Abstract
Ebola virus is a Filoviridae that causes hemorrhagic fever in humans and induces high morbidity and mortality rates. Filoviruses are classified as "Category A bioterrorism agents", and currently there are no licensed therapeutics or vaccines to treat and prevent infection. The Filovirus glycoprotein (GP) is sufficient to protect individuals against infection, and several vaccines based on GP are under development including recombinant adenovirus, parainfluenza virus, Venezuelan equine encephalitis virus, vesicular stomatitis virus (VSV) and virus-like particles. Here we describe the development of a GP Fc fusion protein as a vaccine candidate. We expressed the extracellular domain of the Zaire Ebola virus (ZEBOV) GP fused to the Fc fragment of human IgG1 (ZEBOVGP-Fc) in mammalian cells and showed that GP undergoes the complex furin cleavage and processing observed in the native membrane-bound GP. Mice immunized with ZEBOVGP-Fc developed T-cell immunity against ZEBOV GP and neutralizing antibodies against replication-competent VSV-G deleted recombinant VSV containing ZEBOV GP. The ZEBOVGP-Fc vaccinated mice were protected against challenge with a lethal dose of ZEBOV. These results show that vaccination with the ZEBOVGP-Fc fusion protein alone without the need of a viral vector or assembly into virus-like particles is sufficient to induce protective immunity against ZEBOV in mice. Our data suggested that Filovirus GP Fc fusion proteins could be developed as a simple, safe, efficacious, and cost effective vaccine against Filovirus infection for human use.
1. Introduction
Ebola virus (EBOV) and Marburgvirus (MARV) are members of the Filoviridae, a family of viruses classified as “Category A” bioterrorism agents that cause severe hemorrhagic fever in humans and nonhuman primates with high morbidity and mortality rates up to 90% [1]. After a short incubation period of 4 to 10 days, Filovirus-infected individuals develop an abrupt onset of symptoms that include fever, chills, malaise, and myalgia that are common to many other viral infections. MARV is antigenically stable and exists in only one species, whereas EBOV is more variable and has five species. The Bundibugyo EBOV emerged recently in a late 2007 outbreak in Uganda [2], and is more related to the Ivory Coast than to the Zaire (ZEBOV), Sudan, or Reston EBOV species. ZEBOV is typically associated with the highest lethality. The increased number of outbreaks in Africa and the recent EBOV outbreak in pigs [3], which raised concerns that livestock could transmit the deadly disease to humans, highlighted the urgency for the development of vaccines and rapid diagnostic tests to contain outbreaks. Vaccines based on the Filovirus glycoprotein (GP) are in preclinical and clinical evaluation, and currently there are no licensed therapeutic agents to treat Filovirus infection. Since licensing of safe and effective Filovirus vaccine could take several more years, diagnosis and quarantine of infected individuals is currently the main tool to limit outbreaks.
Filovirus particles contain a negative-strand RNA genome of about 19 kb that encodes seven genes [4]. The envelope glycoprotein (GP) encoded in gene 4 is present as spikes on the virion surface and is responsible for receptor binding, viral entry, and immunity [5, 6]. The Filovirus GP is a class 1 integral membrane glycoprotein derived from gene 4 that undergoes a complex processing involving furin cleavage and disulfide-bond formation between the N-terminus and the membrane proximal portion of the GP. The mature transmembrane GP present on the viral envelope and membrane of infected cells is formed by two subunits: the membrane anchored GP2 that is covalently linked via disulphide linkage to the N-terminus of GP1, which contains a highly O-glycosylated mucin-like domain [7, 8]. A significant amount of GP1 is shed from the cells after release from the GP2 subunit. In addition, a nonstructural soluble glycoprotein (sGP) that shares the amino-terminal 295 amino acids with GP1, lacks a transmembrane anchor, and forms disulphide-linked homodimers, is produced by EBOV but not MARV infected cells [9].
Several Filovirus vaccine candidates containing GP are currently being developed based on recombinant adenovirus [10–12], parainfluenza virus [13], Venezuelan equine encephalitis virus [14], replication-competent [15, 16] and –deficient [17] vesicular stomatitis virus, and virus-like particles [18, 19]. Initial studies using baculovirus-expressed Filovirus GP induced partial protection, which could be attributed to the glycosylation and processing of GP in insect cells [20]. In this work, we analyzed the immunogenicity of a recombinant protein consisting of the extracellular domain of ZEBOV GP fused to the Fc fragment of human IgG1 (ZEBOVGP-Fc) expressed in mammalian cells. We hypothesized that the Fc tag in the ZEBOVGP-Fc would simplify purification of the fusion protein through protein A columns using mild conditions, increase protein stability of the fusion protein, and confer an adjuvant effect in non-human primates (NHP) and humans due to the interactions with Fcγ receptors on antigen presenting cells [21–23]. Here, we show that ZEBOVGP-Fc expressed in mammalian cells undergoes the complex posttranslational modification of the native GP, including the furin cleavage and homotrimer formation. Vaccination with the EBOVGP-Fc protected mice against challenge with a lethal dose of ZEBOV. Our results clearly indicate that the ZEBOVGP-Fc alone and without the need of a viral vector or assembly into virus-like particles is sufficient for inducing protection against ZEBOV infection and suggest that Filovirus GP Fc fusion proteins could be developed into a cost-effective safe and effective subunit vaccine against Filovirus infection.
2. Materials and methods
2.1. Cells lines
Chinese hamster ovary (CHO) cells deficient in the enzyme dihydrofolate reductase (dhfr−) were obtained from the American Type Culture Collection and expanded in growth medium consisting of Iscove’s medium containing 10% fetal bovine serum (FBS) [24]. Human embryonic kidney HEK293-H cells (Invitrogen) were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% FBS. Vero E6 cells were grown in Eagle’s minimal essential medium (MEM) supplemented with 10% FBS. Baby hamster kidney cells (BHK-21) were maintained in DMEM medium supplemented with 5% FBS. The BSR-T7 cells, which are BHK-21 cells that express bacteriophage T7 RNA-polymerase [25], was kindly provided by Dr. K. Conzelmann (Pettenkoffer Institute, Munich, Germany) and maintained in DMEM medium supplemented with 5% FBS and 1 mg/ml geneticin (Invitrogen).
2.2. Mice
C57BL/6 and BALB/c mice were obtained from the National Cancer Institute, Frederick Cancer Research and Development Center (Frederick, MD). All mice were housed in micro-isolator cages and provided standard rodent feed and water ad libitum. Blood samples were obtained from the lateral tail vein. Research was conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the guide for the Care and Use of Laboratory Animals, National Research Council, 1996. The facility where these researches were conducted is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. The animal protocols were approved by CBER-FDA or USAMRIID Institutional Animal Care and Use Committees (IACUC).
2.3. Cloning procedures
The cDNA of the Zaire Ebola virus (ZEBOV) glycoprotein (GP), Mayinga strain (GenBank accession no. AF272001), in pVR-1012-ZEBOV-GP was kindly provided by Dr. Gary Nabel, Vaccine Research Center, NIH, Bethesda, MD [26]. The glycoprotein gene has eight adenosine (A) residues at the RNA editing site needed to produce the full-length ZEBOV GP. The following plasmids were constructed using standard technique of genetic engineering:
2.3.1. pEF1-EBOV-GP
The GP region was excised from pVR-1012-ZEBOV-GP using NcoI and Asp718 (Roche Applied Science) restriction enzymes, filled in with DNA polymerase Klenow (New England Biolabs) enzyme to create blunt ends and subsequently cloned into the EcoRV site of the mammalian expression plasmid pEF1/Myc-His-B (Invitrogen). The resulting plasmid was termed pEF1-EBOV-GP.
2.3.2. pEF-ZEBOVGP-Fc
To construct a plasmid for the expression of an ZEBOV GP Fc fusion protein, a PCR fragment coding for amino acids 1 to 637 (GenBank accession no. U23187) of the ZEBOV GP ectodomain was amplified from pVR-1012-ZEBOV-GP using synthetic oligonucleotides GP/SalI (5’-GTCGACAGTATGGGCGTTACAGGAATATTGCAGTTA-3’), which contains SalI site before the sequence coding for the signal peptide GP, and GP/Flag/SpeI (5’-ACTAGTACTCACCTCCCTTGTCATCGTCGTCCTTGTAGTCTCCACCGCCGTCCGGAAGGGTTTTATCAACAAA-3’), which contains an SpeI site followed by an artificial splicing donor site, the coding sequence for the FLAG tag peptide DTKDDDDK, and nucleotides 1887 to 1911 of GP. This PCR fragment was cloned into the SalI and SpeI sites of pEF-ICAM5(1–2) Fc replacing the ICAM 1 cDNA fragment and in-frame with the Fc fragment of human IgG1 [24]. Silent mutations (GTCGAC to GTAGAC, and CTAGTT to CTCGTT) were introduced into the GP coding sequence to eliminate internal SalI and SpeI restriction sites. The resulting plasmid was termed pEF-ZEBOVGP-Fc.
