiform encephalopathies (TSEs). Although the physiological function of PRNP is largely unknown, its key role in prion infection has been extensively documented. This study examines the functionality of PRNP during the course of embryoid body (EB) differentiation in mouse Prnp-null (KO) and WT embryonic stem cell (ESC) lines. The first feature observed was a new population of EBs that only appeared in the KO line after 5 days of differentiation. These EBs were characterized by their expression of several primordial germ cell (PGC) markers until Day 13. In a comparative mRNA expression analysis of genes playing an important developmental role during ESC differentiation to EBs, Prnp was found to participate in the transcription of a key pluripotency marker such as Nanog. A clear switching off of this gene on Day 5 was observed in the KO line as opposed to the WT line, in which maximum Prnp and Nanog mRNA levels appeared at this time. Using a specific antibody against PRNP to block PRNP pathways, reduced Nanog expression was confirmed in the WT line. In addition, antibody-mediated inhibition of ITGB5 (integrin αvβ5) in the KO line rescued the low expression of Nanog on Day 5, suggesting the regulation of Nanog transcription by Prnp via this Itgb5. mRNA expression analysis of the PRNP-related proteins PRND (Doppel) and SPRN (Shadoo), whose PRNP function is known to be redundant, revealed their incapacity to compensate for the absence of PRNP during early ESC differentiation. Our findings provide strong evidence for a relationship between Prnp and several key pluripotency genes and attribute Prnp a crucial role in regulating self-renewal/differentiation status of ESC, confirming the participation of PRNP during early embryogenesis.
Introduction
Cellular prion protein (PRNP) is a ubiquitous membrane glycoprotein whose abnormal self-replicating, misfolded form is widely believed to cause several central nervous system disorders, collectively known as Transmissible Spongiform Encephalopathies (TSE) [1]. PRNP is expressed in a broad range of vertebrate tissues such as spleen, lymph nodes, lung, heart, kidney, muscle and mammary glands [2], [3]. It is also found in the gonads (spermatogenic cells and ovaries) [4] but most abundantly occurs in the central nervous system. In embryogenesis, mouse Prnp mRNA is first highly expressed, mostly in the differentiating neuroepithelium, between E8.5 and E9, during the transition from anaerobic to oxidative metabolism [5] and thereafter expands to non-neuronal tissues at E13.5 [6]. Coupled to the fact that PRNP is highly conserved across species, these data point to an important role for this protein. However, actual evidences exist that make PRNP an absolute mystery. These evidences are that Prnp-knockout mice [5], [7], [8], cattle [9] and goats [10] show no drastically modified developmental phenotype and only display subtle changes such as synaptic transmission abnormalities [11], [12], disturbed morphology [13], some alterations in circadian rhythm [14] and cognitive deficiency [15], [16], [17], suggesting mechanisms able to withstand the absence of the protein. Similar observations were made when this gene was inhibited in adult neurons [18]. To explain these data, it has been hypothesized that another host-encoded protein is able to compensate for the lack of PRNP [19]. The main candidates for this compensating role are the PRNP-related proteins Doppel (PRND) and Shadoo (SPRN). The postembryonic expression of Doppel is restricted to the male testis, though Prnd mRNA has been detected during embryogenesis. Curiously, the absence of Doppel was observed not to provoke alterations in embryonic and postnatal development, but its deficiency causes male infertility in mice [20]. In contrast, Shadoo shows overlapping mRNA expression with Prnp in the brain and has revealed neuroprotective properties at this site. Shadoo has also been attributed a role in embryogenesis such that its downregulation in a Prnp-null embryo invariably gives rise to a lethal phenotype between E8 and E11 [19]. Hence, both these proteins play a crucial role in mammalian embryogenesis and could explain the lack of a severe phenotype in PRNP-knockout mammals as a step towards deciphering the biological role of this protein.
The expression of Prnp in embryonic stem cells (ESC) and immortalized cells has also been addressed. Thus, the expression of Prnp in mouse ESC is 5 times lower than its expression in the brain, yet is 1.5 times higher than its expression in somatic cells [21]. Moreover, during the immortalization of fibroblast MJ90 cells by Tert transfection, Prnp expression is almost three times greater [22]. In mouse ESC, Prnp is again elevated 1.4 and 6.2 times during the first two weeks of differentiation [23] while during their guided cardiogenic differentiation, the expression of the gene is more than 20-fold higher [24]. Curiously, direct reprogramming of somatic cells (MEFs/B-cells) to a pluripotent state (induced pluripotent stem cells, iPS) increases the expression of Prnp up to 27-fold [25]. Recently, several studies have associated PRNP with pluripotency. For example, PRNP has been demonstrated to be a marker of haematopoietic stem cells, supporting their self-renewal [26]. Other data indicate that PRNP may be implicated in the biology of glioblastoma, breast cancer, prostate and gastric cancer [27], [28] or, in other words, PRNP is involved in long term proliferation as are stem cells. In addition, it has been reported that the absence of Prnp significantly slows the regeneration process of acutely damaged hind-limb tibialis anterior muscles of mice compared to wild-type muscles, affecting the myogenic precursor cells [29]. Collectively, these findings reveal a physiological function of PRNP in immortalized pluripotent ESC.
Despite advances made to date, the real function of PRNP is still unclear with ascribed roles in neuroprotection, the response to oxidative stress, cell proliferation and differentiation, synaptic function and signal transduction [30]. The study of the physiological functions of PRNP seems to be key to understanding both prion diseases and its possible implications in development and cancer biology. Given the widely described similarity of embryoid bodies (EBs) and early postimplantation embryos, we selected EBs as an ideal model [31]. Herein, by comparing stem cell differentiation to EBs in Prnp-KO and WT mouse cell lines, we identify the first non-redundant function of PRNP in ESC differentiation during early embryogenesis along with a relationship between the expression of Prnp and that of several key pluripotency markers.
Materials and Methods
Embryo Collection
Mice were kept on a 14-h light/10-h dark cycle. 10-week-old 129/OLA Prnp knock-out female mice were superovulated by intraperitoneal injections of 10 IU of pregnant mare serum gonadotropin (PMSG) (Folligon; Intervet, Boxmeer, Holland) followed 48 hours later by 10 IU of hCG (Chorulon; Intervet). On the day of hCG injection designated Day 0, the female mice were paired with males of the same strain to allow mating. After disinfecting with 70% alcohol and opening the abdominal wall, the oviducts were excised by clamping, dissecting the peritoneum and fat between the ovary and tube, and cutting the whole oviduct from the proximal end. After washing and flushing the oviduct from the proximal end by incising the whole oviduct with a 30-gauge needle, two-cell embryos were selected by 100x microscopy. All the animals were kept in an animal house and handled using procedures and protocols approved by the Animal Care and Ethical Committee (Informe CEEA 2009/009) of the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA, Madrid), guidelines of the European Union (Directive 86/609/EEC) and current Spanish regulations (BOE 252/34367-91, 2005).
Production of ESC and EBs
For ESC production [32], two-cell embryos were collected 1.5 days after hCG injection and placed in a droplet of tempered KSOM-LIF medium under oil in a 5% CO2 atmosphere at 37°C. After 48 h in KSOM-LIF, the in vitro cultured blastocysts were plated individually in 96-well plates coated with 0.1% gelatine and containing mitomycin-C (Sigma-Aldrich corporation St. Louis, MO, USA) treated mouse embryonic fibroblasts (MEF). They were cultured in ESC medium composed of Dulbecco's modified Eagle medium (DMEM plus 4500 mg/l glucose, glutaMAX, and pyruvate; Invitrogen, Carlsbad, CA, USA) supplemented with 20% FBS (PAA Laboratories Cölbe Germany), 2 mM glutamine, 1 mM MEM nonessential amino acids solution, 1 mM β-mercaptoethanol, 1000 U/ml LIF (leukemia inhibitory factor), and an antibiotic mixture containing 100 U/ml penicillin and 100 µg/ml streptomycin. The blastocysts were allowed to attach to the MEF, until expansion without any further experimental interference for four days. After that time, cell clumps originating from the blastocysts were trypsinized and pipetted directly into a well of a 96-well plate containing MEFs and ESC medium. Approximately 4 days after trypsinization, compact cell colonies resembling ESC colony morphology could be detected and these cells were trypsinized in a 24-well plate containing MEFs and ESC medium. In this process, undifferentiated ESCs were separated from differentiated ones and from old MEF, and also expanded in number. Clones that were not confluent were replated on the same plate. When trypsinizing cells from a 35-mm dish for the first time for a particular clone, half the cells were frozen at −80°C in foetal calf serum with 10% DMSO media. This was considered the first passage. For cell line expansion, cells were trypsinized at 80% of confluency.
