Abstract
The current study has addressed the role of PTEN loss in intrinsic resistance to the BRAF inhibitor PLX4720. Immunohistochemical staining of a tissue array covering all stages of melanocytic neoplasia (n=192) revealed PTEN expression to be lost in >10% of all melanoma cases. Although PTEN expression status did not predict for sensitivity to the growth inhibitory effects of PLX4720, it was predictive for apoptosis, with only limited cell death observed in melanomas lacking PTEN expression (PTEN−). Mechanistically, PLX4720 was found to stimulate AKT signaling in the PTEN− but not the PTEN+ cell lines. Liquid chromatography multiple reaction monitoring mass spectrometry (LC-MRM) was performed to identify differences in apoptosis signaling between the two cell line groups. PLX4720 treatment significantly increased BIM expression in the PTEN+ (>14-fold) compared to the PTEN− cell lines (4-fold). A role for PTEN in the regulation of PLX4720-mediated BIM expression was confirmed by siRNA knockdown of PTEN and through re-introduction of PTEN into cells that were PTEN−. Further studies showed that siRNA knockdown of BIM significantly blunted the apoptotic response in PTEN+ melanoma cells. Dual treatment of PTEN− cells with PLX4720 and a PI3K inhibitor enhanced BIM expression at both the mRNA and protein level and increased the level of apoptosis through a mechanism involving AKT3 and the activation of FOXO3a. In conclusion, we have shown for the first time that loss of PTEN contributes to intrinsic BRAF inhibitor resistance via the suppression of BIM-mediated apoptosis.
Keywords: melanoma, BRAF, resistance, therapy, PTEN, PLX4032
Introduction
One defining moment in our understanding of melanoma initiation and progression was the discovery of activating V600E mutations in BRAF in >50% of melanomas (1, 2). There is now good evidence that mutated BRAF is a bona fide therapeutic target in melanoma (3-5). A number of BRAF specific small molecule kinase inhibitors have been developed that are now undergoing intense pre-clinical and clinical investigation (6, 7). In pre-clinical studies, the BRAF inhibitors PLX4720 and PLX4032 potently inhibited BRAF kinase activity in melanoma cells harboring the BRAF V600E mutation and were cytostatic and cytotoxic in both in vitro cell culture systems and in vivo xenograft melanoma models (5, 6, 8). This promising pre-clinical activity was mirrored by a recent phase I clinical trial of PLX4032 in advanced melanoma in which >80% of patients showed some level of tumor regression (4). Although most patients with BRAF V600E mutated melanoma showed some response to PLX4032, ~20% of those treated did not meet the RECIST criteria threshold for a response (4, 9). Although the mechanisms of intrinsic BRAF inhibitor resistance are not well understood, increased cyclin D1 expression (in ~17% of BRAF mutated melanomas) allows for cell cycle entry when MAPK signaling is abrogated (10, 11). It is also likely that constitutive activity in other pathways, such as phospho-inositide 3-kinase (PI3K)/AKT, may contribute to intrinsic resistance by limiting the apoptotic response (12, 13). One of the most critical negative regulators of AKT activity is the phosphatase and tensin homologue (PTEN), which hydrolyses PI-3,4,5-P3 to PI-4,5-P2, ultimately preventing the phosphorylation of AKT (14). In the present study we identify loss of PTEN expression, observed in >10% of melanoma specimens, as being responsible for increased PI3K/AKT signaling when BRAF is inhibited. We further show that PTEN loss contributes to the intrinsic resistance of BRAF V600E-mutated melanoma cell lines to PLX4720 by suppressing the expression of the pro-apoptotic protein BIM.
Cell culture and MTT assay
Melanoma cell lines were a gift from Dr. Meenhard Herlyn (The Wistar Institute, Philadelphia, PA) and were grown as described in (8). MTT assays were performed as described in (15). The identity of the Wistar Institute cell lines was confirmed using the Coriell Institute (Camden, NJ) cell identity mapping kit. The M233 cell line was derived as described in (16) and its identity confirmed by Biosynthesis Inc (Lewisville, Tx) by STR validation analysis.