2.3.3. pEF-FLAG-Fc
To construct a plasmid for the expression of the Fc fragment of human IgG1, we replaced the ICAM 1 sequence in pEF-ICAM5(1–2)Fc with a cDNA fragment coding for the signal peptide of the ZEBOV GP and a FLAG tag. To do so, we amplified a PCR fragment coding for amino acids 1–32 of the ZEBOV GP signal peptide using pVR-1012-ZEBOV-GP as a template and synthetic oligonucleotides GP/SalI (see above) and SP/Flag/SpeI (5’-ACTAGTACTCACCTCCCTTGTCATCGTCGTCCTTGTAGTCTCCACCGCCGGAAAATGTTCTTTGGAAAAGGAT-3’), which codes for a FLAG tag, nucleotides 73 to 96 of the GP cDNA, and an SpeI restriction site. The amplified PCR fragment was digested with SalI and SpeI restriction enzymes and cloned into pEF-ICAM5(1–2)Fc digested with the same enzymes. The resulting plasmid was termed pEF-FLAG-Fc.
2.3.4. pVSV-EBOVGP
To construct replication-competent VSV-G deleted recombinant Vesicular Stomatitis Virus (VSV) expressing the ZEBOV GP (rVSV-ZEBOVGP), a PCR fragment coding for GP of ZEBOV (amino acids 1–676) amplified from pVR-1012-ZEBOV-GP using oligonucleotides GP/NheI(+) (5’ACTAGTAGTATGGGCGTTACAGGAATATTGCAGTTA-3’), which is identical to GP/SalI except for the SalI site that was substituted for an NheI restriction, and antisense primer GP/NheI (−) (5’-GCTAGCCTAAAAGACAAATTTGCATATACAGAA-3’), was cut with NheI and cloned into NheI cut pVSVΔG. The resulting plasmid was termed pVSV-ZEBOVGP.
2.4. Selection of HEK-293-H stable transfectants expressing ZEBOV GP at the cell surface
HEK293-H cells were transfected with pEF1-ZEBOV-GP or vector pEF1 using Fugene 6 reagent (Roche Applied Science) as suggested by the manufacturer, and stable transfectants were selected with 350 µg/mL geneticin and termed HEK293-ZEBOVGP or HEK293 cells. Single cell clones were produced and analyzed by flow cytometry using anti-ZEBOV GP monoclonal antibody (mAb) 13F6-1-2 [27, 28] to select stable transfectants expressing high levels of ZEBOV GP at the cell surface.
2.5. Production and purification of Fc fusion proteins
Fc fusion proteins were produced in CHO cell transfectants. To do so, CHO dhfr− cells were cotransfected with 0.45 µg of pDHIP and 3.5 µg of pEF-ZEBOVGP-Fc or pEF-Fc using the Fugene 6 reagent as described previously [24]. Briefly, cell transfectants were grown in Iscove's medium containing 10% FBS supplemented with hypoxanthine and thymidine for 48 h at 37°C, split 1:10, and selected in Iscove's medium containing dialyzed FBS without supplements. After 14 days of selection, single cell clones were obtained by end-point dilution in 96-well plates. The supernatants of 56 single-cell clones were assayed for the expression of fusion proteins by a capture ELISA in 96-well plates coated with goat anti-human Fc antibody and staining with anti-GP mAb and HRP-conjugated goat anti-mouse antibody (Ab). Overexpression of the Fc fusion proteins was achieved by a stepwise increase in the concentration of methotrexate (MTX). Cells reached a maximum level expression of ZEBOVGP-Fc and FLAG-Fc at 0.08 and 0.32 µM MTX, respectively.
Fc fusion proteins were purified as described previously [24]. Briefly, CHO cells expressing recombinant proteins were grown in Iscove’s medium containing 10% FBS for 3 days, monolayers were washed twice with Iscove’s medium, and grown in serum-free OptiMEM medium (Invitrogen). Supernatants were harvested 2–3 times at 24 h intervals, clarified at 3,000×g, and stored at −20°C. Fc fusion protein in the OptiMEM supernatant was purified by affinity chromatography in protein A-agarose columns. Eluted fractions were analyzed by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the fractions containing the Fc fusion proteins were pooled, concentrated using Amicon ultra column (Millipore), and washed with PBS.
2.6. Western blot analysis
Purified proteins were fractionated in denaturing SDS-PAGE, transferred to Immobilon-P polyvinylidene difluoride (PVDF) membranes (Millipore), and stained with a 1:1,000 dilution of mouse anti-ZEBOV GP1 13F6-1-2 mAb (USAMRIID), mouse anti-FLAG M2 mAb (Sigma Chemical Co.), or goat anti-human IgG Fc Ab, and a 1:3,000 dilution of phosphatase-labeled goat anti-mouse or rabbit anti-goat Ab. The Western blot was developed with 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium substrate (Kpl Inc.) as recommended by the manufacturer.
2.7. Rescue of rVSV-ZEBOVGP
To generate recombinant VSV carrying the ZEBOV GP (rVSV-ZEBOVGP), we used the reverse genetics system developed by Dr. John Rose (Yale Univ., New Haven, CT), who kindly provided us with plasmid coding for the VSV full-length genome [pVSVFL(+)], VSV-G deleted VSV genome (pVSVΔG), VSV nucleoprotein (pBS-N), VSV phosphoprotein (pBS-P), and VSV-L polymerase (pBS-L) [27]. To do so, BSR-T7 cells grown to 80–90% confluency in 6-well plates were cotransfected with 0.25 µg pBS-N, 0.6 µg pBS-P, 0.13 µg pBS-L, and 1 µg pVSV-ZEBOVGP or positive control pVSVFL(+) using 5 ul Lipofectamine 2000 (Invitrogen) as a facilitator per well [29, 30]. After 48 h of incubation at 37°C, supernatants were harvested and used to infect 50% confluent BHK-21 cells. After monolayers developed cytopathic effect (CPE), supernatants containing rVSV-ZEBOVGP or VSV were collected, titrated in Vero E6 cells, and stored at −80°C.
2.8. Enterokinase cleavage of ZEBOVGP-Fc protein
The Fc fragment of ZEBOVGP-Fc was removed by treatment with 0.4 µg of restriction protease enterokinase (New England Biolabs) per mg of fusion protein, which cleaved the FLAG peptide that was engineered between the GP and Fc fragments. The digestion product was analyzed by FPLC on a Superdex200 column (GE Healthcare) under non-denaturing conditions. The collected fractions were analyzed by denaturing SDS-PAGE and Western blot analysis staining with anti-ZEBOV GP mAb 13F6-1-2 or anti-Fc fragment Ab and appropriate phosphatase-conjugated secondary Ab.
2.9. Vaccination and ZEBOV challenge
For the immunogenicity study, 6–8 week old BALB/c mice were vaccinated intraperitoneally (i.p.) with 100 µg of ZEBOVGP-Fc (n = 4) or with 100 µg of FLAG-Fc (n = 4) in complete Freunds adjuvant. At 21, 45 and 60 days post inoculation, animals were boosted i.p with 25 µg of the corresponding protein in incomplete Freund adjuvant. Serum samples were obtained from each mouse before each boost (bleeds number 1–3) and 8 days after the last boost (bleed number 4).
For the challenge study, 6–8 week old C57BL/6 mice were vaccinated i.p. with 100 µg of ZEBOVGP-Fc (n = 8) or with 100 µg of Fc (n = 8) in complete Freunds adjuvant and boosted i.p with 25 µg of the corresponding protein in incomplete Freund adjuvant 21, 45 and 60 days post inoculation. Serum samples were obtained from each mouse prior to challenge. Mice were challenged 74 days after the primary vaccination (2 weeks after the final vaccination) by i.p. injection with 1,000 pfu of mouse-adapted ZEBOV diluted in PBS [31, 32]. All ZEBOV-infected mice were handled under maximum containment in a biosafety level-4 (BSL4) laboratory at the U.S. Army Medical Research Institute of Infectious Diseases, Frederick, MD.