For embryoid body (EB) differentiation [33], ESC colonies were digested with 0.05% trypsin-EDTA. The dissociated cells were collected in EB medium (DMEM plus (4500 mg/l glucose, glutaMAX, and pyruvate) supplemented with 20% FBS, 2 mM glutamine, 1 mM MEM nonessential amino acids solution, 1 mM β-mercaptoethanol, and an antibiotic mixture containing 100 U/ml penicillin and 100 µg/ml streptomycin and plated on a gelatine-coated dish for 45 min to allow the MEF cells to adhere. Non-adherent cells were collected and plated onto non adherent plastic Petri dishes in the same EB medium. The medium was replaced every 2 days.
RNA Extraction, Reverse Transcription, and mRNA Transcript Quantification
RNA was prepared from ESC or EBs of each experimental group (WT or Prnp KO) using the ULTRASPEC total RNA Isolation Reagent Kit (BIOTECX laboratories, INC) according to the manufacturer's instructions. Immediately after extraction, the RT reaction was carried out following the manufacturer's instructions (Promega, Madrid, Spain) to produce cDNA. Tubes were heated to 70°C for 5 min to denature the secondary RNA structure, allowing Random Primer and Oligo dT attachment, and then the RT mix was completed with the addition of 5 units of Superscript RT enzyme. The tubes were then incubated at room temperature for 10 min and next at 42°C for 60 min to allow the reverse transcription of RNA, followed by 70°C for 10 min to denature the RT enzyme. To detect each transcript, we used 2 µl of the cDNA sample in the RT-PCR.
mRNA transcripts were quantified by real-time qRT-PCR [34]. At least three replicate PCR experiments were conducted for all the genes of interest. Experiments were designed to contrast relative levels of each transcript and histone H2az in each sample. PCR was performed by adding a 2- µl aliquot of each sample to the PCR mix (Quantimix Easy Sig Kit, Biotools) containing the specific primers. Primer sequences, annealing temperatures, and the approximate sizes of the amplified fragments of all transcripts are provided in Table 1. The comparative cycle threshold (CT) method was used to quantify expression levels (User Bulletin, Abi Prism 7700 Sequence Detection System, Applied Biosystems). Quantification was normalized to the endogenous control H2az. Fluorescence was acquired in each cycle to determine the threshold cycle or the cycle during the log-linear phase of the reaction at which fluorescence increased above background for each sample. Within this region of the amplification curve, a difference of one cycle is equivalent to doubling the amplified PCR product. According to the comparative CT method, the CT value was determined by subtracting the H2az CT value for each sample from the CT value of each gene in the sample. CT was calculated using the highest sample CT value (i.e., the sample with the lowest target expression) as an arbitrary constant to be subtracted from all other CT sample values. Fold changes in the relative gene expression of the target were determined using the formula 2–CT.
Table 1. Primers used for RT-PCR.
| Gene | Primer sequence 5′->3′ | Size | GenBank |
| Prnp | GATTATGGGTACCCCCTCCTTGG | 289 | NM_011170.2 |
| ATGGCGAACCTTGGCTACTGGC | |||
| Nanog | AGGGTCTGCTACTGAGATGCTCTG | 363 | M6047 |
| CAACCACTGGTTTTTCTGCCACCG | |||
| Pouf51 (Oct3/4) | GGAGAGGTGAAACCGTCCCTAGG | 391 | M17031 |
| AGAGGAGGTTCCCTCTGAGTTGC | |||
| Genesis (Foxd3) | TCTTACATCGCGCTCATCAC | 171 | M4758 |
| TCTTGACGAAGCAGTCGTTG | |||
| Stat3 | CAGAAAGTGTCCTACAAGGGCG | 340 | U06922 |
| CGTTGTTAGACTCCTCCATGTTC | |||
| Mapk1 | CCTTCAGAGCACTCCAGAAAGT | 74 | NM_011949.3 |
| ACAACACCAAAAAGGCATCC | |||
| Gp130 | ATTTGTGTGCTGAAGGAGGC | 186 | M83336 |
| AAAGGACAGGATGTTGCAGG | |||
| Itgb3 | GTCACATTGGCACCGACAACC | 556 | NM_016780.2 |
| CCACACTCAAAAGTCCCGTTC | |||
| Itgb5 | GGCTGCTGTCTGCAAGGAG | 511 | M96614 |
| TCAAAGGATGCCGTGTCC | |||
| Itga6 | GTGAGGTGTGTGAACATCAG | 540 | NM_008397.3 |
| CATGGTATCGGGGAATGCT | |||
| Shadoo | TTGGCCTGTACAAAGTTGAG | 186 | NM_183147.2 |
| CTGCAATGAGGGAAAAGCCT | |||
| Prnd (Doppel) | GCTCCAAGCTTCAGAGGCCACAGTAGCA | 580 | M1346999 |
| TTACTTCACAATGAACCAAACGAAAC | |||
| Sod1 | GGGATTGCGCAGTAAACATTC | 67 | NM_011434.1 |
| AATGGTTTGAGGGTAGCAGATGA | |||
| Slc2a1 | CCAGCTGGGAATCGTCGTT | 76 | M23384 |
| CAAGTCTGCATTGCCCATGAT | |||
| Irs2 | CTCTGACTATATGAACCTG | 339 | NM_001081212.1 |
| ACCTTCTGGCTTTGGAGGTG | |||
| Gapdh | ACCCAGAAGACTGTGGATGG | 247 | BC102589 |
| ATGCCTGCTTCACCACCTTC | |||
| T (Brachyury) | GCTGTGACTGCCTACCAGCAGAATG | 220 | M913 |
| GAGAGAGAGCGAGCCTCCAAAC | |||
| Hnf3-β | GGACGTAAAGGAAGGGACTCCAC | 174 | M938 |
| AGCCCATTTGAATAATCAGCTCAC | |||
| Nestin | AGTGTGAAGGCAAAGATAGC | 316 | M23742 |
| TCTGTCAGGATTGGGATGGG | |||
| H2AZ | AGGACGACTAGCCATGGACGTGTG | 212 | NM_016750 |
| CCACCACCAGCAATTGTAGCCTTG |
Statistical Analysis
Data were analyzed using the SigmaStat (Jandel Scientific, San Rafael, CA) package by one-way repeated-measures ANOVA with arcsine data transformation.
Semiquantitative PCR
cDNA was obtained as described above and PCR was performed with a PCR mix (Go taq Flexi DNA polymerase Kit, Promega) following the manufacturer's instructions. The PCR products were analyzed on a 2% agarose gel by ethidium bromide staining. The gel was visualized under ultraviolet light to identify the specific band for each gene analyzed.
Antibody Inhibition Experiments
EBs of both ESC lines were re-suspended on Day 5 in EB medium alone or supplemented with SHA31 anti-mouse PRNP antibody (CISA, INIA), with anti-rabbit ITGB5 antibody (ABCAM) or control antibody (mouse IgG) at final concentrations in the medium of 0.2 µg/ml. The EBs were collected 6 or 24 hours later and frozen for mRNA extraction.
