Abstract
A set of shuttle vectors was constructed to facilitate expression of genes for metabolic engineering in Saccharomyces cerevisiae. Selectable markers include the URA3, TRP1, MET15, LEU2-d8, HIS3 and CAN1 genes. Differential expression of genes can be achieved as each marker is available on both CEN/ARS- and 2 μ-containing plasmids. Unique restriction sites downstream of TEF1, PGK1 or HXT7-391 promoters and upstream of the CYC1 terminator allow insertion of open-reading frame cassettes for expression. Furthermore, a fragment appropriate for integration into the genome via homologous recombination can be readily generated in a polymerase chain reaction. Vector marker genes are flanked by loxP recognition sites for the CreA recombinase to allow efficient site-specific marker deletion and recycling. Expression and copy number were characterized for representative high- and low-copy vectors carrying the different marker and promoter sequences. Metabolic engineering typically requires the stable introduction of multiple genes and genomic integration is often preferred. This requires an expanded number of stable expression sites relative to standard gene expression studies. This study demonstrated the practicality of polymerase chain reaction amplification of an expression cassette and genetic marker, and subsequent replacement of endogenous retrotransposons by homologous recombination with flanking sequences. Such reporters were expressed comparably to those inserted at standard integration loci. This expands the number of available characterized integration sites and demonstrates that such sites provide a virtually inexhaustible pool of integration targets for stable expression of multiple genes. Together these vectors and expression loci will facilitate combinatorial gene expression for metabolic engineering.
Keywords: Saccharomyces cerevisiae, vectors, integrants, markers, Cre recombinase, chromosomal sites, metabolic engineering
Introduction
Saccharomyces cerevisiae is an important organism for metabolic engineering. This yeast has been studied extensively, molecular and genetic tools are available for its manipulation and it is a successful industrial microorganism. Sets of low-and high-copy plasmid vectors and integrating vectors have found extensive use in gene function studies. These include plasmid sets that contain multiple cloning sites and are genetically marked with TRP1, HIS3, LEU2, URA3 (Christianson et al., 1992; Gietz and Sugino, 1988; Sikorski and Hieter, 1989), MET15 and ADE2 (Brachmann et al., 1998). These plasmids are ideal for studies of individual genes under their native promoters. Expression vectors include promoters and terminators flanking the cloning site. Examples include the pYES2.0 galactose-inducible URA3-marked vector (Invitrogen, Carlsbad, CA, USA), the tet-on/off vectors (Gari et al., 1997) and a set of vectors with ADH1, TEF1 and GPD1 promoters (Mumberg et al., 1995).
Although plasmid vectors present a useful platform for many individual gene expression studies, high copy plasmids are relatively unstable (Futcher and Carbon, 1986; Mead et al., 1986) and CEN/ARS plasmids can also become unstable in combination (Futcher and Carbon, 1986). Depending on the application, stable maintenance of sequences can be accomplished with genomic integration of the cloned sequences. In S. cerevisiae, this was originally achieved by tagging the gene of interest with a selectable marker and incorporating sequences homologous to the integration target site to allow targeted integration of plasmids (Gietz and Sugino, 1988; Scherer and Davis, 1979; Sikorski and Hieter, 1989). This strategy was later modified by flanking the URA3 marker gene with direct repeats of the 1 kb bacterial hisG sequence from bacteria (‘URA3 blaster’) to facilitate gene knockouts (Alani et al., 1987) or insertions (Lee and Da Silva, 1997) and subsequent marker recycling by homologous recombination between the direct repeats. However, the relatively long hisG sequences used to mediate homologous recombination in that strategy left a significant footprint and thereby risked off-target homologous recombination during subsequent transformations. This complication limited the potential for marker recycling. Site-specific recombination systems were introduced to address this problem. These utilized much shorter direct repeats that could be efficiently recombined upon expression of the appropriate recombinase (Johansson and Hahn-Hagerdal, 2002; Prein et al., 2000; Radhakrishnan and Srivastava, 2005). One example of such a strategy is the CreA–loxP system (Gueldener et al., 2002; Güldener et al., 1996; Sauer, 1994). In addition to vector-based integration, polymerase chain reactions (PCRs) have been used to generate fragments for genomic insertion (Lorenz et al., 1995).
Current vector and PCR-based integration cassettes are particularly well-suited to manipulation of individual genes. Metabolic engineering differs from this undertaking in its specific requirement for testing of multiple genes in order to optimize pathway expression and function. Combinations of plasmids are sometimes useful for initial testing. However, because of issues of plasmid instability, as discussed above, it is ultimately desirable to stabilize pathways by integration of multiple expression cassettes and the number of characterized genomic loci for such integrations is limited.
In order to facilitate metabolic engineering in S. cerevisiae, we have constructed and characterized a toolkit of 28 expression vectors (pXP). This pXP vector set includes high- and low-copy plasmids marked with six standard genetic markers. Cloning sites are flanked by a choice of three promoters and the CYC1 terminator (TCYC). These are adjacent to selectable markers in the vector plasmids, so that the cloned sequence of interest and selectable marker can be amplified together to obtain a fragment suitable for genomic integration. Markers are flanked by loxP sites to allow recycling. Expression of luciferase reporters (McNabb et al., 2005) carried on vectors was compared to expression of the same reporters from previously characterized and novel chromosomal loci. These vector- and genome-based metabolic engineering tools enable plasmid-based testing followed seamlessly by genomic integration for pathway optimization.
Materials and methods
Yeast and bacterial strains and culture conditions
Yeast and bacterial culture methods were standard (Amberg et al., 2005; Ausubel, 2008) except where noted. Escherichia coli strain DH5α [F− φ 80lacZM15Δ (lacZYA-argF) U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ−] (Invitrogen) was used for plasmid preparations. S. cerevisiae BY4741 (MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ) (Open Biosystems, Huntsville, AL, USA) is related to the sequenced strain S288C (Goffeau et al., 1996). Yeast strain yBF1587 was derived from strain BY4741 by deletions of the ADH2, GRE3 and TRP1 open reading frames (ORFs). When using vectors in the trp1 Δ background, it should be kept in mind that the trp1 Δ ORF allele may confer undersirable side-effects, depending upon the experimental conditions (Gonzalez et al., 2008). Strains are described in Table 1. Derivation of reporter strains is described below and derivation of yBF1587 is described in the Materials section of the Supporting information. Yeast were cultured in synthetic minimal medium (SD) with dextrose (2%) as a carbon source and lacking uracil (SD-ura) or specific amino acids (Amberg et al., 2005).
Table 1.