2.10. Antibody titer determination
Anti-ZEBOV specific Ab titers were determined by an endpoint dilution ELISA on 96-well plates coated with sucrose purified inactivated ZEBOV virions as described previously [14] with minor modifications. Two-fold dilutions of sera from vaccinated mice were titrated by duplicate on the inactivated ZEBOV-coated plates and stained with peroxidase-labeled goat anti-mouse IgG Ab. Titers were determined as the highest dilution at which the mean absorbance of the sample was at least two-fold greater than the mean absorbance of the same dilution of control FLAG-Fc sera. Similarly, anti-ZEBOV Ab titers were determined in parallel using 96-well plates coated with rVSV-ZEBOVGP. Plates coated with wild type (wt) VSV were used as specificity control. Sera from vaccinated mice were titrated on the plates, stained with peroxidase-labeled goat anti-mouse IgG Ab, and Ab titers were determined as described above.
For FACS analysis, live HEK293-ZEBOVGP cells expressing ZEBOV GP at the surface or control HEK293 cells transfected with empty vector were stained with 1 µl of sera from vaccinated followed by a PE-conjugated anti-mouse Ab (BD Biosciences). Cells were analyzed in a FACSCanto II flow cytometer (BD Biosciences).
2.11. rVSV-ZEBOVGP plaque reduction assay
Anti-ZEBOV neutralizing antibodies were analyzed by a plaque reduction assay. Five-fold dilutions of sera from vaccinated mice were mixed with 100 pfu of rVSV-EBOVGP or control VSV. Samples were incubated at 37°C for 1 h in the presence of 5% guinea pig serum complement and a standard plaque assay was performed in Vero E6 cells overlayed with medium containing 1% bactoagar (DIFCO). The percent of plaque reduction was calculated by comparing the number of pfu in the neutralized sample versus the input virus [27, 33].
2.12. Intracellular cytokine staining
Intracellular cytokine staining was performed by incubating splenocytes from the BALB/c immunized mice with EBOV GP peptides or ZEBOVGP-Fc as described [34]. Briefly, splenocytes were isolated by passing spleens through a mesh filtration, washed, and cultured in RPMI 1640 medium containing 10% FBS, 2mM L-glutamine, 1mM HEPES, and 0.1 mM nonessential amino acids. Splenocytes were stimulated with 1 µg/ml of three EBOV GP specific peptides (LYDRLASTV, VSTGTGPGAGDFAFHK, and EYLFEVDNL) [34, 35] or an unrelated peptide (KINSTALL) as a negative control. Alternatively, splenocytes were stimulated with 10 µg/ml of EBOVGP-Fc or FC protein. After stimulation and 1 h of incubation at 37°C in the presence of interleukin-2, brefeldin A was added and cells were incubated for 5 h before staining with the APC-conjugated anti-mouse CD8 mAb clone RM4-5 (Invitrogen). Cells were then washed, fixed, and permeabilized using the cytofix/cytoperm kit according to the manufacturer’s instructions (BD Biosciences). Intracellular IFN-γ was stained with PE-conjugated rat anti-mouse IFN-γ mAb clone XMG1.2 (Invitrogen), and analyzed by flow cytometry in a FACSCanto II instrument.
2.13. Statistical analysis
Statistical significance between two means was determined by the unpaired Student’s t-test and calculated using Graph Pad software, and p-values were included in the text and figures.
3. Results
3.1. ZEBOVGP-Fc fusion protein undergoes furin cleavage and the complex processing of native ZEBOV GP
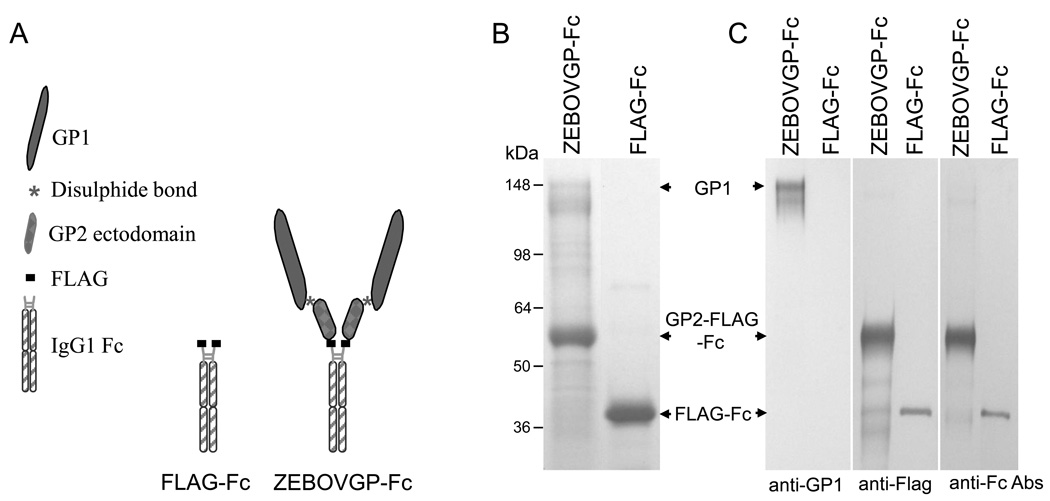
To express large amounts of soluble ZEBOV GP for purification, characterization and immunization purposes, we developed a stable CHO cell line expressing the extracellular domain of ZEBOV GP fused to the human IgG1 Fc fragment. We reasoned that the Fc fragment would increase the yield, simplify purification, and maximize the in vitro and in vivo stability of the recombinant soluble protein [36]. We also included a FLAG tag epitope between the ZEBOV GP and the Fc fragment to monitor the expression of the chimeric protein with anti-FLAG M2 mAb and also to be able to cleave off the Fc fragment using enterokinase, a restriction protease that cuts at the FLAG tag site. To do so, we cotransfected CHO dhfr− cells with pDHIP, a plasmid coding for the dihydrofolate reductase (DHFR) gene, and pEF-EBOVGP-Fc, the construct coding for the ZEBOV GP ectodomain fused to the Fc fragment of IgG1 (Fig. 1A). As a control, we also cotransfected CHO dhfr− cells with pDHIP and pEF-FLAG-Fc, a plasmid coding for the same Fc fragment containing a FLAG tag at the N-terminus. Single cell clones were selected and over expression of the recombinant protein was achieved by increasing the concentration of MTX [24]. A CHO cell clone that produced the highest ZEBOVGP-Fc or FLAG-Fc protein yield as measured by ELISA were used for protein production. Protein A purified proteins were analyzed by SDS-PAGE under reducing conditions followed by Coomassie blue staining (Fig. 1B). Two major bands were observed in ZEBOVGP-Fc: a broad band of approximately 130–150 kDa characteristic of highly glycosylated proteins with the expected molecular weight of GP1, and a smaller band of approximately 60 kDa with the expected molecular weight of GP2-Fc. Additional minor bands corresponding to partially glycosylated or degraded proteins were also observed in the ZEBOVGP-Fc lane. The control FLAG-Fc protein migrated as a 36 kDa band. Western blot analysis probing with anti-GP1 mAb 13F6-1-2, anti-FLAG mAb M2, and anti-Fc Ab confirmed the identity of the GP1, GP2-Fc, and FLAG-Fc bands (Fig. 1C). Our data revealed that the EBOVGP-Fc fusion protein underwent the complex postranslational modifications of the mature GP including the furin cleavage between GP1 and GP2.
Fig. 1.
Schematic representation and purification of the ZEBOVGP-Fc and FLAG-Fc proteins. A) Schematic representation of the fusion proteins. The ZEBOVGP-Fc fusion protein contains the ectodomain of ZEBOV GP tagged at its C terminus with a FLAG peptide and fused to the hinge and Fc regions of human IgG1. The FLAG-Fc fusion protein contains a FLAG tag fused to the hinge and Fc regions of IgG1.
B) SDS-PAGE analysis of fusion proteins. Protein A-purified ZEBOVGP-Fc and FLAG-Fc preparations were analyzed by denaturing SDS-PAGE in a 4–12% gradient gel and stained with Coomassie blue. C) Western blot analysis of ZEBOVGP-Fc and FLAG-Fc. Proteins were resolved by SDS-PAGE under denaturing conditions, transferred to PVDF membranes, and probed with ZEBOV-specific anti-GP1 mAb 13F6-1-2, anti-Flag M2 mAb, or goat anti-human Fc Ab. ZEBOV GP1, GP2-FLAG-Fc, and FLAG-Fc bands are indicated with arrows. Positions and size of molecular weight markers are indicated in kDa.