Terminal Deoxynucleotidyl Transferase-Mediated dUTP Nick End-Labelling (TUNEL) Assay
For the TUNEL method of detecting apoptosis, EBs cultured for 5 days were fixed in 4% paraformaldehyde in PBS, pH = 7.4, for 1 h, and then washed in PBS/1%BSA three times. The fixed EBs were permeabilized with 0.1% Triton X-100 in PBS for 15 min at 37°C and then washed again in PBS/1%BSA three times. TUNEL assays were performed using the In Situ Cell Death Detection Kit, TMR Red (Roche) according to the manufacturer's protocol. As control, it was checked that a lack of TdT in the TUNEL mix completely abolished labelling.
Results
Two distinct types of Prnp-KO EBs exist
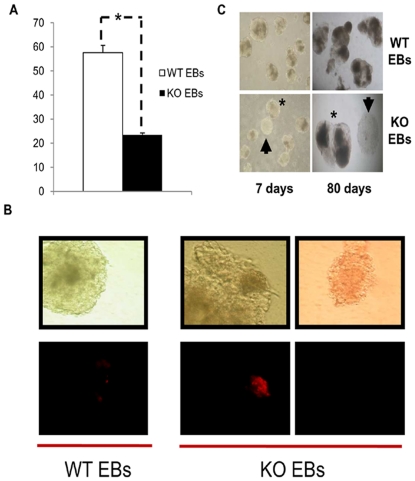
Two lines of Prnp-KO ESC were produced in a 129Ola background (that of the WT control line R1). Since both these lines develop in the same way, the results for each line were collectively considered. Confluent WT and KO ESC lines were trypsinized and grown in a LIF-free medium without the feeder layer to form EBs. From Days 1 to 3, large numbers of unclustered cells could be seen among the KO EBs. This could be the reason of a significantly reduced EB numbers on Day 5 (Fig. 1A). TUNEL apoptosis analysis on Day 5 of ESC differentiation revealed a low number of isolated cells positive for immunostaining in the WT line (Fig. 1B) (2–5% of the WT EB surface). Extensive apoptotic red zones were detected in the KO EBs on Day 5 of differentiation (10–15% of the KO EB surface), which could explain the high number of floating cells observed in the early EB culture supernatants. Interestingly, two types of EBs were observed on Day 5 in the KO group. About 40% of the EBs were white showing no apoptosis signal, and 60% were morphologically similar to the WT EBs. The white EBs persisted after a long period of differentiation (more than 80 days) while in the WT line, only a small number of EBs looked white after 5 days of differentiation (1–2%) and these disappeared over the following 2 days (Fig 1C). In addition, after 30 days of differentiation, mature EBs in both KO populations were plated on a feeder layer in complete stem cell medium (including LIF and bFGF), and after 3 passages, we could only recover cells positive for alkaline phosphatase of the white KO population. mRNA was extracted from the different types of EB and it was noted that Gp130 and Stat3 (proteins involved in maintaining pluripotency) were upregulated only in the white KO population (data not shown).
Figure 1. Apoptotic and macroscopic effects produced by the delection of Prnp.
A) Comparing the numbers of EBs in the WT and KO lines (* P<0.05. Error bars, s.e.m.; “Y” axis represents “number of EBs per plate”) B) TUNEL analysis of wild type (WT) and Prnp-null (KO) embryoid bodies (EBs). C) Morphology of EBs on Day 7 and Day 80 of differentiation. The KO ESC line produced 2 types of EBs: one similar to WT (*) and the other called PGC-like KO EBs (arrowheads).
Prnp–KO ESC and EBs express different pluripotent markers to the WT line during differentiation
The mRNA expression of pluripotency markers was examined in both the WT and Prnp–KO lines. We selected the genes Oct3/4 (Pou5f1), Nanog, and FoxD3 (Genesis) because the feedback regulating cycle of these genes is among the most important for maintaining stem cell pluripotency [35]. Stem cells from the WT line showed significantly higher Nanog and Oct3/4 and lower FoxD3 transcript levels than KO ESC (Fig. 2A and 2B), indicating a different background of pluripotency in the KO and WT lines.
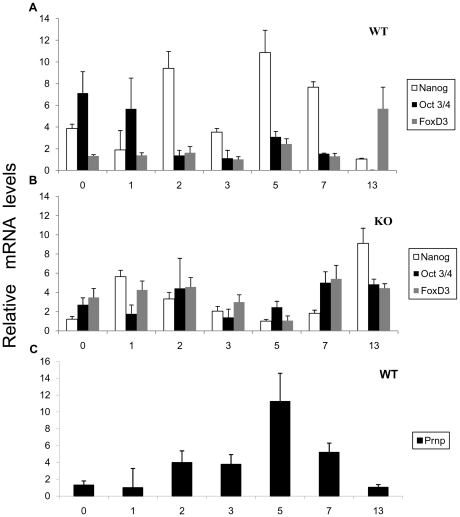
Figure 2. Pluripotency in the WT and Prnp-null cell lines.
WT EB mRNA (A) and KO EB mRNA (B) expression patterns of three pluripotency genes during differentiation. Note the lack in the KO line (B) of an increase in Nanog expression on Day 5. C) WT EB mRNA expression pattern of Prnp during differentiation (Days 0, 1, 2, 3, 5, 7 and 13). Prnp is expressed at its highest levels on Day 5 (Error bars, s.e.m.).
In order to better understand the role of PRNP in differentiation, Prnp mRNA was quantified from the ESC stage (Day 0) to Day 13 EBs. Stem cell lines were cultured in ESC medium without LIF and samples were taken on Days 0, 1, 2, 3, 5, 7 and 13. Next, mRNA was extracted and transcripts were quantified by qRT-PCR. During WT EBs differentiation, Prnp showed increasing levels of expression starting on Day 2 until Day 7, showing peak expression on Day 5 (Fig. 2C). In the WT line, no significant differences could be detected in mRNA levels of FoxD3 until Day 13 when an expression increase was observed (Fig. 2A). However, Prnp expression was negatively correlated with that of the pluripotency marker Oct-4. Thus, higher Oct-4 expression levels were detected on Days 0 and 1 followed by reduced expression until Day 13, following an opposite expression pattern to Prnp and confirming that ES cells had indeed differentiated. Nanog was also downregulated on Day 13, accordingly with the differentiation status. Interestingly, Nanog expression showed a similar pattern to Prnp, with increasing expression levels observed from Day 2 to Day 7 peaking on Day 5 (Fig. 2A). In contrast, in the KO line, Oct-4 and FoxD3 expression levels showed some variation during early differentiation but high mRNA levels persisted on Days 7 and 13 (Fig. 2B). Further, Nanog expression levels were highest on Day 13, indicating the pluripotent state of these cells even after 13 days of differentiation (Fig. 2B).
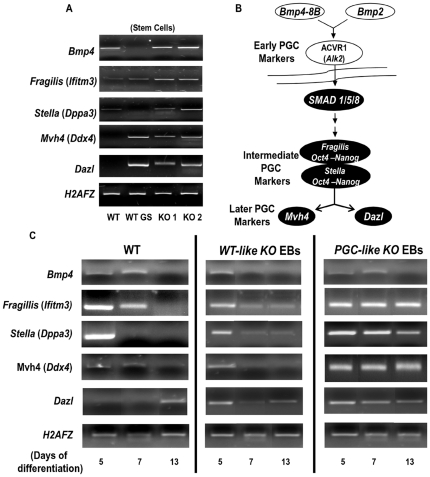
To understand this initial developmental status of the different pluripotency markers in the KO ESC, we examined the expression of bone morphogenetic protein (BMP) genes, which are related to the germ cell program. These genes were chosen because KO EBs show characteristics comparable to those of germ cells, as the only cells in the organism that repress their somatic differentiation program in favour of a germline-specific network of RNA regulation. Indeed, BMPs regulate the differentiation of PGCs and are also able to potentiate pluripotency in the presence of LIF. Our semiquantitative PCR revealed increased expression levels of all the PGC markers in the KO stem cells, regardless of the stem cell line (Fig. 3A). The expression of two later PGC markers, Mvh4 (Ddx4) and Dazl (Fig. 3B), was lacking in the WT line but was detected in WT cells grown in a GS medium and in the KO lines.