Generation WM793TR-PTEN cell lines
Wild-type and G129E PTEN (phosphatase deficient) human cDNAs were a gift from Dr. Bill Sellers (Dana-Farber Cancer Institute) (17). WM793TR-PTEN-wt, WM793TR-PTEN-G129E and WM793 cells overexpressing wild-type BAD were a kind gift from Dr Andrew Aplin (Kimmel Cancer Center, Philadelphia, PA). Inducible expression of PTEN was obtained by treatment of cultures with doxycycline at a final concentration of 100ng/ml. The WM793 cells stably expressing wild-type BAD were generated as described in (18).
Western blotting
Proteins were blotted for as described in (15). The antibodies to phospho-AKT (Ser473 and T308), total AKT, phospho-BAD (S75 and S99), Bcl-2, BIM, BRAF, FOXO3a, phospho-PDK1, total PDK1, PTEN, phospho-S6 and total S6 were from Cell Signaling Technology (Beverly, MA).
Flow cytometry
Cells were treated with 3 or 10μM PLX4720 for 24 or 48 hr or treated with PLX4720 (3 or 10μM) in the absence or presence of GDC-0941 (3 μM, Selleck Chemical Co.) and harvested after 48 hr. Annexin-V/TMRM staining was performed as described in (19).
RNA interference
Cells were grown overnight in RPMI complete media. The following day, complete media was replaced with Opti-MEM (Invitrogen) and one of the following siRNA sequences in complex with Lipofectamine 2000 (Invitrogen): 50nM BRAF (Dharmacon), 20nM PTEN, 25nM BIM (Cell Signaling Technology). Scrambled siRNA’s at each concentration were also added as non-targeting controls. A final concentration of 5% FBS in complete RPMI was added the next day. Cells were transfected for a total of 48-72 hr prior to treatment with PLX4720 (3-10μM).
Quantitative real-time PCR
Total RNA was isolated using Qiagen’s RNeasy mini kit. The following TaqMan® Gene Expression Assays primer/probes were used: Hs00197982_m1 (BIM), P/N 4319413E (18S) and Hs99999905_m1 (GAPDH). The 18S + GAPDH data were used for normalizing BIM. After a 2-min incubation at 50°C, AmpliTaq Gold was activated by a 10-min incubation at 95°C, followed by 40 PCR cycles consisting of 15 s of denaturation at 95°C and hybridization of probe and primers for 1 min at 60°C. All standards and samples were tested in triplicate wells and data were analyzed using SDS software version 2.3.
Immunofluorescence staining
Cells were plated onto coverslips and treated with PLX4720 for 48hrs before being fixed and permeabilized as previously described (15) and imaged with a Leica confocal microscope at 40X magnification.
3D spheroid assays
Collagen implanted spheroids were prepared using the liquid overlay method (12) and were treated with 3μM of PLX4720, 10μM LY294002 or both drugs in combination for 72hr before being analyzed by fluorescence microscopy as described in (11). In other studies, spheroids were treated for 72hrs, washed 3X in fresh media and allowed to recover for 120hrs before analysis.
Immunohistochemical staining
A melanoma tissue array was generated from de-identified formalin-fixed paraffin-embedded tissue samples from the Moffitt Pathology archives under a protocol approved by the Institutional Review Board of the University of South Florida. Slides were stained using the Ventana Discovery XT automated system (Ventana Medical Systems, Tucson, AZ) as per manufacturer’s protocol. The PTEN antibody (1:200, #E4250, Spring Bioscience, Pleasanton, CA) was incubated for 32 min and the pAKT antibody (1:20, #4051, Cell Signaling, Danvers, MA) was incubated for 16 min. Slides were analyzed by two independent observers and consensus scored on a scale from (0 to +3).
Liquid chromatography, multiple reaction monitoring mass spectrometry (LC-MRM) analysis
Whole cell proteins extracts were separated by SDS-PAGE, visualized with Coomassie Brilliant Blue G-250 (Bio-Rad, Hercules, CA) and selected bands were excised. Following digestion, the internal standard peptides were added in 2% acetonitrile. LC-MRM analysis was performed as described in (20) with three replicate analyses for each peptide. Quantification was achieved by using the sum of the peak areas for all detected transitions using Xcalibur QuanBrowser (Thermo, San Jose, CA). Relative protein expression is determined using the ratio of peak area of the native peptide to corresponding internal standard (Supplemental Table 2).