3.2. ZEBOV GP in ZEBOVGP-Fc assembles into homotrimers
Analysis of purified ZEBOVGP-Fc (50 µg) by size exclusion chromatography through a Superdex 200 10/300 GL column using an AKTA FPLC system (GE) revealed that this fusion protein migrated as a main peak of approximately 1,000 kDa with a broad shoulder consistent with the highly glycosylated nature of GP as observed by SDS-PAGE analysis (Fig. 2A). Since GP forms homotrimers at the virus and cell surface, we hypothesized that the large ZEBOVGP-Fc complexes could be due to GP homotrimer formation, and that three copies of the ZEBOVGP-Fc (as shown in Fig. 1A, each copy of ZEBOVGP-Fc contains two ZEBOVGP monomers linked by the Fc fragment) could form two ZEBOVGP homotrimers. The expected size of the two homotrimer complex formed by three copies of ZEBOVGP-Fc would be approximately 1,000–1,200 kDa. To test our hypothesis, we digested the ZEBOVGP-Fc (100 µg) with the restriction protease enterokinase (40 ng), which cleaves at the FLAG peptide inserted between GP2 and the Fc fragment. Size exclusion chromatography of this digested product revealed 3 main peaks of approximately 600, 140, 50 kDa (Fig. 2A). We determined that the 140 kDa peak corresponded to a carrier protein present in the enterokinase enzyme preparation because size exclusion chromatography of the enterokinase preparation alone revealed a single protein that co-migrated with the 140 kDa protein found in the enterokinase digested ZEBOVGP-Fc. It should be pointed out that the amount of enterokinase (40 ng) used in the digestion is bellow the limit of detection of the FPLC instrument. The 600 kDa peak was consistent with the migration of a soluble ZEBOVGP homotrimer formed by GP1 (130–150 kDa) and GP2 (25 kDa) containing a deletion of the transmembrane domain and cytoplasmic tail. The 50 kDa peak had the expected size of an Fc fragment (2 chains of 25 kDa associated by disulfide bonds), and comigrated with the Fc fragment ran under the same conditions (data not shown). Western blot analysis of the 600, 140, and 50 kDa peaks probed with anti-GP1 and anti-Fc antibodies confirmed that the 600 kDa peak corresponded to the ZEBOVGP extracellular domain, the 140 kDa peak was neither ZEBOVGP nor Fc, and the 50 kDa peak corresponded to the Fc fragment (Fig. 2B). The lack of reaction of the 140 kDa peak (fraction 28) with the anti-GP1 and anti-Fc antibodies further supported the notion that the 140 kDa peak corresponded to a carrier protein included in the enterokinase preparation. These results clearly show that the GP in EBOVGP-Fc formed homotrimers resembling the conformation of the natural form of GP expressed on the viral particle and cell surface [37].
Fig. 2.
Characterization of ZEBOVGP-Fc fusion protein. A) FPLC analysis of protein A-purified ZEBOVGP-Fc. Undigested ZEBOVGP-Fc (green), enterokinase-digested ZEBOVGP-Fc (blue), or enterokinase alone (gray) were run on a Superdex 200 size exclusion column under non-denaturing conditions, and absorbance at 280 nm was recorded for each of the 38 collected fractions. Peaks representing ZEBOVGP-Fc, ZEBOV GP, and Fc are marked with arrows. A peak of approximately 150 kDa represents a carrier protein in the enterokinase preparation. The migration of molecular weight standards is shown as arrowheads and their molecular weight is expressed in kDa. B) Western blot analysis of enterokinase-digested ZEBOVGP-Fc protein. Gel filtration peak fractions (20, 28 and 32) were fractionated in denaturing SDS-PAGE, transferred to a PVDF membrane, probed with ZEBOV-specific anti-GP1 mAb 13F6-1-2 or goat anti-human anti-Fc antibody. Undigested (U) or enterokinase digested (D) ZEBOVGP-Fc, and FLAG-Fc (Fc) were included in the gel as markers. Arrows indicate the migration of GP1, GP2-FLAG-Fc, and FLAG-Fc. The migration of the molecular weight markers and their sizes are indicated in kDa.
3.3. ZEBOVGP-Fc elicits anti-ZEBOV humoral response in immunized mice
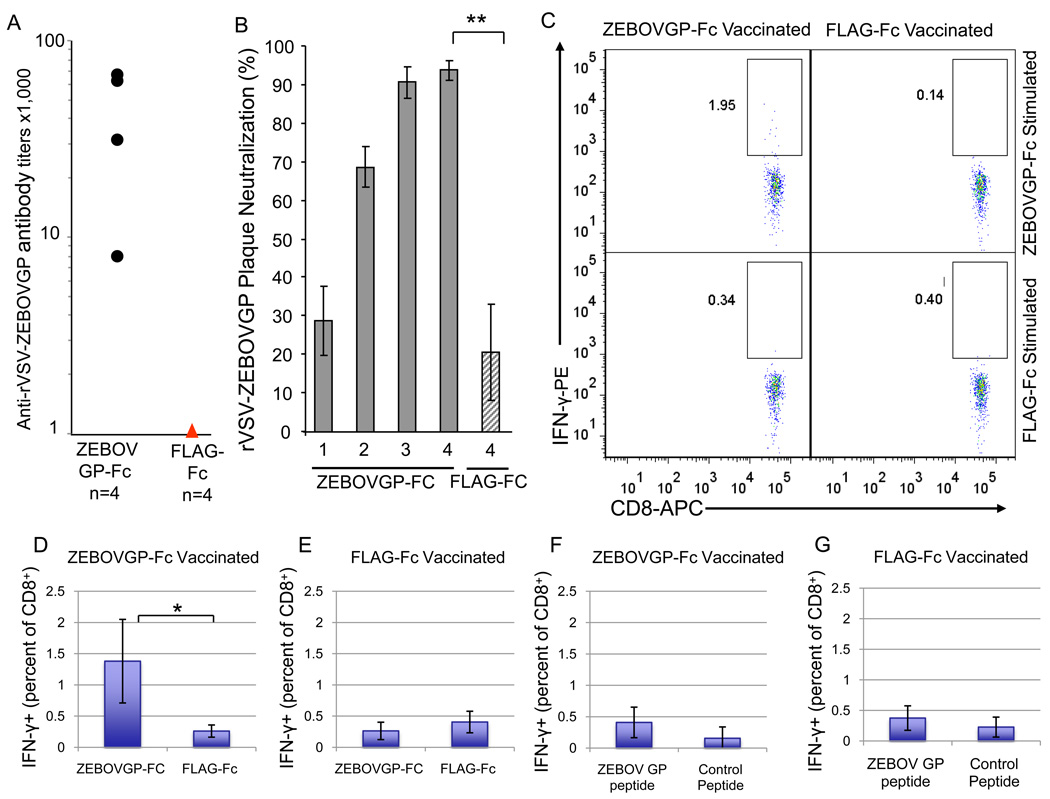
To determine whether ZEBOVGP-Fc was immunogenic, we vaccinated four BALB/c mice i.p. with 100 µg of ZEBOVGP-Fc or control FLAG-Fc in complete Freunds adjuvant followed by three i.p. boosts with 25 µg protein in incomplete Freund adjuvant at 21, 45 and 60 days post inoculation. Mice were bleed before each boost (bleeds number 1–3) and 8 days after the last boost (bleed number 4). Anti-ZEBOV GP antibodies in the sera from the last bleed of the vaccinated mice were analyzed by ELISA. The four mice vaccinated with ZEBOVGP-Fc developed anti-ZEBOV GP antibody titers of 1:64,000 to 1:8,000 as measured by an ELISA in 96-well plates coated with purified rVSV-ZEBOVGP, a replication-competent VSV-G-deleted recombinant VSV containing the ZEBOV GP that infect cells via the ZEBOV GP [30] (Fig. 3A). As expected, the control mice vaccinated with FLAG-Fc did not develop anti-ZEBOV GP antibodies.
Fig. 3.