Yeast strains constructed in this study
| Strain* | Genomic integration |
|---|---|
| yFF1743 | ura3::PPGK1-Rluc-loxP-URA3-loxP |
| yFF1744 | met15:: PPGK1-Rluc-loxP-URA3-loxP |
| yFF1745 | leu2:: PPGK1-Rluc-loxP-URA3-loxP |
| yFF1746 | trp1:: PPGK1-Rluc-loxP-URA3-loxP |
| yFF1748 | yilmty3-1:: PPGK1-Rluc-loxP-URA3-loxP |
| yFF1749 | ylrcty1-1:: PPGK1-Rluc-loxP-URA3-loxP |
| yFF1750 | ymlwty1-1::PPGK1-Rluc-loxP-URA3-loxP |
| yFF1751 | ydrcty1-2:: PPGK1-Rluc-loxP-URA3-loxP |
| yFF1752 | yblwty1-1::PPGK1-Rluc-loxP-URA3-loxP |
| yFF1753 | ydrwty1-5::PPGK1-Rluc-loxP-URA3-loxP |
| yFF1754 | ymlwty1-2::PPGK1-Rluc-loxP-URA3-loxP |
| yFF1755 | yprcty1-2::PPGK1-Rluc-loxP-URA3-loxP |
| yFF1756 | ymlwty1-1::PPGK1-Rluc-loxP-URA3-loxP (PPGK1 is proximal to tRNA region) |
| yFF1757 | ymlwty1-2::PPGK1-Rluc-loxP-URA3-loxP (PPGK1 is proximal to tRNA gene) |
| yBF1587 | MATa his3 Δ 1 leu2 Δ 0 met15 Δ 0 ura3 Δ 0 adh2 Δ 0 gre3 Δ 0 trp1 Δ 0 his3 |
| yFF1683 | MATa his3 Δ 1 leu2 Δ 0 met15 Δ 0 ura3 Δ 0 adh2 Δ 0 gre3 Δ 0 trp1 Δ 0 his3::PTK-Fluc-loxP |
Strains yFF1743–yFF1757 are derivatives of yFF1683. yBF1587 and yFF1683 are derived from BY4741.
pXP series plasmid constructions
Recombinant manipulations were conducted using standard molecular techniques (Ausubel, 2008). Sequences of fragments amplified in PCR were verified by DNA sequence analysis (data not shown) (GeneWiz, South Plainfield, NJ, USA). Strains with genomic integrations were identified as prototrophs based on marker gene activity and were verified by PCR generation of diagnostic fragments of appropriate size using primers within the integrated sequence and within flanking DNA (data not shown). Details of constructions, oligonucleotide primer sequences, and sequences of plasmids are provided in the Supporting information. Plasmid constructs and source DNAs are described in Table 2.
Table 2.
List of plasmids created in this study
| Plasmid name | Base vector | Description |
|---|---|---|
| pXP1 | pCR Blunt II | PPGK1 with 53 SspI and 33 SpeI |
| pXP2 | pCR Blunt II | TCYC1 with 53 SpeI-N12-XhoI and 33 SspI |
| pXP3 | pCR Blunt II | 2 μ with EcoRI flanking sites |
| pXP4 | pCR Blunt II | CEN/ARS ori with EcoRI flanking sites |
| pXP5 | pCR Blunt II | TRP1 marker with loxP repeats and SmaI flanking sites |
| pXP6 | pCR Blunt II | URA3 marker with loxP repeats and SmaI flanking sites |
| pXP7 | pCR Blunt II | CAN1 marker with loxP repeats and SmaI flanking sites |
| pXP8 | pCR Blunt II | MET15marker with loxP repeats and SmaI flanking sites |
| pXP9 | pCR Blunt II | PPGK1-TCYC1 SspI cassette |
| pXP13 | pUC18 | PPGK1-TCYC1 SspI cassette |
| pXP16 | pCR Blunt II | PTEF1 with 53 SspI and 33 SpeI |
| pXP17 | pUC18 | PTEF1-TCYC1 SspI cassette |
| pXP100 | pXP13 | CEN/ARS, PPGK1 |
| pXP200 | pXP13 | 2 μ, PPGK1 |
| pXP300 | pXP17 | CEN/ARS, PTEF1 |
| pXP400 | pXP17 | 2 μ, PTEF1 |
| pXP112 | pXP100 | PPGK1−, CAN1+, CEN/ARS− |
| pXP212 | pXP200 | PPGK1−, CAN1+, 2 μ− |
| pXP312 | pXP300 | PTEF1+, CAN1−, CEN/ARS+ |
| pXP412 | pXP400 | PTEF1+, CAN1+, 2 μ− |
| pXP114 | pXP100 | PPGK1−, MET15+, CEN/ARS− |
| pXP214 | pXP200 | PPGK1−, MET15+, 2 μ− |
| pXP314 | pXP300 | PTEF1+, MET15−, CEN/ARS+ |
| pXP414 | pXP400 | PTEF1+, MET15−, 2 μ+ |
| pXP116 | pXP100 | PPGK1−, TRP1+, CEN/ARS− |
| pXP216 | pXP200 | PPGK1−, TRP1+, 2 μ− |
| pXP316 | pXP300 | PTEF1+, TRP1+, CEN/ARS+ |
| pXP416 | pXP400 | PTEF1+, TRP1+, 2 μ+ |
| pXP118 | pXP100 | PPGK1−, URA3−, CEN/ARS− |
| pXP218 | pXP200 | PPGK1−, URA3−, 2 μ− |
| pXP318 | pXP300 | PTEF1+, URA3+, CEN/ARS+ |
| pXP418 | pXP400 | PTEF1+, URA3+, 2 μ+ |
| pXP120 | pXP100 | PPGK1−, HIS3−, CEN/ARS− |
| pXP220 | pXP200 | PPGK1−, HIS3+, 2 μ− |
| pXP320 | pXP300 | PTEF1+, HIS3+, CEN/ARS+ |
| pXP420 | pXP400 | PTEF1+, HIS3+, 2 μ− |
| pXP122 | pXP100 | PPGK1−, LEU2-d8−, CEN/ARS− |
| pXP222 | pXP200 | PPGK1−, LEU2-d8−, 2 μ− |
| pXP322 | pXP300 | PTEF1+, LEU2-d8+, CEN/ARS+ |
| pXP422 | pXP400 | PTEF1+, LEU2-d8+, 2 μ+ |
| pXP518 | pBF3055 | PHX7–391+, URA3+, CEN/ARS− |
| pXP618 | pBF3055 | PHX7–391+, URA3+, 2 μ+ |
| pXP522 | pBF3055 | PHX7–391+, LEU2-d8+, CEN/ARS− |
| pXP622 | pBF3055 | PHX7–391+, LEU2-d8+, 2 μ− |
| pXP114-Rluc | pXP114 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP214-Rluc | pXP214 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP116-Rluc | pXP116 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP216-Rluc | pXP216 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP118-Rluc | pXP118 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP218-Rluc | pXP218 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP318-Rluc | pXP318 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP418-Rluc | pXP418 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP518-Rluc | pXP518 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP618-Rluc | pXP618 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP120-Rluc | pXP120 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP220-Rluc | pXP220 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP122-Rluc | pXP122 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pXP222-Rluc | pXP222 | 6A-Rluc was inserted into SpeI and XhoI sites |
| pBF3037 | pCRBluntII | URA3 was cloned into NheI and MluI flanking by two codon-optimized 200 bp GAG fragments: one was cloned into AscI and NheI sites and the other was cloned into MluI and SalI sites |
| pKN2735 | pYES2 | LEU2 replaced URA3 |
| pKN2736 | pKN2735 | The MCS was modified by introducing NdeI and XmaI |
| pNB3045 | pXP100 | LEU2-d8 was flanked by SmaI-loxP on both sides |
| pBF3038 | pKN2736 | creA was cloned between NcoI and XmaI |
| pBF3052 | pXP100 | PPGK1-TCYC1 was deleted by SspI digestion |
| pBF3055 | pBF3052 | PHXT7–391-TCYC1was inserted into NdeI and HindIII sites |
| pBF3060 | pBF3038 | URA3 replaced LEU2 |
| pBF3187 | pXP522 | PTK-Fluc was inserted into NdeI and XhoI sites |
Sequence orientation in the pUC18 vector, proceeding 5′ to 3′ clockwise around the map shown in Figure 1.