Statistical analysis
Data show the mean of at least three independent experiments ± the S.E. mean, unless stated otherwise. GraphPad Prism 5 statistical software was used to perform the Student’s T-test where *indicates P≤ 0.05.
Results
The role of PTEN loss in the response to PLX4720
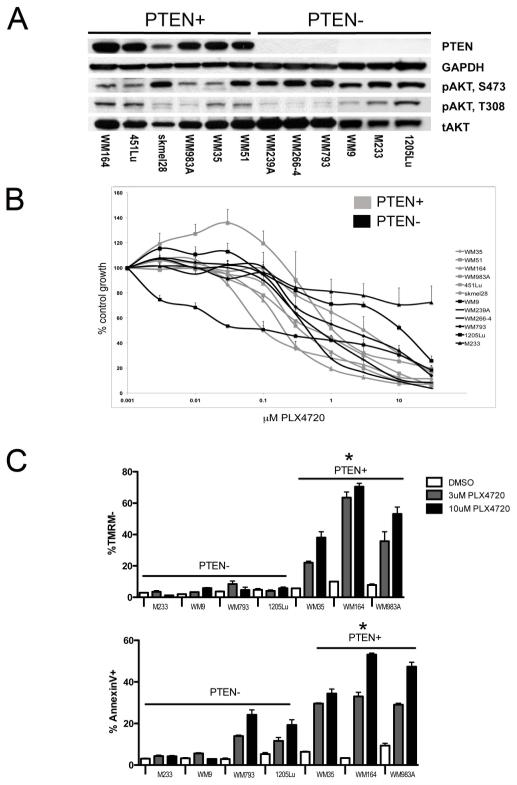
Initial studies identified 6 BRAF mutated melanoma cell lines that retained PTEN expression (PTEN+; WM35, WM51, WM164, WM983A, 451Lu, SK-Mel-28) and 6 that lacked PTEN expression (PTEN−; WM239A, WM266-4, WM793, 1205Lu, WM9, M233) (Figure 1A; Supplemental Table 1). Genomic analysis showed the WM9 and M233 cell lines to be homozygously deleted for PTEN and the WM793 and 1205lu cell lines be hemizygously deleted for PTEN in conjunction with a PTEN mutation (Supplemental Table 1). The PTEN+ cell lines had lower constitutive levels of pAKT (Ser473) compared to the PTEN− (Figure 1A). Similar levels of pAKT (Thr308) were observed in the PTEN− and PTEN+ cell lines. Analysis of the growth inhibitory effects of PLX4720 by the MTT and Alamar Blue assays (Figure 1B, Supplemental Figure 1) did not reveal any statistically significant differences in the GI50 values between the PTEN+ and PTEN− cell lines (P=0.48, Supplemental Figure 2).
Figure 1. PTEN predicts for PLX4720-induced apoptosis.
(A) Basal PTEN and phospho-AKT (pAKT) (S473, T308) expression in PTEN+ (WM164, 451Lu, SK-mel-28, WM983A, WM35, WM51) and PTEN− (WM239A, WM266-4, WM793, M233, WM9, 1205Lu) melanoma cell lines. (B) MTT assay of PTEN+ (gray) expressing versus PTEN− (black) cell lines. (C) PTEN+ cells are more sensitive than PTEN− cells to PLX4720 mediated apoptosis. Cells treated for 48hrs with 3 or 10μM PLX4720 before being stained for TMRM and Annexin-V. Apoptosis was measured by flow cytometry. Data shows mean, +/− S.E. mean of three independent experiments. * Indicates PTEN+ cohort significantly different from PTEN− cohort (P<0.05).
As increased PI3K/AKT signaling is known to limit apoptosis, we next measured PLX4720-induced apoptosis in our PTEN−/PTEN+ melanoma cell line panel (Figures 1C). Here we observed that following PLX4720 treatment (3 and 10μM, 48 hrs), the PTEN− melanoma cell lines showed significantly less apoptosis than the PTEN+ (*P<0.05: Figure 1C). PLX4720 mediated apoptosis was blocked by high doses (>75μM) of the capase inhibitor z-vad-fmak (Supplemental Figure 3).