Figure 3. Consequences of Prnp absence in the BMP pathway.
A) Detection of PGC markers (Bmp4, Fragillis, Stella, Mvh4 and Dazl) in WT ESC, WT GS and the two different Prnp KO ESC lines (KO 1 and KO 2) by PCR. Knock out cells show all the PGC markers analyzed in stem cell state. B) Diagram representing the BMP pathway showing the early, intermediate and later PGC markers (figure adapted from Young et al. 2009). C) Detection of primordial germ cell markers (Bmp4, Fragillis, Stella, Mvh4 and Dazl) by PCR during differentiation in WT EBs, WT-like KO EBs and PCG-like KO EBs.
When we assessed the same markers during the time course of EB differentiation, it was found that early (Bmp4) and intermediate [Fragillis (Ifitm3) and Stella (Dppa3)] PGC markers (Fig. 3B) were upregulated on Day 5 in WT EBs and that these were gradually replaced with the later expressed Mvh4 and Dazl from Day 7 to 13 (Fig. 3C). The same occurred in normal-appearance KO EBs, but in the white EB population all these markers persisted at all the time points. For this reason, we called the white EBs derived from the KO ES cells PGC-like to differentiate them from the KO EBs of similar morphological appearance to WT EBs (designated WT-like KO EBs).
Characterizing the WT and Prnp-KO lines during differentiation to EBs
The appearance of two EB populations in the KO line could be the consequence of two different ways of compensation and expression of Shadoo and Doppel, two genes whose functions are redundant to those of Prnp in some tissues and developmental stages. Accordingly, we then examined mRNA levels of these genes during the time course of EB differentiation. In addition, to determine if the differences between the three types of EBs were due to differences in tissue differentiation, we analyzed the mRNA expression of early markers of ectoderm (Nestin), mesoderm (Brachury -T-) and endoderm (Hnf3) (Fig. 4). We considered it might be also interesting to determine the metabolic state of these EB populations since PRNP is thought to be involved in protection from oxidative stress [30]. Thus, we examined mRNA levels of glucose capturing genes (Slc2a and Irs2) and glycolysis genes (Gapdh) (Fig. 5) giving information about oxidative metabolism, and mRNA levels of the SOD1 gene (Sod1), which codes for a protein whose antioxidant activity is regulated by PRNP [36], [37].
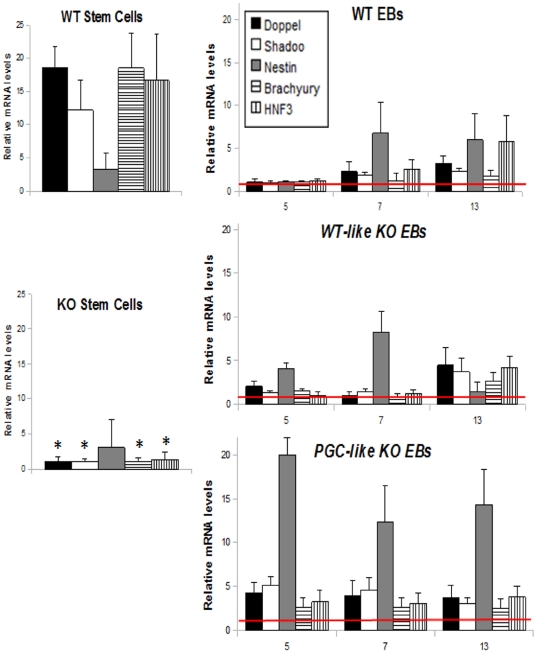
Figure 4. Differentiation transcription behaviour of the WT and Prnp-null cell lines.
mRNA expression patterns of PRNP related proteins (Doppel and Shadoo) and early markers of endoderm (Hnf3), mesoderm (Brachyury -T-) and ectoderm (Nestin) in WT and KO stem cells and in WT, WT-like KO and PGC-like KO EBs during differentiation (Days 5, 7 and 13). (* indicates statistical differences for the transcription of each gene between WT and KO ESC at P<0.05) (Error bars, s.e.m).
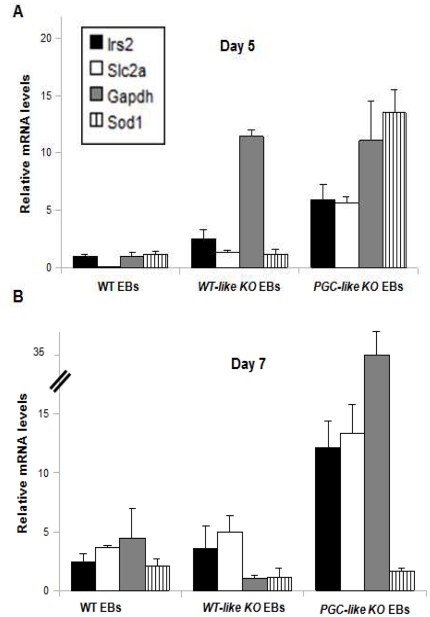
Figure 5. Metabolic transcription behaviour of the WT and Prnp-null cell lines.
WT, WT-like KO EB and PGC-like KO EB mRNA expression patterns for Irs2 (Insulin Receptor Substrate 2), Slc2a (Glucose Transporter 1 - Glut1 -), Gapdh (Glyceraldehyde 3-phosphate Dehydrogenase) and Sod1 (Superoxide Dismutase 1) on Days 5 (A) and 7 (B). (Error bars, s.e.m.).
WT ESC expressed all the transcripts except Nestin in higher amounts than the KO ESC (Fig. 4, Day 0). Interestingly, during differentiation, mRNA levels of all markers were significantly raised and maintained from Day 5 in the PGC-like KO EBs (Fig. 4) compared to WT and WT-like KO EBs, which showed similar expression among each other. However, in the WT-like KO EBs the absence of PRNP resulted in non-expression of Nestin on Day 13 (Fig 4) suggesting a role for PRNP in neural cell differentiation during early embryogenesis. This phenomenon was not observed in the PGC-like KO EBs since these cells maintained similar transcription levels during the differentiation time examined.
On the other hand, the mRNAs of Irs2, Slc2a, Gapdh and Sod1 were augmented in the PGC-like KO EBs on Days 5 and 7 (Fig. 5). Gapdh mRNA was also elevated on Day 5 in WT-like KO EBs but reached baseline levels on Day 7, suggesting a transition feature. This metabolic situation was maintained until Day 13 in both kinds of KO EB (data not shown) and indicates enhanced glycolytic and oxidative metabolism in the PGC-like KO EBs.
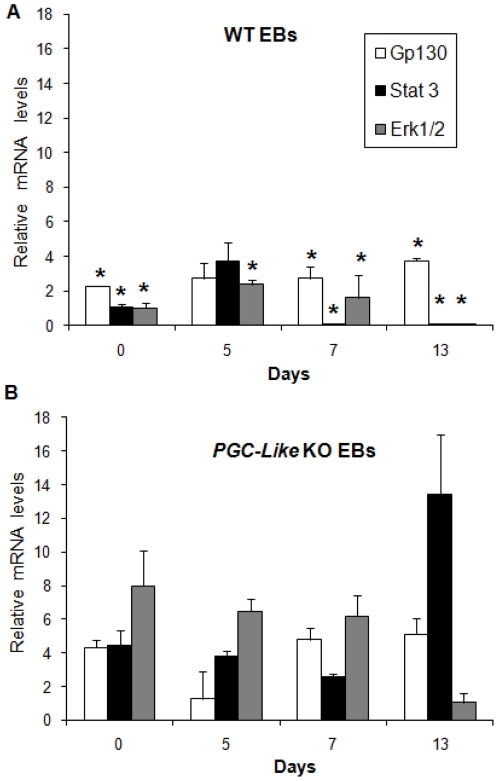
Stat3 mRNA is upregulated in the later stages of differentiation
Our experimental protocol includes the use of LIF-starved ES medium, so the receptor (Gp130) and main effector genes (Erk1/2 and Stat3) of the LIF pathway were analysed. ERK 1/2 is known to be involved in the loss of pluripotency and is also a downstream component of the PRNP biochemical matrix, while STAT3 contributes to the maintenance of self-renewal. WT EBs showed lower expression of Erk1/2 mRNA than PGC-like KO EBs, disappearing on Day 13 (Fig. 6). The same lower levels were observed for Stat3 in the WT EBs, but disappearing on day 7. However, in the PGC-like KO EBs, increased transcription of this gene was observed on Day 13, indicating and supporting the previously described pluripotent state of these EBs. In contrast, Gp130 expression failed to vary or only showed subtle variations during the time course of differentiation, indicating that the upregulation of Stat3 transcription is not related to activation of the LIF pathway.