Sequence described in Supporting information S3.
Sequence described in Supporting information S3, orientated with 5′ end distal to the SspI site.
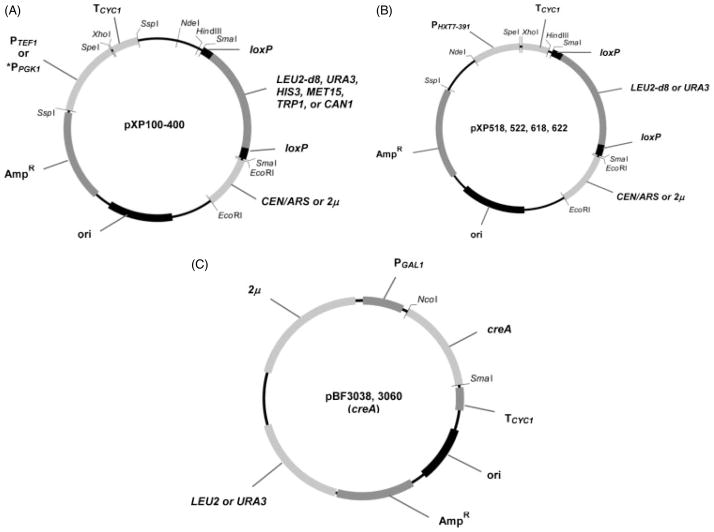
Plasmid pUC18 (Yanisch-Perron et al., 1985) provided the backbone for the pXP vector series. The pUC18 plasmid is 2686 bp in length and contains a multiple cloning site (MCS), the pMB1 replication origin mutated to ensure high-copy number in bacteria and the bla ampicillin resistance marker to enable selection. Two different promoters (PPGK 1 and PTEF1) coupled with TCYC 1 for transcription termination were inserted into the unique SspI site. Promoter and terminator sequences were separated by unique SpeI and XhoI sites to allow directional cloning of ORFs to be expressed. CEN6/ARS4 or 2 μ sequences were inserted into the EcoRI site of the MCS and six different marker genes were individually inserted into the SmaI site of the MCS in the respective high-and low-copy vectors. Altogether these manipulations yielded a set of 24 plasmids (Figure 1A, Table 2). The 391 bp HXT7 promoter (PHXT 7–391) coupled with TCYC 1 was inserted at the unique NdeI and HindIII sites in the MCS in high- and low-copy plasmids with URA3 and LEU2-d8 marker genes to create an additional four plasmids for a final total of 28 plasmids (Figure 1B, Table 2). The LEU2-d8 allele encodes LEU2 truncated by 8 bp and complements LEU2Δ.
Figure 1.
Schematic representation of pXP plasmid series and creA plasmids. (A) The pXP plasmids contain CEN/ARS or 2 micron sequences, unique SpeI–XhoI restriction sites separating PPGK1/TEF1 and TCYC1 located at the unique SspI site (note that PPGK1-TCYC1 cassette is in the negative orientation, opposite to that of PTEF1-TCYC1), and six alternative markers flanked by loxP sites cloned into the unique SmaI site. (B) The pXP plasmids contain CEN/ARS or 2 micron sequences, unique SpeI–XhoI restriction sites separating PHXT7–391 and TCYC1 cloned into the unique NdeI and HindIII sites in the multiple cloning site (MCS), and URA3- or LEU2-d8 markers as described in (A). (C) The pYES2.0-based creA expression plasmid marked with URA3 or LEU2 marker
Individual fragments for the construction of these vectors were derived by PCR amplification of genomic DNA or plasmid templates, using Pfu Ultra II polymerase (Stratagene, La Jolla, CA, USA) or KOD polymerase (Novagen, Gibbstown, NJ, USA) as suggested by the manufacturer. Each fragment was cloned into the pCR Blunt II TOPO cloning vector (Invitrogen).
CreA plasmid construction
The gene encoding bacteriophage P1 CreA recombinase was recoded for expression in S. cerevisiae using computationally optimized DNA assembly (CODA) (Hatfield and Roth, 2007; Larsen et al., 2008) and the optimized creA sequence (see Supporting information S1). The assembled fragment was amplified using PCR and primers containing NcoI and SmaI sites at the 5′ and 3′ ends, respectively. The creA fragment was ligated into the pCR-Blunt II TOPO cloning vector. Plasmid pKN2736 is similar to pYES2.0, but is marked with LEU2 and contains a SmaI restriction site in the MCS (K. Nguyen, personal commmunication). The creA NcoI–SmaI fragment was cloned into the NcoI and SmaI sites of plasmid pKN2736 to create plasmid pBF3038. In order to express CreA from a URA3-marked vector, the ScaI fragment containing LEU2 was replaced with a fragment containing URA3 to produce plasmid pBF3060 (Figure 1C). Recoded CreA recombinase was confirmed to be active based on the deletion of marker genes flanked by direct repeats of the loxP recognition sequence. In a typical experiment ~200 cells were plated onto YPD, allowed to grow into colonies and replica-plated onto SD-ura or -leu. Typically 10–20% of cells had lost the marker even in the absence of galactose induction of the CreA promoter. Appropriate auxotrophs were then screened by PCR for loss of the marker between the loxP sites using primers in the flanking genomic target DNA. Typically four colonies were tested and all four had the expected deletion. Up to five sequential rounds of marker deletion have been performed in the same strain (data not shown).
Luciferase reporter plasmid construction
Gene expression from vectors and integration sites was measured as activity of Renilla luciferase (RLuc) relative to activity of an integrated copy of Firefly luciferase (FLuc) (described below). The Rluc gene templated from phRL-null (Promega, Madison, WI, USA) was amplified with flanking SpeI and XhoI sites and cloned under the control of PPGK 1 and TCYC 1 in the SpeI and XhoI sites of low-copy pXP100 and high-copy pXP200 derivatives containing MET15, TRP1, LEU2-d8, HIS3 and URA3 genetic markers. This fragment was similarly cloned under control of PTEF1 and PHXT 7–391 and TCYC 1 in URA3-marked low- and high-copy vectors.