Humoral and cellular immune responses in BALB/c mice vaccinated with ZEBOVGP-Fc. Four BALB/c mice were immunized with ZEBOVGP-Fc or control FLAG-Fc. After a primary inoculation with 100 µg of each protein, animals were boosted 3 times with 25 µg of the corresponding proteins. A) Analysis of anti-ZEBOV GP specific antibodies in immunized BALB/c mice by viral particle ELISA. Sera samples collected 8 days after the last boost were titrated by ELISA on plates coated with rVSV-ZEBOVGP. The endpoint dilution titer for each mouse sera is represented as a black dot. The titers of the sera from FLAG-Fc immunized mice were bellow the detection limit and are represented as a read triangle. B) Analysis of neutralizing antibodies in sera samples from BALB/c mice immunized with ZEBOVGP-Fc. Sera samples from ZEBOVGP-Fc immunized mice were collected 2 weeks after the primary immunization (1), first boost (2), and second boost (3). Sera samples were also collected 8 days after the third boost (4). Sera samples from FLAG-Fc vaccinated mice were collected 8 days after the third boost (4). Values in the graph represent mean percent neutralization of rVSV-ZEBOVGP with 1/10 dilution of sera (n=4), and bars represent the standard deviation. The difference between neutralization of sera from ZEBOVGP-Fc- and FLAG-Fc-vaccinated mice is statistically significant (**, p≤0.01). C) Splenocytes from immunized BALB/c mice were collected eight days after the third boost, stimulated with fusion proteins or peptides, and stained at the cell surface with APC-labeled anti-CD8 mAb and intracellularly with PE-labeled anti-IFN-γ mAb. The percent of IFN-γ- producing CD8+ T-cells was determined by flow cytometry gating on CD8+ splenocytes. Dot plot analysis of CD8+ splenocytes from a representative mouse vaccinated with ZEBOVGP-Fc or FLAG-Fc and stimulated with ZEBOVGP-Fc or FLAG-Fc. Double-positive splenocytes expressing CD8 and IFN-γ are shown within a rectangular box and numbers represent the percentage of IFN-γ-positive CD8+ cells within the box. D–G) Bar graph representation of the percentage of IFN-γ-positive CD8+ cells in splenocytes obtain in (C) from mice vaccinated with ZEBOVGP-Fc (D,F) or FLAG-Fc (E,G) and stimulated with ZEBOVGP-Fc or FLAG-Fc (D,E), or synthetic ZEBOV GP or negative control peptides (F,G). Values in the graphs are the mean percentage of total CD8+ cells that express IFN-γ (n=4 mice), and the bars represent the standard deviations. The difference between the percentage of IFN-γ-positive CD8+ splenocytes in ZEBOVGP-Fc-vaccinated mice stimulated with ZEBOVGP-Fc and FLAG-Fc is statistically significant (*, p=0.01).
Since the vaccinated mice developed anti-ZEBOV GP antibodies, it was of interest to determine whether these antibodies were capable of neutralizing virus. To do so, we performed a plaque-reduction test based on the neutralization of rVSV-EBOVGP. Because neutralizing antibodies against ZEBOV are directed towards GP, the only envelope glycoprotein at the cell surface of the virus, neutralization of rVSV-EBOVGP mimics neutralization of ZEBOV. We analyzed neutralization levels in sera from each inoculation (Fig. 3B). Minimal levels of neutralizing antibodies were elicited by the first inoculation. Neutralization levels increased to approximately 70% after the second immunization (first boost) and to approximately 90% after the third (second boost) and fourth (third boost) immunizations. Since the forth inoculation did not increase significantly the neutralization level compared to the third inoculation, we concluded that production of neutralizing antibodies reached the plateau. Sera from FLAG-Fc vaccinated mice showed background levels of neutralization of approximately 20%. Taken together, these data clearly showed that ZEBOVGP-Fc induced an antibody response against ZEBOVGP in immunized mice that contained neutralizing antibodies.
3.4. ZEBOVGP-Fc induces a CD8+ T-cell response in vaccinated mice
Since protection against Filovirus infection is mediated by humoral and cellular immunity [1], we analyzed T-cell immunity in mice vaccinated with ZEBOVGP-Fc. We used the BALB/c mice immunized with ZEBOVGP-Fc or FLAG-Fc described above in section 3.3 to perform the studies. Eight days after the final immunization, mice were euthanized, spleens were harvested, and splenocytes were isolated from ZEBOVGP-Fc and FLAG-Fc immunized mice and stimulated with ZEBOVGP-Fc, FLAG-Fc, ZEBOV GP peptides, or control peptides. Splenocytes were stained at the cell surface with APC-labeled anti-mouse CD8 mAb and intracellularly with PE-labeled anti-mouse IFN-γ mAb and analyzed by flow cytometry, which showed the presence of 1.95% IFN-γ-positive CD8+ cells in splenocytes of ZEBOVGP-Fc-vaccinated mice stimulated with ZEBOVGP-Fc as shown in Figure 3C for a representative mouse. Statistical analysis of four animals per group revealed a significant increase in IFN-γ-positive CD8+ cells in splenocytes of ZEBOVGP-Fc-vaccinated mice stimulated with ZEBOVGP-Fc compared to FLAG-Fc (Fig. 3D, E). No increase in IFN-γ-positive CD8+ cells was observed in splenocytes of ZEBOVGP-Fc- or FLAG-Fc-vaccinated mice stimulated with ZEBOV GP or control peptides (Fig. 3F, G). These data indicated that ZEBOVGP-Fc induced a significant increase in the level of anti-ZEBOV GP IFN-γ-positive CD8+ cells. It should be pointed out that the Fc fragment was a weak inducer of IFN-γ-positive CD8+ cells and resulted in a background level activation of T-cells from both ZEBOVGP-Fc and FLAG-Fc vaccinated mice. Unexpectedly, the ZEBOV GP specific peptides did not activate IFN-γ-positive CD8+ cells in ZEBOVGP-Fc-vaccinated mice (Fig. 3F) compared to FLAG-Fc-vaccinated mice (Fig. 3G).
3.5. ZEBOVGP-Fc protect mice against ZEBOV challenge
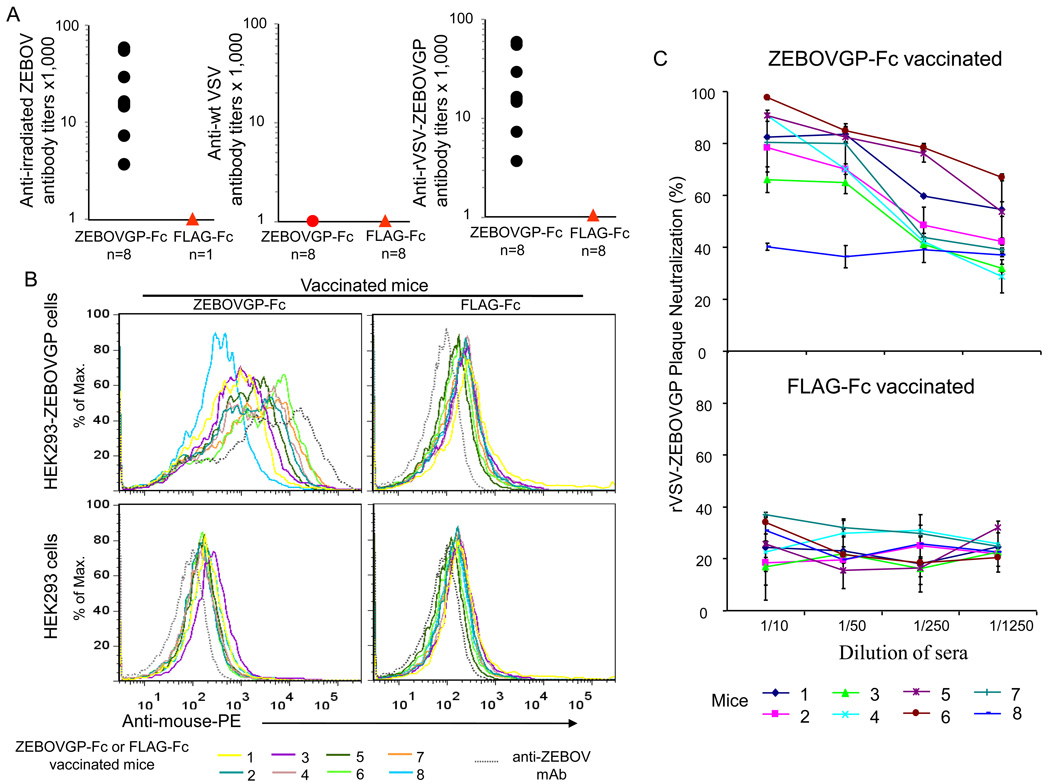
Our results showed that ZEBOVGP-Fc elicited humoral and cellular immune responses in vaccinated BALB/c mice. We hypothesized that these immune response against ZEBOV GP was not restricted to the strain of mice and that vaccination with ZEBOVGP-Fc would protect mice against a lethal ZEBOV challenge. To test our hypotheses, we vaccinated C57BL/6 mice with the same regime used for the BALB/c mice, which showed a plateau in the production of neutralizing antibodies after 3 boosts (Fig. 3B). Eight C57BL/6 mice were vaccinated 100 µg of ZEBOVGP-Fc or control FLAG-Fc in complete Freunds adjuvant followed by three boosts with 25 µg protein in incomplete Freund adjuvant. Anti-ZEBOV GP antibodies in the sera of the vaccinated mice was analyzed by ELISA two weeks after the final boost. The 8 mice vaccinated with ZEBOVGP-Fc developed anti-ZEBOV GP antibody titers of 1:64,000 to 1:4,000 as measured by an ELISA in 96-well plates coated with rVSV-EBOVGP whereas the mice vaccinated with FLAG-Fc did not develop anti-ZEBOV GP antibody titers (Fig. 4A). The antibodies were specific to the ZEBOV GP because sera from the vaccinated animals did not react with wild type VSV in an ELISA using plates coated with wt VSV. More interestingly, the anti-ZEBOV GP titers determined using plates coated with irradiated ZEBOV were identical to those obtained using rVSV-EBOVGP coated indicating that ZEBOV GP on the surface of ZEBOV and rVSV-ZEBOVGP expose similar epitopes. The sera of the vaccinated mice were also tested by FACS analysis on live HEK293 cells expressing EBOV GP protein (HEK293-ZEBOVGP) at the cell surface or vector-transfected cells (HEK293). Sera from ZEBOVGP-Fc immunized mice reacted with the HEK293-ZEBOVGP cells but not with HEK293 cells (Fig. 4B). It should be pointed out that one of the immunized animals reacted poorly with the ZEBOV GP (animal #8). As expected, sera from the FLAG-Fc-vaccinated animals did not react with HEK293-ZEBOVGP or HEK293 cells, and anti-ZEBOV GP mAb 13F6-1-2 reacted only with HEK293-ZEBOVGP cells. These data are consistent with the ELISA data and demonstrated that anti-ZEBOV GP antibodies in the ZEBOVGP-Fc vaccinated animals react with native ZEBOV GP.