Figure 6. LIF pathway regulation in the WT and Prnp-null cell lines.
WT EB mRNA (A) and PGC-like KO EB mRNA (B) expression patterns of some LIF pathway genes during differentiation (Days 0 [(Stem Cell], 5, 7 and 13). (*indicates significant differences for a particular gene between WT and PGC-like KO EBs (P<0.05)) (Error bars, s.e.m.).
Prnp regulates Nanog mRNA expression
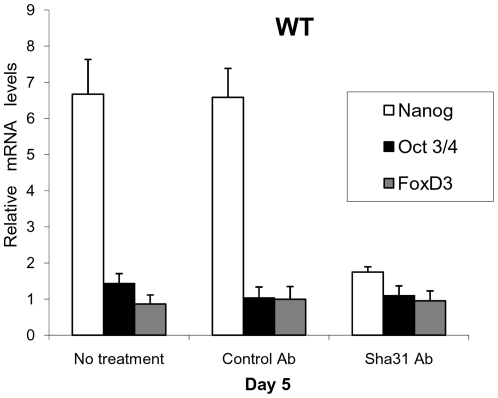
On Day 5 of EB differentiation, the highest levels of Prnp and Nanog were detected in the WT EBs. Similar waves in their mRNA expression pattern were also observed suggesting Prnp-dependent Nanog transcription (Fig. 2A and Fig. 2C). To confirm this relationship, we conducted Prnp inhibition experiments. Several studies have shown that the addition of specific PRNP antibody to the culture medium inactivates this protein's signal transduction pathways [38], [39] so we proceeded to add the monoclonal antibody Sha31 to WT EBs on Day 5. Interestingly, a significant reduction in Nanog mRNA levels was detected, even over the shorter time interval (6 h) (Fig. 7), and this effect persisted for 24 hours without refreshing the antibody (data not shown). Further, mRNA expression patterns of important PRNP related markers on Day 5 (e.g. integrins αvβ5 and α6 and the metabolic gene Sod1), which were differentially expressed between the PGC-like KO EBs and WT EBs, became similar (data not shown). This inhibition of Nanog expression confirms the involvement of Prnp during EB differentiation.
Figure 7. Blocking of PRNP by the monoclonal antibody Sha31.
On Day 5 and after 6 hours of antibody treatment, it is revealed that Nanog mRNA transcription is PRNP-dependent during differentiation and independent of feedback from the pluripotency cluster genes Oct3/4 and FoxD3. (Error bars, s.e.m.).
Nanog expression on Day 5 is blocked by integrin beta5 in Prnp-KO EBs
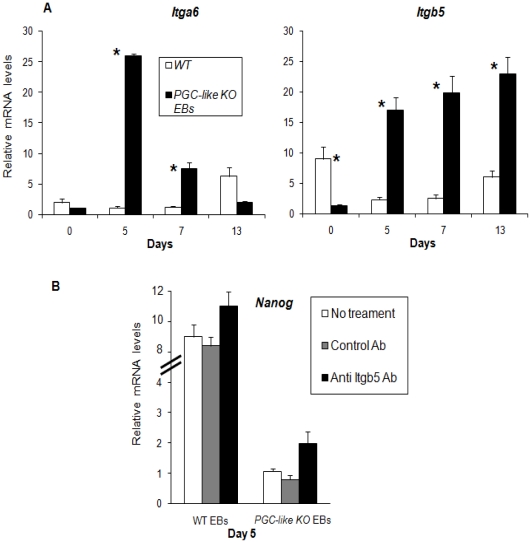
Integrins are essential for PGC functionality [40], [41] and it has been reported that the absence of Prnp may be compensated by these proteins [42], [43]. Since integrin signalling is crucial in promoting mouse ESC self-renewal [44], we speculated that Prnp expression might also be related to this integrin expression during ESC differentiation. So we examined by real time PCR the transcription levels of three genes coding for integrins αvβ3 (Itgb3), αvβ5 (Itgb5), and α6 (Itga6) in stem cells and during early EB differentiation (Days 5, 7, and 13). We found that Itgb3 was only detected in PGC-like KO EBs on Day 5. Similar transcription levels of Itga6 were found in the KO stem cells and in the WT line. During differentiation, significantly elevated expression was observed on Day 5 in the Prnp-KO background population; this transcription gradually fell from Day 7 to 13. In the WT, expression was increased only on Day 13 (Fig. 8A). Interestingly, mRNA for Itgb5, described to be a gonadal tissue marker [41], appeared in higher amounts in the WT ESC than in the KO ESC; however, expression was higher in the PGC-like KO EBs at all the differentiation times (Fig. 8A).
Figure 8. Influence of Prnp in the integrin matrix.
A) Detection of PRNP-related integrins (Itga6 and Itgb5) in WT and KO stem cells on Day 0, and in WT EBs and PGC-like KO EBs on Days 5, 7 and 13. B) Blocking of ITGB5 by a polyclonal antibody shows that Nanog mRNA transcription is ITGB5-dependent during differentiation (*indicates significant differences between WT and PGC-like KO EBs on a particular day (P<0.05)) (Error bars, s.e.m.).
Because the expression of Itgb5 on Day 5 differed significantly between WT and KO, we designed an Itgb5 antibody mediated inhibition experiment to analyse its effect on the transcription patterns observed on Day 5 of EB differentiation. Surprisingly, when ITGB5 was blocked at the membrane level in the PGC-like KO EBs, Nanog mRNA transcription was significantly augmented, while in the WT line, this treatment had no significant effect (Fig. 8B). These results indicate that in the absence of Prnp, the expression of Nanog on Day 5 of EB differentiation may be partially mediated by the inhibition of this integrin.
Correlated Prnp and Nanog expression in WT mice points to a role for both genes in the development of the foetal gonads
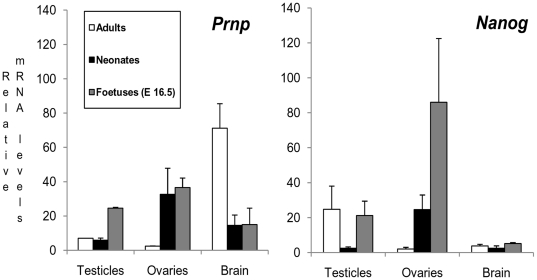
We quantified the in vivo transcription of Prnp and Nanog in the brains, testicles and ovaries of E16.5 WT mouse foetuses, neonates and adults. These tissues were selected because Prnp-KO ESC and their derived EBs showed an ectodermal gene expression pattern. In brain, as expected, Prnp was highly expressed, with no Nanog expression detected in any case (Fig. 9). In the testicles, a clear increase in Prnp mRNA levels was seen during the foetal period; and in the ovaries, this was observed in the foetal and neonatal period. Nanog expression in these two tissues was upregulated at the same time points as Prnp except in adult testicles, where Nanog showed clear upregulation contrary to the behaviour of Prnp. However, Doppel was upregulated at this point (data not shown) and could explain the absence of Prnp expression in the adult testicle. These results indicate that correlative Nanog-Prnp mRNA expression, possibly due to Nanog Prnp-dependant transcription as was seen in the “in vitro” experiments, occurs in the gonads but not in neuronal tissues.
Figure 9. Prnp and Nanog expression in testicles, ovaries and brain in adult, neonates and foetal mice at embryonic day 16.5 (E16.5).