Construction of yeast reporter strains
In order to monitor expression from a subset of vectors and from test chromosomal sites, luciferase assays were used. RLuc reporter activity was normalized to FLuc activity expressed from an integrated copy of Fluc. Fluc was expressed under control of the human thymidine kinase promoter (PTK ) and TCYC 1 (PTK -Fluc-TCYC 1). PTK was amplified in a PCR using pRL-TK (Promega) as a template and primers with NdeI and SpeI sites. This fragment was used to replace PHXT 7–391 in pXP522. The Fluc ORF was amplified in a PCR using pGL3-Basic (Invitrogen) as a template and primers to incorporate a SpeI restriction site followed by A6 and a XhoI site at the 5′ and 3′ ends, respectively. It was then inserted downstream of PTK between the unique SpeI and XhoI sites to give plasmid pBF3187.
PTK -Fluc-TCYC 1 was introduced into yeast strain yBF1587 by two-fragment transformation (Nielsen et al., 2007). PCR was used to generate fragments containing PTK -Fluc-TCYC 1 and loxP-LEU2-d8-loxP with 50 bp of overlapping sequence at the 3′ end of the former and the 5′ end of the latter and 50 bp on the non-overlapping ends of each with homology to genomic sequences flanking HIS3. The two PCR fragments were co-transformed (Gietz et al., 1995) into yeast strain yBF1587. The LEU2-d8 marker was deleted by introduction of plasmid pBF3060 from which CreA was expressed. The resulting strain was designated yFF1683.
To compare gene expression from different chromosomal sites, the PPGK 1-Rluc-loxP-URA3-loxP cassette was introduced at test loci in strain yFF1683 containing the integrated PTK -Fluc-TCYC 1 reporter. This was accomplished by amplification of PPGK 1-Rluc-TCYC 1 and loxP-URA3-loxP fragments and co-transformation as described for Fluc. Integrations into chromosomal loci were confirmed by PCR analysis using primers that annealed upstream and downstream of these chromosomal sites.
Dual luciferase assay
Luciferase activity was assayed as previously described (McNabb et al., 2005), using the dual luciferase reporter (DLR) assay (Sherf et al., 1996; Promega) in which activity of the test reporter, Rluc, was normalized to activity of the integrated control reporter, Fluc. Luminescence measurements were performed with a Sirius Single Tube Luminometer (Berthold Detection Systems GmbH, Pforzheim, Germany). Data are expressed as the ratio of RLuc to FLuc activity (Rluc/Fluc) within the linear range. Cells were sampled in the range OD600 nm = 0.8–1.0.
Copy-number determination
Low- and high-copy plasmids were transformed into yeast strain yFF1683 containing the integrated PTK - Fluc-TCYC 1 reporter. Cells were grown to exponential phase in the appropriate dropout medium at 30 °C, suspended in breaking buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 100 mM NaCl, 2% Triton X-100, 1% SDS) and vortexed vigorously with glass beads and phenol : chloroform. Extracted nucleic acid was precipitated in ethanol, redissolved and incubated with RNase (Hoffman and Winston, 1987). DNA was digested with HindIII and subjected to Southern blot analysis using standard procedures (Ausubel, 2008). Plasmid copy number was determined based on hybridization of a 32P-radiolabelled PPGK 1-specific probe. Intensity of hybridization signals was determined using Quantity One software (Bio-Rad, Richmond, CA, USA). Intensity of the band from the vector-based promoter PPGK 1 signal was normalized to the intensity of the chromosomal promoter PPGK 1 signal.
Results and discussion
Vector design strategy
Genetic engineering of metabolic pathways often involves expression of multiple genes. In this study, a shuttle vector series based on pUC18 was constructed for use in E. coli and S. cerevisiae. Six markers that can be selected in commonly-used strains of S. cerevisiae (CAN1, MET15, TRP1, URA3, LEU2-d8 and HIS3 ) are represented in the series. In addition, each marker was cloned in vectors with either 2 μ or CEN/ARS sequences to allow high- or low-copy level expression, respectively. Plasmids were constructed with three S. cerevisiae promoters including, for all six markers, two strong promoters (PPGK 1 and PTEF1) (Figure 1A) and, in addition, PHXT 7–391, for URA3 and LEU2-d8 (Figure 1B). Should a particular marker not be available as an ORF deletion allele in the background strain of choice, it is possible to amplify a deletion cassette from any of the plasmid markers. The vector markers are flanked by loxP sites so that marker and flanking loxP sites can be amplified and used to replace chromosomal sequences, resulting in generation of deletion alleles. The marker gene can then be removed following CreA expression and recombination of the loxP repeats.
Although plasmids provide a useful testbed for the initial characterization of gene expression, pathway construction and optimization typically requires stable expression and therefore chromosomal integration of expression cassettes. To facilitate this process, the pXP vectors were designed not only to include a wide choice of marker genes but also with compact arrangement of expression cassette and marker gene to allow straightforward amplification of a PCR fragment for homology-targeted chromosomal integration. Thus, the expression cassette and flanking marker can be amplified for transformation using primers whose outside ends have ~50 bp of homology to the target genomic sequence. Introduction of a pathway typically involves integration of multiple genes. An advantage of the current collection of markers is that genes can be integrated for preliminary testing without need for time-consuming sequential deletion of markers for re-use. However, most applications ultimately require deletion of markers to allow recycling or other strain manipulation. One method of marker gene elimination is to flank the marker with direct repeats of sufficient length to insure gene loss via homologous recombination (Alani et al., 1987; Wilson et al., 2000). Alternatively, a site-specific recombinase can be employed, such as the bacteriophage loxP– CreA (Hoess and Abremski, 1984) or the yeast FRT–FLP (Radhakrishnan and Srivastava, 2005) systems. For marker deletion to allow recycling, we chose CreA-mediated site-specific recombination. Marker genes were flanked with direct repeats of loxP, the 34 bp recognition sequence of the CreA recombinase, which is active in yeast (Sauer, 1987). In the direct repeat configuration of loxP sites, CreA-mediated recombination deletes the marker gene and leaves one copy of loxP (Güldener et al., 1996; Sauer, 1994). Advantages of this approach include the small size of the recognition sequence (avoiding inappropriate targeting of subsequent integrations), the efficiency of the marker excision and the ability to remove multiple markers simultaneously (Delneri et al., 2000) or sequentially (Güldener et al., 1996). CreA was expressed from PGAL1 on high-copy plasmids marked with URA3 or LEU2 (Figure 1C). Even uninduced expression of the recombinase resulted in sufficient recombination to allow cells from which the marker is lost to be readily identified on appropriate drop-out media in previous studies (Gueldener et al., 2002) and in our study involving different CreA expression vectors (data not shown).
Comparison of plasmid copy number
Plasmid copy number is an important basal determinant of cloned gene expression level. Copy number was determined for the PPGK 1 vectors, marked with MET15 and URA3, for both the empty and Rluc-expression derivatives. DNA was isolated from cells and Southern blot analysis was performed using hybridization of vector and chromosomal DNA to a PPGK 1-specific probe. As shown in Figure 2 and Table 3, this ratio for empty low-copy CEN/ARS vectors with either marker was 1.5. The empty 2 μ vector showed differences between the MET15- and URA3-marked versions with ratios of 7.5 and 11.6, respectively, resulting in ratios of high- to low-copy empty vectors marked with MET15 and URA3 of 5.2 and 7.8, respectively. A caveat in these experiments is that they measure average copy number and the 2 μ plasmid in particular is known to be relatively unstable (Mead et al., 1986).