Fig. 4.
Humoral immune response in C57BL/6 mice vaccinated with ZEBOVGP-Fc. A) Analysis of anti-ZEBOV GP specific antibodies by viral particle ELISA. C57BL/6 mice (n=8) were vaccinated with ZEBOVGP-Fc or FLAG-Fc and sera samples were obtained 2 weeks after the final vaccination. Sera were titrated on 96-well plates coated with rVSV-ZEBOVGP, control wt VSV, or sucrose-purified irradiated ZEBOV particles. The endpoint dilution titer for each mouse sera is represented as a black dot. The anti-wt VSV titers of sera from the ZEBOVGP-Fc vaccinated mice were bellow the detection limit and are represented as a read dot. The titers of the sera from FLAG-Fc immunized mice were also bellow the detection limit and are represented as read triangles. B) Flow cytometry analysis of binding of the mouse sera to ZEBOV GP expressed at the cell surface of HEK293-ZEBOVGP cells. HEK293-ZEBOVGP or control vector-transfected HEK293 cells were stained with mouse sera (1 µl) and PE-conjugated goat anti-mouse IgG Ab and analyzed by flow cytometry. The histogram of each mouse sera is shown in a different color, and the histogram of the positive control anti-GP mAb 13F6-1-2 is shown as a dashed grey line. To simplify the graph, the same color was used for sera from two different mice, one vaccinated with ZEBOVGP-Fc (left panels) and the other with FLAG-Fc (right panels). C) Neutralization of rVSV-ZEBOVGP with sera from ZEBOVGP-Fc vaccinated mice. A plaque reduction assay was performed in Vero E6 cells using 100 pfu of rVSV-ZEBOVGP treated with five-fold dilutions of sera from mice vaccinated with ZEBOVGP-Fc or FLAG-Fc. Percent neutralization of each mouse serum was calculated as the reduction in the number of plaques compared to untreated rVSV-ZEBOVGP. Values in the graphs are the mean percent neutralization in duplicate samples for each mouse sera dilution, and bars represent the standard deviations.
We also analyzed anti-ZEBOV GP neutralizing antibodies using the rVSV-ZEBOVGP plaque reduction assay. Interestingly, all but one C57BL/6 mouse (number 8) vaccinated with ZEBOVGP-Fc developed neutralizing antibodies (Fig. 4C), and neutralization was antibody-dilution dependent. Mouse number 8 serum also showed low ELISA titers (Fig. 4A) and bound poorly to HEK293-ZEBOVGP cells as assessed by FACS analysis (Fig. 4B). Treatment with sera of mice vaccinated with FLAG-Fc did not neutralize rVSV-EBOVGP and resulted in background titer reductions of 40% or lower that were independent of the antibody dilution. These data show that all the mice vaccinated with ZEBOVGP-Fc except number 8 developed neutralizing antibodies against ZEBOV. The ELISA and rVSV-ZEBOVGP neutralization data clearly showed that immunization with ZEBOVGP-Fc elicited similar levels of anti-ZEBOV GP response in the BALB/c and C57BL/6 mice indicating that this response is not restricted to the strain of mouse. The C57BL/6 mice vaccinated with ZEBOVGP-Fc or FLAG-Fc were challenged with 1,000 pfu of mouse-adapted ZEBOV 74 days after the primary inoculation (two weeks after the third boost) (Fig. 5). Seven out of eight mice vaccinated with EBOVGP-Fc survived the ZEBOV challenge whereas seven out of the eight mice that received the FLAG-Fc vaccine died within 10 days after challenge. We performed daily checks for outward appearance of the mice. For the ZEBOGP-Fc vaccinated group, 6 out of 7 survivors did not show any outward symptoms of disease. One survivor appeared to be sick (with ruffled fur and hunched posture), but recovered and remained healthy through the end of the study.
Fig. 5.
EBOVGP-Fc protected mice against a ZEBOV lethal challenge. C57BL/6 mice vaccinated with ZEBOVGP-Fc (filled triangle) or FLAG-Fc (open squares) were challenged with 1,000 pfu of mouse adapted ZEBOV 74 days after the primary vaccination (2 weeks after the final vaccination). Results are plotted as percent survival for each vaccination group (n = 8 mice per group).
These data showed that ZEBOVGP-Fc elicited a protective immune response in mice against challenge with ZEBOV.
4. Discussion
Here we show that the GP ectodomain in the ZEBOVGP-Fc fusion protein underwent the complex posttranslational modification, including the furin cleavage and disulfide bond formation between the GP1 and GP2 subunits, of the membrane-bound full-length GP. Moreover, our FPLC analysis of enterokinase-digested ZEBOVGP-Fc showed that the ZEBOVGP fragment of the Fc fusion protein formed trimers that resemble the natural GP expressed at the viral membrane and cell surface. Vaccination with ZEBOVGP-Fc elicited a humoral immune response that recognized membrane bound GP at the cell and viral particle surface. Interestingly, the anti-GP antibodies reacted similarly with inactivated ZEBOV and rVSV-EBOVGP, a replication-competent recombinant VSV particle carrying the ZEBOV GP at the viral surface that induces a protective response in NHP [15, 16]. The anti-ZEBOVGP-Fc antibodies neutralized rVSV-EBOVGP suggesting that this fusion protein may also elicit protective neutralizing antibodies in NHP.
VSVG-deleted VSV pseudotypes complemented with the Filovirus GP have been widely used to study cell entry of Filoviruses [6, 38–40]. Replication-competent VSVG-deleted recombinant VSV containing the Filovirus GP (rVSV-FiloGP) [30]has been used to select mAb-resistant mutants to map neutralizing epitopes in GP [41]. These rVSV-FiloGP have been used to develop attenuated vaccines [15, 16, 30, 42–48]. However, Filovirus neutralizing antibodies in vaccinated animals have been mainly assessed under BSL-4 conditions using plaque reduction assays, especially in animals vaccinated with recombinant VSV construct where neutralization of rVSV-FiloGP could be attributed to the immune response against non-GP components in the vaccine. Our data showed that rVSV-EBOVGP is a practical tool to evaluate total and neutralizing antibodies under BSL-2 conditions and suggested that rVSV-FiloGP could be used to assess vaccine potency, evaluate consistency in vaccine production, and as a surrogate marker of vaccine efficacy in clinical trials. Further research will be required to test whether rVSVFiloGP could be used to assess Filovirus neutralizing antibodies in animals vaccinated with recombinant VSV constructs.