Note that correlative Prnp-Nanog expression only occurs in gonadal tissues. (Error bars, s.e.m.).
Discussion
The key role of prion protein in infection has been expansively reported, yet its physiological function remains elusive. To address this issue, we designed a comparative analysis during differentiation of WT and Prnp-KO ESC. In the current study, we employed ESC to assess the potential role of PRNP in early embryo development, examining how this locus becomes transcriptionally activated when potential transient compensatory mechanisms are still not established. We here present the first evidence that Prnp is involved in regulating the self-renewal/differentiation status of stem cells. Its expression was found to affect that of several pluripotent genes, including Nanog, a protein that drives constitutive stem cell self-renewal and safeguards pluripotency [45], [46] through its involvement in the Sox2-Oct3/4 transcription factor network [47], [48], [49], [50], [51].
The main pathway through which ESC-derived cells become differentiated cells involves the formation of embryoid bodies, thus denoted because of their similarity to the early postimplantation embryo. Embryoid bodies are formed “in vitro” after several days of growth of ESC in the absence of LIF (leukemia inhibitory factor). These bodies are the final ESC population that has the potential to give rise to all the cell types of adult tissues and have been described as the developmental equivalent of the egg cylinder-stage mouse embryo, with an outer endodermal layer and a core of differentiating cells, often comprised of epithelial-lined cavities [52]. Hence the data obtained in this work may represent what happens “in vivo” during embryo development.
Our results indicate that despite the widespread expression of key genes of pluripotency in WT and Prnp-KO ESC lines (e.g., Oct4) and their characteristic undifferentiated cell morphology, functional ESC may represent only a small fraction of the total number of cells grown under self-renewal conditions. Effectively, in our KO ESC, a large PGC-like stem cell population was identified. Further, mRNA levels of early endoderm and mesoderm markers were significantly lower, while early neuronal ectoderm marker mRNA levels were fairly similar to those in WT ESC. Taken together with the fact that PGC markers were also expressed, an ectodermal stem cell type was defined in the KO ESC, according to the simple absence of Prnp. Indeed, a heterogeneity in the stem cell population leading to different transcription behavior has been previously described based on Nanog mRNA levels [53]. This could explain the appearance of two different kinds of EB when LIF was removed from the culture medium in the KO line, based on the detected absence of Nanog transcription in this EB population. Thus, phenotypically “undifferentiated” cells may consist of a heterogeneous population of functionally distinct cell types, and small changes in gene expression could lead to the predominance of one of these types over the others.
Interestingly, in Prnp-KO ESC, Doppel and/or Shadoo were clearly downregulated. However, on Day 5, PGC-like KO EBs overexpressed Doppel and Shadoo, showing the altered expression of early embryo layer markers and also enhanced glucose and oxidative metabolism. This last feature has been related to a high rate of proliferation in cancer or stem cells (Warburg effect) [54], [55] and to increased protection against oxidative stress, according to the high pluripotency level of these PGC-like KO EBs and to compensation for the high sensitivity to oxidants of the KO lines, respectively. Until now, Doppel and Shadoo, ascribed to the PRNP family, were known to compensate for the absence of PRNP, having redundant (in the case of Shadoo in brain [19]) or exclusive (in the case of Doppel in male testicles [20]) functions. However, we noted that WT-like KO EBs exhibited similar expression patterns of these genes with subtle differences in comparison to WT EBs. Thus, we could speculate that an ESC, lacking Prnp yet showing increased Doppel and Shadoo gene expression during differentiation, will give rise to a living system closely resembling PGCs, morphologically different to WT cells, more metabolically active in terms of differentiation [55] and with a high degree of pluripotency. Conversely, if Doppel and Shadoo were not overexpressed, KO EBs would be practically identical to their WT counterparts. Our results therefore suggest that the absence of PRNP is not counteracted by the presence of Doppel and Shadoo, as previously thought. The two different populations of KO EBs with a completely different expression pattern for these genes, further supports the idea of an irreplaceable role of PRNP in this process, most likely in coordinating the expression of indispensable proteins for the appropriate behaviour of the stem cells.
Recently, it has been indicated that during certain embryogenesis processes some redundancy for PRNP interactions resides within the integrin pathway [43], and that integrin signalling is crucial for promoting mouse ESC self-renewal [44]. Here, we analysed the expression of three integrins during PRNP-null stem cell differentiation to investigate if these integrins play some sort of role in maintaining the stemness of these ESC. Integrin αvβ3 (ITGB3) and integrin αvβ5 (ITGB5) are expressed in mouse foetal developing gonads (from Days 10.5 to 13.5); ITGB3 is specific to primordial germ cells [41] and ITGB5 plays important roles in maintaining stemness in undifferentiated mouse ESC [56]. The observation of Itgb3 transcription only on Day 5 in the KO EB population indicated primordial germ cell transcription activity at this stage. Moreover, the higher expression of Itgb5 on Days 5, 7 and 13 was in agreement with the presence of the primordial germ cell markers identified in the PGC-like KO EBs. It has been reported that integrin-α6 (ITGA6) function is required for the early stages of lens cell differentiation [57]. The higher expression of Itga6 we identified on Day 5 in KO EBs, and on Day 13 in the WT EBs, could point to the cell differentiation activity of PGCs in the KO EBs, and of the three embryonic layers in the WT line.
Our results also indicate a relationship between PRNP and integrins in ES cells and during differentiation. It has been shown that PRNP binds some extra cellular matrix (ECM) proteins [43], [58], [59], and other important ECM proteins, such as integrins, could also interact or cooperate with PRNP in this process. Integrins have been the focus of several studies over the past few years, not only because of their capacity to bind ECM, but also because of their ability to activate a number of cell signaling pathways, including those of differentiation and pluripotency [44], [56]. It is also known that Prnp downregulates Itgb3 mRNA expression in Mesenchymal Embryonic Cells (MEC) [42], highlighting the importance of integrins in the PRNP biochemical matrix. Other authors have determined that PGCs contain subpopulations showing low or null integrin-α6 (ITGA6) expression with a greater ability to develop into pluripotent stem cells [40]. The fact that several authors have also related the upregulation of certain integrin transcription with a loss of pluripotency and self-renewal [44], suggest a pivotal link between PRNP or integrins and these processes. Our results revealed that Itga6, Itgb3 and Itgb5 were deregulated in the KO line. Specifically, Itgb3 (data not shown) and Itgb5 mRNAs appeared in high quantities in the PGC-like KO EBs on Day 5 and, only for the last gene, this population of EBs was labelled by this integrin for longer times (Days 7 and 13). Since, as mentioned earlier, these integrins are confirmed PGC and gonadal tissue markers, respectively, this suggests the idea of arrested EBs in that ectoderm cell line, also consistent with the loss of self-renewal in a situation of high integrin expression [44].
In addition, the expression of Erk1/2, previously incriminated in differentiation, was significantly downregulated on Day 13 in the PGC-like KO EBs, at the same time an increase in Stat3 transcription and downregulation of Itga6 were observed. These results suggest that PGC-like KO EBs seemed to acquire pluripotent and PGC self-renewal characteristics on Day 13. Further, in our antibody-mediated inhibition experiments, when ITGB5 was blocked on Day 5 at the membrane level in the PGC-like KO EBs, the important pluripotency mRNA transcript Nanog was significantly elevated, while this treatment had no effect in the WT line, though a non-significant upward trend was observed. Similar results were reported when mouse ESC cultured on laminin or fibronectin (activators of integrin pathways) in ESF7 medium showed low immunostaining for alkaline phosphatase, weak immunostaining in immunocytochemical experiments detecting NANOG and a low population of NANOG positive cells in flow cytometric analysis [44]. However, when the mouse ESC was treated with an anti-β1 integrin antibody (blocking the interaction ECM-integrins) these findings were reversed [44].