Figure 2.
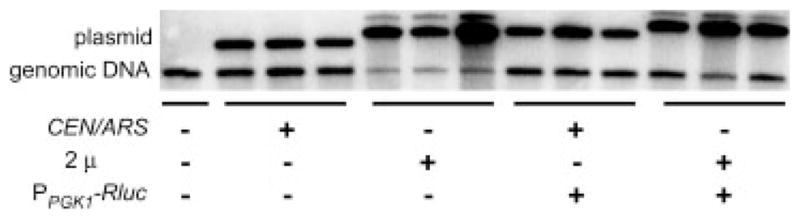
CEN/ARS and 2 μ plasmid copy number. Southern blot analysis of cells containing URA3-marked plasmids. DNA was prepared from logarithmic phase cultures containing low- and high-copy URA3-marked vectors and restricted with HindIII. Triplicate samples of 20 μg total DNA for low-copy plasmids and 2 μg total DNA for high-copy plasmids were analysed by Southern blot analysis using a probe specific for PPGK1, as described in Materials and methods. Band intensity was quantitated in the linear range for plasmid and chromosomal fragment hybridization using Quantity One Software (Bio-Rad)
Table 3.
Low- and high-copy MET15- and URA3-marked plasmid copy number. The ratio of plasmid-borne PPGK1 hybridization was normalized to chromosomal PPGK1 hybridization and the average of three measurements is shown
| Marker | Relative copy number |
|||||
|---|---|---|---|---|---|---|
| Plasmids |
Plasmids with Rluc expression |
|||||
| CEN/ARS/genome | 2 μ/genome | 2 μ/CEN/ARS | CEN/ARS/genome | 2 μ/genome | 2 μ/CEN/ARS | |
| MET15 | 1.4 ± 0.2 | 7.5 ± 1.0 | 5.2 ± 1.0 | 1.9 ± 0.4 | 4.2 ± 0.4 | 2.2 ± 0.6 |
| URA3 | 1.5 ± 0.03 | 11.6 ± 0.8 | 7.8 ± 0.6 | 1.6 ± 0.1 | 5.6 ± 0.9 | 3.4 ± 0.6 |
Expression of cloned genes can affect plasmid copy number. In order to explore this variable, DNA from cells containing low- and high-copy vectors carrying the PPGK 1-RLuc-TCYC 1 expression cassette was hybridized to the PPGK 1 probe. The copy numbers of the low-copy Rluc reporter plasmids were 1.9 and 1.6 for the MET15- and URA3-marked vectors, respectively; these values were similar to the empty CEN/ARS vectors. In contrast, the copy numbers of the high-copy PPGK 1-RLuc-TCYC 1 reporter plasmids were lower than those for the empty vectors, dropping from 7.5 to 4.2 and from 11.6 to 5.6 for the plasmids marked with MET15 and URA3, respectively. The copy-number ratios of 2 μ to CEN/ARS for MET15-and URA3-marked plasmids actively expressing the reporter under the PGK1 promoter were therefore 2.2 and 3.4, respectively. Overall, these ratios are lower than what might have been anticipated based on published reports of vector copy number and suggest that effects of cloned gene expression on copy number should be a consideration in planning vector-based experiments. These findings also underscore the potential advantage of integrating multiple copies of genes rather than relying on vector-based strategies.
Luciferase assays for evaluation of vector-based gene expression
Because expression level is critical for metabolic engineering, but is affected by multiple variables, vectors were also directly compared using reporter assays. Luciferase reporter systems offer the advantages of a rapid and quantitative assay. Reporters configured so that activity can be measured as the ratio of Rluc test activity to Fluc control activity have the advantage of being internally controlled. PTK displays weak activity in S. cerevisiae (Moriyoshi, 2009) and was therefore used to express Fluc at a useful baseline level for normalization. PTK -Fluc-TCYC 1 was integrated into yeast strain yBF1587 to create strain yFF1683. The Rluc ORF was cloned under PPGK 1 in the high-and low-copy MET15-, TRP1-, HIS3-, LEU2-d8-, and URA3-marked vectors. RLuc reporter plasmid transformants were grown in logarithmic phase in selective medium for several doublings and the dual luciferase assay (McNabb et al., 2005; Sherf et al., 1996) was performed as described in Materials and methods. To determine the linear range of the assay, RLuc/Fluc was determined in cells expressing the URA3-marked high- and low-copy plasmids with amounts of lysate representing 500–10 000 cells (R2 = 0.99). Within this range, the mean RLuc : FLuc ratio in cells expressing the low-copy plasmid was 33 (±3) × 103 (Figure 3A). Similarly, activity was relatively independent of cell number for measurements from cells containing the high-copy plasmid between 500 and 10 000 cells, with some loss of linearity between 5000 and 10 000. With <5000 cells, the mean Rluc : Fluc ratio was 141 (±6) × 103 (Figure 3A). Thus, Rluc activity in cells expressing the high-copy URA3 reporter was about four-fold greater than activity in cells expressing the low-copy URA3 reporter. This is consistent with the 3.4 ratio of high- to low-copy plasmid number observed for the URA3 reporter (Figure 2B).
Figure 3.
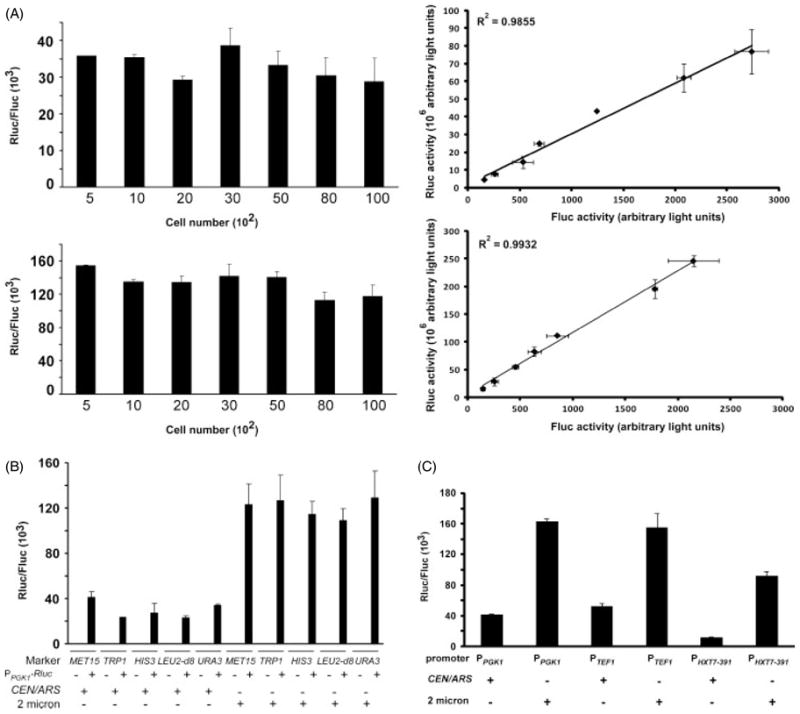
Analysis of vector expression by dual luciferase assay (DLR). (A) Luciferase assay for cells containing CEN/ARS and 2 μ URA3-marked plasmids. PPGK1-Rluc reporter in plasmids pXP118 and pXP218 were transformed into yeast strain yFF1683 and cells were grown in SD – ura medium to logarithmic phase. Cell number was determined by OD at A600 and the indicated number of cells was used to assay plasmid-borne Rluc and chromosomal Fluc activities. Data represent the mean of three independent assays and the bars show one standard deviation (SD). The plasmids without the Rluc reporter served as negative controls. Upper panel, cells contain pXP118-Rluc (CEN/ARS plasmid with URA3 marker). Lower panel, cells contain pXP218-Rluc (2 μ plasmid with URA3 marker). Left panels, RLuc : FLuc ratios; right panels, scatter plots of Rluc vs. Fluc for the same number of cells as shown in left panels. (B) Comparison of expression from differently marked pXP vectors. PPGK1-Rluc-TCYC1 reporters in the CEN/ARS and 2 μ plasmids with MET15, TRP1, HIS3, LEU2-d8 and URA3 were transformed individually into yeast strain yFF1683. Activities were analysed as described in (A). (C) Comparison of expression from pXP CEN/ARS and 2 μ vectors using different promoters. Cells containing URA3-marked vectors with PPGK1, PTEF1 and PHXT7–391 controlling expression of Rluc were analysed as described in (A)
RLuc/FLuc activity ratios were also determined for cells transformed with low- and high-copy vectors carrying five different selectable markers and PPGK 1-Rluc-TCYC 1. As observed for cells containing the URA3-marked vectors, cells containing vectors carrying other marker genes showed three- to five-fold greater activity for high-copy than low-copy versions (Figure 3B). The MET15 marker showed the least and the TRP1 and LEU2-d8 marked plasmids the greatest differential between high- and low-copy vector transformants.