Cellular immunity characterized by the production of TNF-α and INF-γ plays a significant role in the protection against Filovirus infection ([49] and references therein). ZEBOVGP-Fc induced a T-cell response in mice as evidenced by the activation of IFN-γ-positive CD8+ cells in splenocytes stimulated with ZEBOVGP-Fc. Interestingly, stimulation with FLAG-Fc elicited a background level response indicating that the Fc fragment contributed minimally to the cellular response against the fusion protein. In NHP and humans, the human IgG1 Fc fragment present in ZEBOVGP-Fc is likely to be recognized as a self-antigen and may enhance the immunogenicity of the ZEBOVGP-Fc fusion protein by interacting with Fcγ receptors on antigen presenting cells [21–23]. It is not clear why the GP synthetic peptides were ineffective in activating GP-specific T-cells in ZEBOVGP-Fc vaccinated mice. In many studies, these peptides were shown to activate ZEBOV GP-specific T cells in mice under similar conditions used in our assay [34, 35]. However, one of the peptides (LYDRLASTV) failed to activate IFN-γ-positive CD8+ cells in BALB/c mice vaccinated with replication-deficient EbolaΔVP30 virus [17]. Further research will be required to determine whether ZEBOVGP-Fc vaccination elicited a cellular response against a different set of T-cell epitopes. Taken together, our results clearly showed that ZEBOVGP-Fc induced both cellular and humoral immunity against the ZEBOV GP ectodomain.
One mouse vaccinated with EBOVGP-Fc did not survive the lethal challenge with ZEBOV. Unfortunately, we did not use individual identifiers at challenge and cannot unequivocally correlate the serology and the survival data. However, it is likely that the EBOVGP-Fc vaccinated animal that did not survive the challenge was the only mouse in the group that developed low levels of anti-ZEBOV GP antibodies that failed to neutralize rVSV-ZEBOVGP. Studies are being planned to determine whether the level of neutralizing antibodies against the replication-competent rVSV-ZEBOVGP correlates with protection against lethal challenge with ZEBOV.
Our data suggested that ZEBOVGP-Fc could be used as a subunit Filovirus vaccine for human use. Although the Freunds adjuvant and the i.p. route used in our mouse studies are not suitable for human use, it is possible that other adjuvants licensed for human use could enhance the immunogenicity of ZEBOVGP-Fc. Further experimentation will be required to determine the best inoculation route and the need of adjuvant to enhance the immunogenicity of ZEBOV-GP in vaccines. Previous attempts to develop Filovirus subunit vaccines based on soluble forms of Filovirus GP expressed in insect cells provided limited protection against lethal challenge in the guinea pig model [20, 50]. It is possible that glycosylation in insect cells may have affected epitope structure, homotrimer formation, and stability of the soluble GP protein, factors that could account for the poor performance of the soluble GP vaccine. The EBOVGP-Fc used in our study was produced in mammalian cells and most likely resembles the native GP better than the soluble GP expressed in insect cells. In addition, the Fc tag in the ZEBOVGP-Fc conferred several advantages to our vaccine strategy including the ease of purification through protein A columns using mild conditions, an increased protein stability conferred by the Fc fragment. The Fc tag may also have adjuvant effect in NHP and humans due to the interactions with Fcγ receptors on antigen presenting cells [21–23]. However, we do not know whether the Fc tag increased the stability of the antigen or functioned as an adjuvant in the mouse model described in this paper. Further research will be required to determine whether the Fc tag played any role in the immunogenicity of ZEBOVGP-Fc in mice.
Our results demonstrated that vaccination with the EBOVGP-Fc fusion protein elicited a significant level of protection against challenge with ZEBOV and suggested that a subunit vaccine based on Filovirus GP-Fc fusion proteins could be developed to protect against viral infection. This Filovirus GP-Fc could be used as a stand-alone vaccine or in combination with other strategies such as DNA vaccines, virus-like particles, and viral vector vaccines that are currently under development. A subunit vaccine based on Filovirus GP-Fc fusion proteins will be simple to produce, easy to purify, and cost-effective and will likely result in limited adverse events. Additional experiments in the guinea pig and monkey models will be needed to further determine the feasibility of developing a Filovirus subunit vaccine based on GP-Fc fusion proteins.
Acknowledgements
This work was supported by CBER/FDA - NIAID/NIH Interagency Agreements Y1-AI-6153-01 and Y1-AI-0664-01, grants from the Biomedical Advanced Research and Development Authority (BARDA) to GGK, intramural funding from the Food and Drug Administration to GGK, and USAMRIID to SB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare that they have no competing interests.
"The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any determination or policy".
References
- 1.Sanchez A, Geisbert TW, Feldmann H. Filoviridae: Marburg and Ebola viruses. In: Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, Straus SE, editors. Fields Virology. 5th ed. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 1409–1448. [Google Scholar]
- 2.Towner JS, Sealy TK, Khristova ML, Albarino CG, Conlan S, Reeder SA, et al. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008 Nov;4(11):e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Normile D. Emerging infectious diseases. Scientists puzzle over Ebola-Reston virus in pigs. Science. 2009 Jan 23;323(5913):451. doi: 10.1126/science.323.5913.451a. [DOI] [PubMed] [Google Scholar]
- 4.Volchkov VE, Volchkova VA, Dolnik O, Feldmann H, Klenk HD. Polymorphism of filovirus glycoproteins. Adv Virus Res. 2005;64:359–381. doi: 10.1016/S0065-3527(05)64011-0. [DOI] [PubMed] [Google Scholar]
- 5.Feldmann H, Volchkov VE, Volchkova VA, Stroher U, Klenk HD. Biosynthesis and role of filoviral glycoproteins. J Gen Virol. 2001 Dec;82(Pt 12):2839–2848. doi: 10.1099/0022-1317-82-12-2839. [DOI] [PubMed] [Google Scholar]
- 6.Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, et al. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci U S A. 1997 Dec 23;94(26):14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci U S A. 1998 May 12;95(10):5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeffers SA, Sanders DA, Sanchez A. Covalent modifications of the ebola virus glycoprotein. J Virol. 2002 Dec;76(24):12463–12472. doi: 10.1128/JVI.76.24.12463-12472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldmann H, Kiley MP. Classification, structure, and replication of filoviruses. Curr Top Microbiol Immunol. 1999;235:1–21. doi: 10.1007/978-3-642-59949-1_1. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan NJ, Geisbert TW, Geisbert JB, Shedlock DJ, Xu L, Lamoreaux L, et al. Immune protection of nonhuman primates against Ebola virus with single low-dose adenovirus vectors encoding modified GPs. PLoS Med. 2006 Jun;3(6):e177. doi: 10.1371/journal.pmed.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan NJ, Geisbert TW, Geisbert JB, Xu L, Yang ZY, Roederer M, et al. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature. 2003 Aug 7;424(6949):681–684. doi: 10.1038/nature01876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan NJ, Sanchez A, Rollin PE, Yang ZY, Nabel GJ. Development of a preventive vaccine for Ebola virus infection in primates. Nature. 2000 Nov 30;408(6812):605–609. doi: 10.1038/35046108. [DOI] [PubMed] [Google Scholar]
- 13.Bukreyev A, Rollin PE, Tate MK, Yang L, Zaki SR, Shieh WJ, et al. Successful topical respiratory tract immunization of primates against Ebola virus. J Virol. 2007 Jun;81(12):6379–6388. doi: 10.1128/JVI.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hevey M, Negley D, Pushko P, Smith J, Schmaljohn A. Marburg virus vaccines based upon alphavirus replicons protect guinea pigs and nonhuman primates. Virology. 1998 Nov 10;251(1):28–37. doi: 10.1006/viro.1998.9367. [DOI] [PubMed] [Google Scholar]
- 15.Feldmann H, Jones SM, Daddario-DiCaprio KM, Geisbert JB, Stroher U, Grolla A, et al. Effective post-exposure treatment of Ebola infection. PLoS Pathog. 2007 Jan;3(1):e2. doi: 10.1371/journal.ppat.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones SM, Feldmann H, Stroher U, Geisbert JB, Fernando L, Grolla A, et al. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat Med. 2005 Jul;11(7):786–790. doi: 10.1038/nm1258. [DOI] [PubMed] [Google Scholar]