We also observed here that the disruption of PRNP by knocking out through transgenesis or knocking down through antibody inhibition led to diminished Nanog mRNA levels (especially on Day 5, when peak Prnp and Nanog were observed in the WT line), accompanied by high levels of Itgb5 mRNA. These data thus indicate the possible Prnp regulation of Nanog transcription via Itgb5 during early differentiation to EBs. Moreover, the results of our “in vivo” experiments suggest that this relationship during early differentiation between Prnp and Nanog specifically occurs in gonadal but not in brain tissue. In summary, our data point to the first known relationship between PRNP and NANOG, one of the most important proteins needed to maintain pluripotency, with a major role in the self-renewal of ESC and their differentiation into gonadal cells.
A loss of PRNP functionality was largely proposed as one of the main causes of a prion disease physiopathology. There is a lack of a premortem way of detecting them and bearing in mind that Nanog levels remain low or disappear during the first stages of differentiation of Prnp-KO ESC, the use of this gene as a marker of prion disease and, more importantly, as an early detection method could be investigated. Otherwise, PRNP is not essential for ESC survival since other pathways could support its absence, although a significant percentage of cells die in the first stages of differentiation to EBs. In contrast, Prnp knockout or downregulation modifies the normal macroscopic and transcriptional behaviour of the stem cell during differentiation, confirming the participation of PRNP in early embryogenesis [19]. The association between Prnp and pluripotency marker expression also provides evidence of this contribution of Prnp to stem cell differentiation (e.g. the role of PRNP in neural cell differentiation [60]). This feature is mediated by Nanog and not compensated by the PRNP-related proteins Doppel and Shadoo, suggesting for the first time a non-redundant function of PRNP in maintaining pluripotency and differentiation during early embryogenesis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by Grant AGL2009-11358 from the Spanish Ministry of Science and Innovation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Castilla J, Gutierrez Adan A, Brun A, Pintado B, Ramirez MA, et al. Early detection of PrPres in BSE-infected bovine PrP transgenic mice. Arch Virol. 2003;148:677–691. doi: 10.1007/s00705-002-0958-4. [DOI] [PubMed] [Google Scholar]
- 2.Horiuchi M, Yamazaki N, Ikeda T, Ishiguro N, Shinagawa M. A cellular form of prion protein (PrPC) exists in many non-neuronal tissues of sheep. J Gen Virol. 1995;76(Pt 10):2583–2587. doi: 10.1099/0022-1317-76-10-2583. [DOI] [PubMed] [Google Scholar]
- 3.Castilla J, Gutierrez-Adan A, Brun A, Pintado B, Parra B, et al. Different behavior toward bovine spongiform encephalopathy infection of bovine prion protein transgenic mice with one extra repeat octapeptide insert mutation. J Neurosci. 2004;24:2156–2164. doi: 10.1523/JNEUROSCI.3811-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujisawa M, Kanai Y, Nam SY, Maeda S, Nakamuta N, et al. Expression of Prnp mRNA (prion protein gene) in mouse spermatogenic cells. J Reprod Dev. 2004;50:565–570. doi: 10.1262/jrd.50.565. [DOI] [PubMed] [Google Scholar]
- 5.Bueler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, et al. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 6.Hajj GN, Santos TG, Cook ZS, Martins VR. Developmental expression of prion protein and its ligands stress-inducible protein 1 and vitronectin. J Comp Neurol. 2009;517:371–384. doi: 10.1002/cne.22157. [DOI] [PubMed] [Google Scholar]
- 7.Manson JC, Clarke AR, Hooper ML, Aitchison L, McConnell I, et al. 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Mol Neurobiol. 1994;8:121–127. doi: 10.1007/BF02780662. [DOI] [PubMed] [Google Scholar]
- 8.Sakaguchi S, Katamine S, Nishida N, Moriuchi R, Shigematsu K, et al. Loss of cerebellar Purkinje cells in aged mice homozygous for a disrupted PrP gene. Nature. 1996;380:528–531. doi: 10.1038/380528a0. [DOI] [PubMed] [Google Scholar]
- 9.Richt JA, Kasinathan P, Hamir AN, Castilla J, Sathiyaseelan T, et al. Production of cattle lacking prion protein. Nat Biotechnol. 2007;25:132–138. doi: 10.1038/nbt1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu G, Chen J, Xu Y, Zhu C, Yu H, et al. Generation of goats lacking prion protein. Mol Reprod Dev. 2009;76:3. doi: 10.1002/mrd.20960. [DOI] [PubMed] [Google Scholar]
- 11.Collinge J, Whittington MA, Sidle KC, Smith CJ, Palmer MS, et al. Prion protein is necessary for normal synaptic function. Nature. 1994;370:295–297. doi: 10.1038/370295a0. [DOI] [PubMed] [Google Scholar]
- 12.Mallucci GR, Ratte S, Asante EA, Linehan J, Gowland I, et al. Post-natal knockout of prion protein alters hippocampal CA1 properties, but does not result in neurodegeneration. Embo J. 2002;21:202–210. doi: 10.1093/emboj/21.3.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colling SB, Khana M, Collinge J, Jefferys JG. Mossy fibre reorganization in the hippocampus of prion protein null mice. Brain Res. 1997;755:28–35. doi: 10.1016/s0006-8993(97)00087-5. [DOI] [PubMed] [Google Scholar]
- 14.Tobler I, Gaus SE, Deboer T, Achermann P, Fischer M, et al. Altered circadian activity rhythms and sleep in mice devoid of prion protein. Nature. 1996;380:639–642. doi: 10.1038/380639a0. [DOI] [PubMed] [Google Scholar]
- 15.Walz R, Amaral OB, Rockenbach IC, Roesler R, Izquierdo I, et al. Increased sensitivity to seizures in mice lacking cellular prion protein. Epilepsia. 1999;40:1679–1682. doi: 10.1111/j.1528-1157.1999.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 16.Criado JR, Sanchez-Alavez M, Conti B, Giacchino JL, Wills DN, et al. Mice devoid of prion protein have cognitive deficits that are rescued by reconstitution of PrP in neurons. Neurobiol Dis. 2005;19:255–265. doi: 10.1016/j.nbd.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Coitinho AS, Roesler R, Martins VR, Brentani RR, Izquierdo I. Cellular prion protein ablation impairs behavior as a function of age. Neuroreport. 2003;14:1375–1379. doi: 10.1097/01.wnr.0000078541.07662.90. [DOI] [PubMed] [Google Scholar]
- 18.Fuhrmann M, Bittner T, Mitteregger G, Haider N, Moosmang S, et al. Loss of the cellular prion protein affects the Ca2+ homeostasis in hippocampal CA1 neurons. J Neurochem. 2006;98:1876–1885. doi: 10.1111/j.1471-4159.2006.04011.x. [DOI] [PubMed] [Google Scholar]
- 19.Young R, Passet B, Vilotte M, Cribiu EP, Beringue V, et al. The prion or the related Shadoo protein is required for early mouse embryogenesis. FEBS Lett. 2009;583:3296–3300. doi: 10.1016/j.febslet.2009.09.027. [DOI] [PubMed] [Google Scholar]
- 20.Behrens A, Genoud N, Naumann H, Rulicke T, Janett F, et al. Absence of the prion protein homologue Doppel causes male sterility. Embo J. 2002;21:3652–3658. doi: 10.1093/emboj/cdf386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grskovic M, Chaivorapol C, Gaspar-Maia A, Li H, Ramalho-Santos M. Systematic identification of cis-regulatory sequences active in mouse and human embryonic stem cells. PLoS Genet. 2007;3:e145. doi: 10.1371/journal.pgen.0030145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiyama K, Otani K, Ohtaki M, Satoh K, Kumazaki T, et al. Differentially expressed genes throughout the cellular immortalization processes are quite different between normal human fibroblasts and endothelial cells. Int J Oncol. 2005;27:87–95. [PubMed] [Google Scholar]