Effect of promoter on gene expression
In addition to copy number, promoter activity is a major determinant of expression level. Therefore, it was of interest to determine the RLuc : FLuc ratios for plasmids carrying different promoters but a common marker. PPGK 1, PTEF1 and PHXT 7–391 (Lai et al., 2007) were chosen because they are relatively strong promoters under most growth conditions. Expression from these promoters on URA3-marked low- and high-copy plasmids in logarithmic growth was compared using the luciferase assay (Figure 3C). In cells containing low-copy plasmids, the Rluc : Fluc ratio was 41 (±1) × 103 with PPGK 1 and 52 (±4) × 103 with PTEF1. This suggests that PTEF1 may be slightly stronger than PPGK 1. This would be consistent with previous reports (Nacken et al., 1996). In cells expressing RLuc from the high-copy plasmids, the RLuc/FLuc ratios for PPGK 1 and PTEF1 were similar [163 (±4) × 103 vs. 155 (±19) × 103]. However, cells expressing RLuc from these promoters had significantly higher RLuc : FLuc ratios than cells expressing RLuc from PHXT 7–391. The RLuc : FLuc ratios in cells with vectors utilizing the PHXT 7–391 promoter were 12 (±1) × 103 for the low-copy plasmid and 92 (±6) × 103 for the high-copy plasmid.
The difference in RLuc expression among PPGK 1 and PTEF1 and PHXT 7–391 indicated that PHXT 7–391 is the weakest promoter in the context tested. In the case of PPGK 1-Rluc-TCYC 1, on URA3-marked plasmids there is an approximately four-fold difference in activity between high- and low-copy number plasmids. Similarly, in the case of PTEF1-Rluc-TCYC 1 the ratio between high- and low-copy plasmids is about 3. In contrast, for PHXT 7–391-Rluc, this ratio is about 8 (Figure 3C). These results suggest that in the case of the 2 μ plasmid, a weaker promoter suppresses copy number less than a strong promoter. Mumberg et al. (1995) compared the effect of promoter strength on relative activity of beta-galactosidase in cells carrying a lacZ reporter on high- or low-copy pRS vectors. These investigators found that the ratio of β-gal between cells with 2 μ and CEN/ARS vectors was three-fold when lacZ was driven by the strong PGPD1 promoter but 30-fold when it was expressed under the weaker PADH 1. Different versions of the PGAL1 showed similar patterns. Four-fold higher β-gal activity was observed in cells expressing the reporter from PGAL1 on a high-copy plasmid compared to a low-copy plasmid, but if the UAS was truncated the ratio increased to 30-fold (Mumberg et al., 1994). These data are consistent with our results.
Comparison of expression from marker and Ty genomic loci
For the introduction of multiple pathway genes, chromosomal integration of expression cassettes is often desired to both control and stabilize expression levels. However, there are relatively few quantitative comparisons of expression from different chromosomal integration sites. Furthermore, the number of frequently used sites is limited so that in the case of pathway engineering it may be necessary to integrate multiple genes into one site or into uncharacterized sites. In order to compare expression from various genomic loci, the expression cassette PPGK 1-Rluc-TCYC 1-loxP-URA3-loxP was amplified using PCR, and integrated at several frequently used loci in the yFF1683 PTK -Fluc-TCYC 1 reporter strain. The amplification used primers with outside ends of 50 bp of sequence flanking the target promoter and downstream end of the ORF (see Supporting information S2), so that PPGK 1-Rluc- TCYC 1-loxP-URA3-loxP replaced the target loci (Figure 4A). Replacement integrations targeted the URA3, MET15, LEU2 and TRP1 loci using the homologous sequences upstream of promoter and downstream of terminator regions. Comparison of RLuc : FLuc activity ratios in strains with different integration sites showed that they varied from 17 (±1) × 103 to 25 (±3) × 103, but overall were similar (Figure 4B). This included expression at the TRP1 locus, which is peri-centromeric (CENIV ). Comparison of activity of integrated reporters to activity in a strain transformed with pXP100, (CEN/ARS, URA3-marked vector) carrying PPGK 1-Rluc-TCYC 1 grown under selection for the plasmid, showed about 1.5-fold higher activity in the strain expressing the low-copy vector PPGK 1-Rluc-TCYC 1 (Figure 4B). This is consistent with the copy number slightly greater than one for the CEN/ARS plasmids determined in this study and previously reported (Amberg et al., 2005; Singh and Weil, 2002).
Figure 4.