- 17.Halfmann P, Ebihara H, Marzi A, Hatta Y, Watanabe S, Suresh M, et al. Replication-deficient ebolavirus as a vaccine candidate. J Virol. 2009 Apr;83(8):3810–3815. doi: 10.1128/JVI.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warfield KL, Bosio CM, Welcher BC, Deal EM, Mohamadzadeh M, Schmaljohn A, et al. Ebola virus-like particles protect from lethal Ebola virus infection. Proc Natl Acad Sci U S A. 2003 Dec 23;100(26):15889–15894. doi: 10.1073/pnas.2237038100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warfield KL, Swenson DL, Olinger GG, Kalina WV, Aman MJ, Bavari S. Ebola virus-like particle-based vaccine protects nonhuman primates against lethal Ebola virus challenge. J Infect Dis. 2007 Nov 15;196 Suppl 2:S430–S437. doi: 10.1086/520583. [DOI] [PubMed] [Google Scholar]
- 20.Mellquist-Riemenschneider JL, Garrison AR, Geisbert JB, Saikh KU, Heidebrink KD, Jahrling PB, et al. Comparison of the protective efficacy of DNA and baculovirus-derived protein vaccines for EBOLA virus in guinea pigs. Virus Res. 2003 Apr;92(2):187–193. doi: 10.1016/s0168-1702(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 21.Zhang MY, Wang Y, Mankowski MK, Ptak RG, Dimitrov DS. Cross-reactive HIV-1-neutralizing activity of serum IgG from a rabbit immunized with gp41 fused to IgG1 Fc: possible role of the prolonged half-life of the immunogen. Vaccine. 2009 Feb 5;27(6):857–863. doi: 10.1016/j.vaccine.2008.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guyre PM, Graziano RF, Goldstein J, Wallace PK, Morganelli PM, Wardwell K, et al. Increased potency of Fc-receptor-targeted antigens. Cancer Immunol Immunother. 1997 Nov–Dec;45(3–4):146–148. doi: 10.1007/s002620050418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Xu X, Jones IM. Immunogenicity of the outer domain of a HIV-1 clade C gp120. Retrovirology. 2007;4:33. doi: 10.1186/1742-4690-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silberstein E, Dveksler G, Kaplan GG. Neutralization of hepatitis A virus (HAV) by an immunoadhesin containing the cysteine-rich region of HAV cellular receptor-1. J Virol. 2001 Jan;75(2):717–725. doi: 10.1128/JVI.75.2.717-725.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999 Jan;73(1):251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang ZY, Duckers HJ, Sullivan NJ, Sanchez A, Nabel EG, Nabel GJ. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat Med. 2000 Aug;6(8):886–889. doi: 10.1038/78645. [DOI] [PubMed] [Google Scholar]
- 27.Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, et al. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000 Mar 3;287(5458):1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- 28.Lee JE, Kuehne A, Abelson DM, Fusco ML, Hart MK, Saphire EO. Complex of a protective antibody with its Ebola virus GP peptide epitope: unusual features of a V lambda × light chain. J Mol Biol. 2008 Jan 4;375(1):202–216. doi: 10.1016/j.jmb.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawson ND, Stillman EA, Whitt MA, Rose JK. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci U S A. 1995 May 9;92(10):4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garbutt M, Liebscher R, Wahl-Jensen V, Jones S, Moller P, Wagner R, et al. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J Virol. 2004 May;78(10):5458–5465. doi: 10.1128/JVI.78.10.5458-5465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bray M, Davis K, Geisbert T, Schmaljohn C, Huggins J. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis. 1998 Sep;178(3):651–661. doi: 10.1086/515386. [DOI] [PubMed] [Google Scholar]
- 32.Bradfute SB, Swanson PE, Smith MA, Watanabe E, McDunn JE, Hotchkiss RS, et al. Mechanisms and consequences of ebolavirus-induced lymphocyte apoptosis. J Immunol. 2010 Jan 1;184(1):327–335. doi: 10.4049/jimmunol.0901231. [DOI] [PubMed] [Google Scholar]
- 33.Warfield KL, Swenson DL, Negley DL, Schmaljohn AL, Aman MJ, Bavari S. Marburg virus-like particles protect guinea pigs from lethal Marburg virus infection. Vaccine. 2004 Sep 3;22(25–26):3495–3502. doi: 10.1016/j.vaccine.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 34.Olinger GG, Bailey MA, Dye JM, Bakken R, Kuehne A, Kondig J, et al. Protective cytotoxic T-cell responses induced by venezuelan equine encephalitis virus replicons expressing Ebola virus proteins. J Virol. 2005 Nov;79(22):14189–14196. doi: 10.1128/JVI.79.22.14189-14196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warfield KL, Olinger G, Deal EM, Swenson DL, Bailey M, Negley DL, et al. Induction of humoral and CD8+ T cell responses are required for protection against lethal Ebola virus infection. J Immunol. 2005 Jul 15;175(2):1184–1191. doi: 10.4049/jimmunol.175.2.1184. [DOI] [PubMed] [Google Scholar]
- 36.Chamow SM, Ashkenazi A. Immunoadhesins: principles and applications. Trends Biotechnol. 1996 Feb;14(2):52–60. doi: 10.1016/0167-7799(96)80921-8. [DOI] [PubMed] [Google Scholar]
- 37.Hood CL, Abraham J, Boyington JC, Leung K, Kwong PD, Nabel GJ. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J Virol. 2010 Mar;84(6):2972–2982. doi: 10.1128/JVI.02151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito H, Watanabe S, Sanchez A, Whitt MA, Kawaoka Y. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J Virol. 1999 Oct;73(10):8907–8912. doi: 10.1128/jvi.73.10.8907-8912.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito H, Watanabe S, Takada A, Kawaoka Y. Ebola virus glycoprotein: proteolytic processing, acylation, cell tropism, and detection of neutralizing antibodies. J Virol. 2001 Feb;75(3):1576–1580. doi: 10.1128/JVI.75.3.1576-1580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, et al. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6(9) doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takada A, Feldmann H, Stroeher U, Bray M, Watanabe S, Ito H, et al. Identification of protective epitopes on ebola virus glycoprotein at the single amino acid level by using recombinant vesicular stomatitis viruses. J Virol. 2003 Jan;77(2):1069–1074. doi: 10.1128/JVI.77.2.1069-1074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geisbert TW, Daddario-Dicaprio KM, Geisbert JB, Reed DS, Feldmann F, Grolla A, et al. Vesicular stomatitis virus-based vaccines protect nonhuman primates against aerosol challenge with Ebola and Marburg viruses. Vaccine. 2008 Dec 9;26(52):6894–6900. doi: 10.1016/j.vaccine.2008.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geisbert TW, Daddario-Dicaprio KM, Lewis MG, Geisbert JB, Grolla A, Leung A, et al. Vesicular stomatitis virus-based ebola vaccine is well-tolerated and protects immunocompromised nonhuman primates. PLoS Pathog. 2008 Nov;4(11):e1000225. doi: 10.1371/journal.ppat.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geisbert TW, Daddario-DiCaprio KM, Williams KJ, Geisbert JB, Leung A, Feldmann F, et al. Recombinant vesicular stomatitis virus vector mediates postexposure protection against Sudan Ebola hemorrhagic fever in nonhuman primates. J Virol. 2008 Jun;82(11):5664–5668. doi: 10.1128/JVI.00456-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geisbert TW, Geisbert JB, Leung A, Daddario-DiCaprio KM, Hensley LE, Grolla A, et al. Single-injection vaccine protects nonhuman primates against infection with marburg virus and three species of ebola virus. J Virol. 2009 Jul;83(14):7296–7304. doi: 10.1128/JVI.00561-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geisbert TW, Hensley LE, Geisbert JB, Leung A, Johnson JC, Grolla A, et al. Postexposure treatment of Marburg virus infection. Emerg Infect Dis. 2010 Jul;16(7):1119–1122. doi: 10.3201/eid1607.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daddario-DiCaprio KM, Geisbert TW, Geisbert JB, Stroher U, Hensley LE, Grolla A, et al. Cross-protection against Marburg virus strains by using a live, attenuated recombinant vaccine. J Virol. 2006 Oct;80(19):9659–9666. doi: 10.1128/JVI.00959-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daddario-DiCaprio KM, Geisbert TW, Stroher U, Geisbert JB, Grolla A, Fritz EA, et al. Postexposure protection against Marburg haemorrhagic fever with recombinant vesicular stomatitis virus vectors in non-human primates: an efficacy assessment. Lancet. 2006 Apr 29;367(9520):1399–1404. doi: 10.1016/S0140-6736(06)68546-2. [DOI] [PubMed] [Google Scholar]
- 49.Hensley LE, Mulangu S, Asiedu C, Johnson J, Honko AN, Stanley D, et al. Demonstration of cross-protective vaccine immunity against an emerging pathogenic Ebolavirus Species. PLoS Pathog. 2010 May;6(5):e1000904. doi: 10.1371/journal.ppat.1000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hevey M, Negley D, Geisbert J, Jahrling P, Schmaljohn A. Antigenicity and vaccine potential of Marburg virus glycoprotein expressed by baculovirus recombinants. Virology. 1997 Dec 8;239(1):206–216. doi: 10.1006/viro.1997.8883. [DOI] [PubMed] [Google Scholar]