- 23.Hailesellasse Sene K, Porter CJ, Palidwor G, Perez-Iratxeta C, Muro EM, et al. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faustino RS, Behfar A, Perez-Terzic C, Terzic A. Genomic chart guiding embryonic stem cell cardiopoiesis. Genome Biol. 2008;9:R6. doi: 10.1186/gb-2008-9-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang CC, Steele AD, Lindquist S, Lodish HF. Prion protein is expressed on long-term repopulating hematopoietic stem cells and is important for their self-renewal. Proc Natl Acad Sci U S A. 2006;103:2184–2189. doi: 10.1073/pnas.0510577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Y, Zhao L, Liang J, Liu J, Shi Y, et al. Cellular prion protein promotes invasion and metastasis of gastric cancer. Faseb J. 2006;20:1886–1888. doi: 10.1096/fj.06-6138fje. [DOI] [PubMed] [Google Scholar]
- 28.Mehrpour M, Codogno P. Cancer Lett; 2009. Prion protein: From physiology to cancer biology. [DOI] [PubMed] [Google Scholar]
- 29.Stella R, Massimino ML, Sandri M, Sorgato MC, Bertoli A. Cellular prion protein promotes regeneration of adult muscle tissue. Mol Cell Biol. 2010;30:4864–4876. doi: 10.1128/MCB.01040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westergard L, Christensen HM, Harris DA. The cellular prion protein (PrP(C)): its physiological function and role in disease. Biochim Biophys Acta. 2007;1772:629–644. doi: 10.1016/j.bbadis.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin J, Guo X, Cui GH, Zhou YC, Zhou DR, et al. Cluster characterization of mouse embryonic stem cell-derived pluripotent embryoid bodies in four distinct developmental stages. Biologicals. 2009;37:235–244. doi: 10.1016/j.biologicals.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez MA, Fernandez-Gonzalez R, Perez-Crespo M, Pericuesta E, Gutierrez-Adan A. Effect of stem cell activation, culture media of manipulated embryos, and site of embryo transfer in the production of F0 embryonic stem cell mice. Biol Reprod. 2009;80:1216–1222. doi: 10.1095/biolreprod.108.075044. [DOI] [PubMed] [Google Scholar]
- 33.Ramirez MA, Pericuesta E, Fernandez-Gonzalez R, Pintado B, Gutierrez-Adan A. Inadvertent presence of pluripotent cells in monolayers derived from differentiated embryoid bodies. Int J Dev Biol. 2007;51:397–407. doi: 10.1387/ijdb.062255mr. [DOI] [PubMed] [Google Scholar]
- 34.Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A. Epigenetic differences between male and female bovine blastocysts produced in vitro. Physiol Genomics. 2008;32:264–272. doi: 10.1152/physiolgenomics.00234.2007. [DOI] [PubMed] [Google Scholar]
- 35.Hatano SY, Tada M, Kimura H, Yamaguchi S, Kono T, et al. Pluripotential competence of cells associated with Nanog activity. Mech Dev. 2005;122:67–79. doi: 10.1016/j.mod.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Brown DR, Besinger A. Prion protein expression and superoxide dismutase activity. Biochem J . 1998;334(Pt 2):423–429. doi: 10.1042/bj3340423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klamt F, Dal-Pizzol F, Conte da Frota MJ, Walz R, Andrades ME, et al. Imbalance of antioxidant defense in mice lacking cellular prion protein. Free Radic Biol Med. 2001;30:1137–1144. doi: 10.1016/s0891-5849(01)00512-3. [DOI] [PubMed] [Google Scholar]
- 38.Monnet C, Gavard J, Mege RM, Sobel A. Clustering of cellular prion protein induces ERK1/2 and stathmin phosphorylation in GT1-7 neuronal cells. FEBS Lett. 2004;576:114–118. doi: 10.1016/j.febslet.2004.08.076. [DOI] [PubMed] [Google Scholar]
- 39.Mouillet-Richard S, Ermonval M, Chebassier C, Laplanche JL, Lehmann S, et al. Signal transduction through prion protein. Science. 2000;289:1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- 40.Morita-Fujimura Y, Tokitake Y, Matsui Y. Dev Growth Differ; 2009. Heterogeneity of mouse primordial germ cells reflecting the distinct status of their differentiation, proliferation and apoptosis can be classified by the expression of cell surface proteins integrin alpha6 and c-Kit. [DOI] [PubMed] [Google Scholar]
- 41.Ishii M, Tay TW, Matsui T, Kidokoro T, Mizukami T, et al. Expression pattern of alphavbeta3 and alphavbeta5 integrin mRNA in mouse fetal gonads. J Reprod Dev. 2006;52:461–468. doi: 10.1262/jrd.17111. [DOI] [PubMed] [Google Scholar]
- 42.Muras AG, Hajj GN, Ribeiro KB, Nomizo R, Nonogaki S, et al. Prion protein ablation increases cellular aggregation and embolization contributing to mechanisms of metastasis. Int J Cancer. 2009;125:1523–1531. doi: 10.1002/ijc.24425. [DOI] [PubMed] [Google Scholar]
- 43.Hajj GN, Lopes MH, Mercadante AF, Veiga SS, da Silveira RB, et al. Cellular prion protein interaction with vitronectin supports axonal growth and is compensated by integrins. J Cell Sci. 2007;120:1915–1926. doi: 10.1242/jcs.03459. [DOI] [PubMed] [Google Scholar]
- 44.Hayashi Y, Furue MK, Okamoto T, Ohnuma K, Myoishi Y, et al. Integrins regulate mouse embryonic stem cell self-renewal. Stem Cells. 2007;25:3005–3015. doi: 10.1634/stemcells.2007-0103. [DOI] [PubMed] [Google Scholar]
- 45.Chambers I, Colby D, Robertson M, Nichols J, Lee S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 46.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Rao S, Chu J, Shen X, Levasseur DN, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 48.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, et al. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 49.Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 50.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 51.Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 52.O'Shea KS. Self-renewal vs. differentiation of mouse embryonic stem cells. Biol Reprod. 2004;71:1755–1765. doi: 10.1095/biolreprod.104.028100. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka TS. Transcriptional heterogeneity in mouse embryonic stem cells. Reprod Fertil Dev. 2009;21:67–75. doi: 10.1071/rd08219. [DOI] [PubMed] [Google Scholar]
- 54.Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, et al. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9:293–299. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- 55.Kondoh H. Cellular life span and the Warburg effect. Exp Cell Res. 2008;314:1923–1928. doi: 10.1016/j.yexcr.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Lee ST, Yun JI, Jo YS, Mochizuki M, van der Vlies AJ, et al. Engineering integrin signaling for promoting embryonic stem cell self-renewal in a precisely defined niche. Biomaterials. 2010;31:1219–1226. doi: 10.1016/j.biomaterials.2009.10.054. [DOI] [PubMed] [Google Scholar]
- 57.Walker JL, Zhang L, Zhou J, Woolkalis MJ, Menko AS. Role for alpha 6 integrin during lens development: Evidence for signaling through IGF-1R and ERK. Dev Dyn. 2002;223:273–284. doi: 10.1002/dvdy.10050. [DOI] [PubMed] [Google Scholar]
- 58.Graner E, Mercadante AF, Zanata SM, Forlenza OV, Cabral AL, et al. Cellular prion protein binds laminin and mediates neuritogenesis. Brain Res Mol Brain Res. 2000;76:85–92. doi: 10.1016/s0169-328x(99)00334-4. [DOI] [PubMed] [Google Scholar]
- 59.Schmitt-Ulms G, Legname G, Baldwin MA, Ball HL, Bradon N, et al. Binding of neural cell adhesion molecules (N-CAMs) to the cellular prion protein. J Mol Biol. 2001;314:1209–1225. doi: 10.1006/jmbi.2000.5183. [DOI] [PubMed] [Google Scholar]
- 60.Steele AD, Emsley JG, Ozdinler PH, Lindquist S, Macklis JD. Prion protein (PrPc) positively regulates neural precursor proliferation during developmental and adult mammalian neurogenesis. Proc Natl Acad Sci U S A. 2006;103:3416–3421. doi: 10.1073/pnas.0511290103. [DOI] [PMC free article] [PubMed] [Google Scholar]