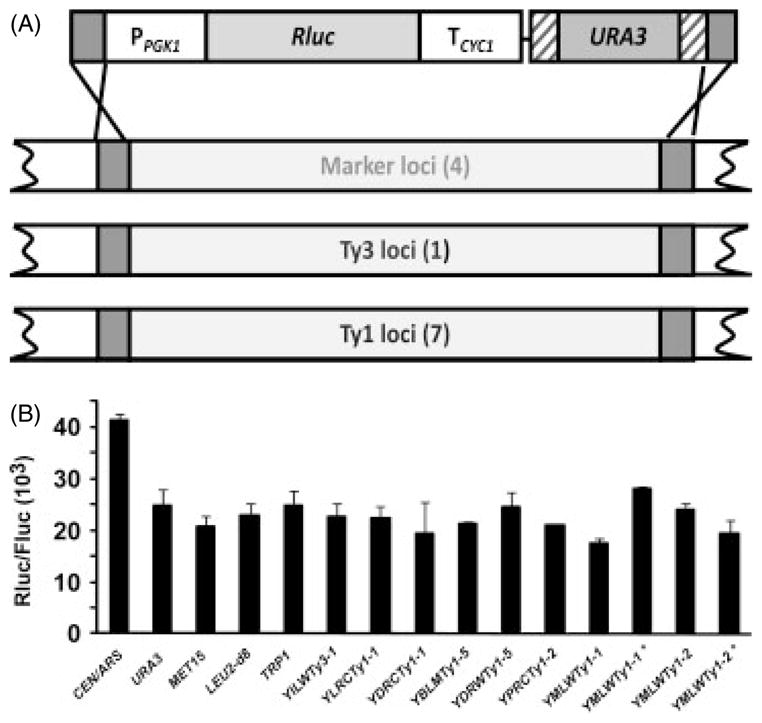
Comparison of PPGK1-Rluc-TCYC1 expression from different chromosomal loci. (A) Schematic representation of integration strategy. PPGK1-Rluc-TCYC1 and URA3 marker cassette were used to replace genomic loci in strain yFF1683. (B) Rluc/Fluc is shown for PPGK1-Rluc-TCYC1 expression from a CEN/ARS vector (pXP118) and from genomic sites in strain yFF1683. Assays were as described in Figure 3 legend. Asterisks indicate that the PPGK1-Rluc-TCYC1 promoter is proximal to the tRNA in the Ty replacement
Current S. cerevisiae strain engineering typically targets genes to well-characterized chromosomal loci, such as the ones described above. However, this strategy can be ultimately limited by availability of appropriate replacement sites, as relatively few such sites have been thoroughly characterized. An approach that potentially addresses this limitation is replacement of endogenous retrotransposons with heterologous sequences. Ty retrotransposons exist in multiple copies in most strains and ones that are represented in current genomes are likely well tolerated (Sikorski and Boeke, 1991). Ty1 and Ty3 long terminal repeat (LTR) retrotransposons in S. cerevisiae are present in 50 and two copies, respectively, in the sequenced genome. Ty1 LTRs or δ elements are present in several hundred copies and individual Ty3 LTRs are present in approximately 40 copies (Kim et al., 1998). Ty1 and Ty3 elements insert preferentially into the region upstream of RNA polymerase III-transcribed genes (Chalker and Sandmeyer, 1990; Devine and Boeke, 1996). tRNA genes have been associated with reduced expression of associated RNA polymerase II promoters (Hull et al., 1994; Ji et al., 1993; Kinsey and Sandmeyer, 1991). Nonetheless, expression of marker genes associated with Ty1 and Ty3 elements can be readily detected at levels expected for single gene copies (Chalker and Sandmeyer, 1990; Lee and Da Silva, 1996; Wang and Da Silva, 1996). Cumulatively, Ty1 RNAs have been shown to account for approximately 10% of the total poly(A) RNA in the yeast cells and the individual expression of 31 Ty1 elements has been characterized using lacZ reporter fusions (Lesage and Todeschini, 2005). A δ sequence fused to a linearized URA3 blaster cassette (Alani et al., 1987) has been successfully used to reiteratively target expression cassettes to multiple Ty1 LTRs in the genome (Lee and Da Silva, 1997). Indeed, expression from such insertions is very consistent and nearly linearly correlated with gene copy number (Lee and Dasilva, 2006). Nonetheless, because of the large number of potential targets, this strategy is somewhat unpredictable. Furthermore, stable gene insertion via double-crossover integration is limited; typically only one copy of the same gene can be inserted as gene replacements are obtained rather than new targeted integrations. The feasibility of completely replacing Ty1 and Ty3 elements with expression cassettes was therefore explored.
In order to test the utility of a Ty1 replacement strategy, Ty1 elements previously shown to have different levels of expression (Lesage and Todeschini, 2005) were completely replaced by the PPGK 1-Rluc-TCYC 1 reporter in the PTK -Fluc-TCYC 1 reporter strain yFF1683. The PPGK 1-Rluc-TCYC 1 and loxP-URA3-loxP were amplified as two overlapping fragments with collective outside ends homologous to target genome sequences flanking selected Ty1 elements (for details, see Supporting information S4). Seven Ty1 elements, including four loci that showed high Ty1–lacZ fusion expression and three that represented relatively low Ty1–lacZ fusion expression (Lesage and Todeschini, 2005), and one Ty3 (YILWTy3-1) element were replaced with the PPGK 1-Rluc-TCYC 1 expression cassette (Figure 4A). Successful integration demonstrated that the PCR-based strategy could be used to exchange sequences of about 3 kb for genomic sequences of 5–6 kb. In order to test the effect of flanking tRNA genes on reporter expression, the YMLWTy1-2 and the YMLWTy1-1 loci were replaced in both orientations, with PPGK 1 either proximal or distal to the tRNA genes.
Three isolates with PPGK 1-Rluc-T CYC 1-loxP-URA3-loxP integrated into each locus were grown to logarithmic phase and Rluc : Fluc ratios determined. Protein expression levels from all loci were similar and similar to that of the four standard loci tested (Figure 4B). Thus, differences in expression among these Ty1 elements previously seen by Lasage and Todeschini (2005) most likely reflect differences in promoter strength among individual elements, rather than influences of flanking chromatin structure on Ty1 expression. Furthermore, in the YMLWTy1-1 locus, PPGK 1 is located only 201 bp from a tRNA, and in the YMLWTy1-2 locus PPGK 1 is only 161 bp away from the adjacent tRNA. Nonetheless, in contrast to the prediction that tRNA gene proximity might interfere with expression, PPGK 1-Rluc-TCYC 1 activity was similar whether PPGK 1 was proximal or distal to the tRNA gene at both the YMLWTy1-2 and YMLWTy1-1 loci (Figure 4B). The relative insensitivity of the expression cassette to tRNA gene-based repression could be due to the strength of the PGK1 promoter.
Summary
The system described here is designed to facilitate metabolic engineering in S. cerevisiae. The first element of this system is a set of plasmids with alternative strong promoters present on low-and high-copy plasmids to allow initial testing of pathway genes. Alternative promoters can be easily inserted via unique flanking restriction sites; currently the plasmid set is being expanded to include the GAL1, CUP1 and ADH2 promoters. The second element of the system is ease of PCR amplification of the expression cassette flanked by the marker gene embedded in the plasmid for integration into genomic sites of choice. Multiple marked expression cassettes can be thus be readily integrated to optimize pathway functions. The third element of the system is demonstration that it is practical to replace loci including redundant Ty1 elements with expression-marker amplicons and to subsequently delete the associated markers using CreA recombinase. Together these steps constitute a practical and rapid method for assembly of metabolic pathways in S. cerevisiae.
Supplementary Material
Acknowledgments
This material is based upon work supported by the National Science Foundation under Award Nos EEC-0813570 (to N.A.D. and S.B.S.) and BES-0422684 (to N.A.D.), by UC Discovery Grant No. Bio06-10630 (to G.W.H., S.B.S., N.A.D. and K.S.) and Public Health Services Grant No. GM33281 (to S.B.S.). We thank K. Nguyen for sharing plasmids prior to publication.
Footnotes
Supporting information may be found in the online version of this article.
Methods - strain and plasmid construction
S1. Sequence for codon-optimized creA
S2. Oligonucleotide primers used in this study
S3. Sequences for promoters, genetic markers and copy-number control
S4. Four strongly expressed and three weakly expressed Ty1 elements in S. cerevisiae S288C
References
- Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Burke D, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. 2005. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2005. [Google Scholar]
- Ausubel FM. Current Protocols in Molecular Biology. Wiley; Hoboken, NJ: 2008. [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chalker DL, Sandmeyer SB. Transfer RNA genes are genomic targets for de novo transposition of the yeast retrotransposon Ty3. Genetics. 1990;126:837–850. doi: 10.1093/genetics/126.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, et al. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Delneri D, Tomlin GC, Wixon JL, et al. Exploring redundancy in the yeast genome: an improved strategy for use of the cre–loxP system. Gene. 2000;252:127–135. doi: 10.1016/s0378-1119(00)00217-1. [DOI] [PubMed] [Google Scholar]
- Devine SE, Boeke JD. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- Futcher B, Carbon J. Toxic effects of excess cloned centromeres. Mol Cell Biol. 1986;6:2213–2222. doi: 10.1128/mcb.6.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13 :837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS–DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, et al. Life with 6000 genes. Science. 1996;274:546, 563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Larroy C, Biosca JA, Arino J. Use of the TRP1 auxotrophic marker for gene disruption and phenotypic analysis in yeast: a note of warning. Fems Yeast Res. 2008;8:2–5. doi: 10.1111/j.1567-1364.2007.00315.x. [DOI] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, et al. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fielder T, et al. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield GW, Roth DA. Optimizing scaleup yield for protein production: computationally optimized DNA assembly (CODA) and translation engineering. Biotechnol Annu Rev. 2007;13:27–42. doi: 10.1016/S1387-2656(07)13002-7. [DOI] [PubMed] [Google Scholar]
- Hoess RH, Abremski K. Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc Natl Acad Sci USA. 1984;81:1026–1029. doi: 10.1073/pnas.81.4.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hull MW, Erickson J, Johnston M, Engelke DR. tRNA genes as transcriptional repressor elements. Mol Cell Biol. 1994;14:1266–1277. doi: 10.1128/mcb.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Moore DP, Blomberg MA, et al. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- Johansson B, Hahn-Hagerdal B. Overproduction of pentose phosphate pathway enzymes using a new CRE–loxP expression vector for repeated genomic integration in Saccharomyces cerevisiae. Yeast. 2002;19:225–231. doi: 10.1002/yea.833. [DOI] [PubMed] [Google Scholar]
- Kim JM, Vanguri S, Boeke JD, et al. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- Kinsey PT, Sandmeyer SB. Adjacent pol II and pol III promoters: transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res. 1991;19:1317–1324. doi: 10.1093/nar/19.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MT, Liu DY, Hseu TH. Cell growth restoration and high level protein expression by the promoter of hexose transporter, HXT7, from Saccharomyces cerevisiae. Biotechnol Lett. 2007;29:1287–1292. doi: 10.1007/s10529-007-9397-3. [DOI] [PubMed] [Google Scholar]
- Larsen LS, Wassman CD, Hatfield GW, Lathrop RH. Computationally optimised DNA assembly of synthetic genes. Int J Bioinform Res Appl. 2008;4:324–336. doi: 10.1504/IJBRA.2008.019578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FW, Da Silva NA. Ty1-mediated integration of expression cassettes: host strain effects, stability, and product synthesis. Biotechnol Prog. 1996;12:548–554. doi: 10.1021/bp9600288. [DOI] [PubMed] [Google Scholar]
- Lee FW, Da Silva NA. Improved efficiency and stability of multiple cloned gene insertions at the delta sequences of Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1997;48:339–345. doi: 10.1007/s002530051059. [DOI] [PubMed] [Google Scholar]
- Lee W, Da Silva NA. Application of sequential integration for metabolic engineering of 1,2-propanediol production in yeast. Metab Eng. 2006;8:58–65. doi: 10.1016/j.ymben.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Lesage P, Todeschini AL. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet Genome Res. 2005;110:70–90. doi: 10.1159/000084940. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Muir RS, Lim E, et al. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- McNabb DS, Reed R, Marciniak RA. Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1539–1549. doi: 10.1128/EC.4.9.1539-1549.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead DJ, Gardner DC, Oliver SG. The yeast 2 μ plasmid: strategies for the survival of a selfish DNA. Mol Gen Genet. 1986;205:417–421. doi: 10.1007/BF00338076. [DOI] [PubMed] [Google Scholar]
- Moriyoshi K. pBT, a novel vector for tetracycline-regulated yeast three-hybrid assay. Nucleic Acids Res. 2009;37:e11. doi: 10.1093/nar/gkn969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Nacken V, Achstetter T, Degryse E. Probing the limits of expression levels by varying promoter strength and plasmid copy number in Saccharomyces cerevisiae. Gene. 1996;175:253–260. doi: 10.1016/0378-1119(96)00171-0. [DOI] [PubMed] [Google Scholar]
- Nielsen ML, de Jongh WA, Meijer SL, et al. Transient marker system for iterative gene targeting of a prototrophic fungus. Appl Environ Microbiol. 2007;73:7240–7245. doi: 10.1128/AEM.01839-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prein B, Natter K, Kohlwein SD. A novel strategy for constructing N-terminal chromosomal fusions to green fluorescent protein in the yeast Saccharomyces cerevisiae. FEBS Lett. 2000;485:29–34. doi: 10.1016/s0014-5793(00)02179-7. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan P, Srivastava V. Utility of the FLP–FRT recombination system for genetic manipulation of rice. Plant Cell Rep. 2005;23:721–726. doi: 10.1007/s00299-004-0876-x. [DOI] [PubMed] [Google Scholar]
- Sauer B. Functional expression of the cre–lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B. Recycling selectable markers in yeast. Biotechniques. 1994;16:1086–1088. [PubMed] [Google Scholar]
- Scherer S, Davis RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherf BA, Navarro SL, Hannah RR, Wood KV. Dual-luciferase reporter assay: an advanced coreporter technology integrating firefly and Renilla luciferase assays. Promega Notes. 1996;57 :2–9. [Google Scholar]
- Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MV, Weil PA. A method for plasmid purification directly from yeast. Anal Biochem. 2002;307:13–17. doi: 10.1016/s0003-2697(02)00018-0. [DOI] [PubMed] [Google Scholar]
- Wang X, Da Silva NA. Site-specific integration of heterologous genes in yeast via Ty3 retrotransposition. Biotechnol Bioeng. 1996;51:703–712. doi: 10.1002/(SICI)1097-0290(19960920)51:6<703::AID-BIT9>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Enloe BM, Mitchell AP. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast. 2000;16:65–70. doi: 10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.