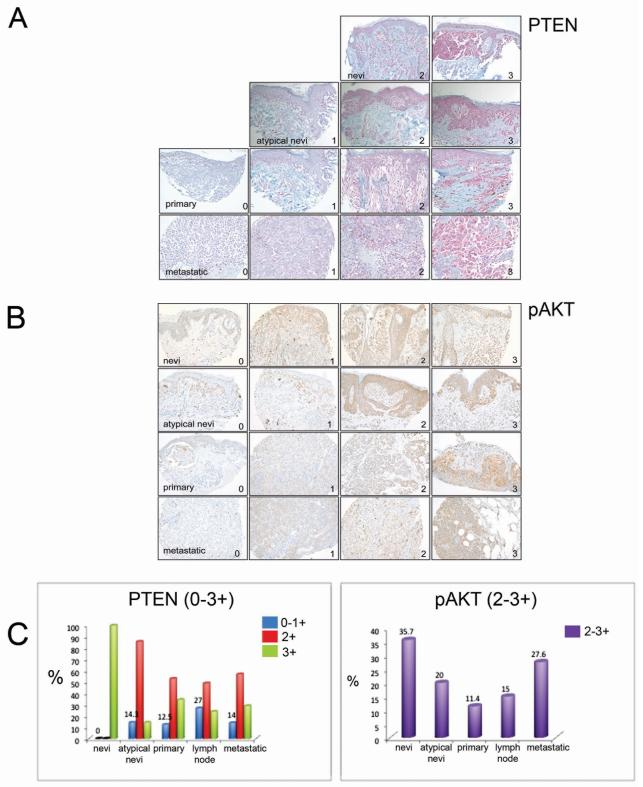
Loss of PTEN expression is independent of melanoma stage
We confirmed the incidence of PTEN loss in a tissue microarray containing a large sample of melanocytic neoplasms (n=192) drawn from all stages of tumor progression (Figures 2A-C). Results of immunohistochemical staining were graded from 0-3+ based on strength of the staining. It was observed that while non-atypical nevi rarely demonstrated loss of PTEN, >10% of atypical nevi and every stage of melanoma demonstrated loss of PTEN expression (either 0 or +1). Significantly, primary melanoma, lymph node metastases and distant metastases melanoma demonstrated loss of PTEN in 12.5%, 27% and 14% of cases each (Figure 2A and 2C). Staining of the same TMA for pAKT demonstrated an increase in AKT activation as the tumors progressed from primary melanoma to distant metastasis (Figure 2B). The level of pAKT positivity only partially correlated with PTEN expression status (Figure 2C).
Figure 2. PTEN expression is lost at all stages of melanoma progression.
(A) Images showing representative immunohistochemical staining of PTEN and (B) pAKT expression in a tissue array of nevi, atypical nevi, primary and metastatic melanoma patient tumor samples. 0-1 indicates no to low PTEN expression and +3 indicates the highest expression while +2-3 relates to high expression of pAKT. Magnification x 200. (C) Left panel shows percentage loss of PTEN expression in each subset of patient samples as indicated in blue while the right panel shows AKT activity in matched samples.
PLX4720 and BRAF siRNA leads to AKT signaling in BRAF V600E-mutated/PTEN− melanoma cell lines
Treatment of the PTEN+/− cell line panels with PLX4720 increased pPDK1 and pAKT signaling only in the melanoma cell lines lacking PTEN expression (Figure 3A). In contrast, PLX4720 inhibited BRAF activity in both PTEN− and PTEN+ cell lines with a similar potency (Supplemental Figure 4) and prevented BrdU uptake in both PTEN+ and PTEN− cell lines (Supplemental Figure 5). Addition of PLX4720 also led to the inhibition of mTOR activity in the PTEN+ cell lines only (Figure 3A) and was associated with stimulation of LKB1 and AMPK signaling (Supplemental Figure 6). The requirement for PTEN in the increased AKT signaling was confirmed by studies showing that PLX4720 stimulated pAKT in WM164 cells (PTEN+) when PTEN was knocked down by siRNA (Figure 3B). The effects of PLX4720 upon pAKT signaling were BRAF specific, with BRAF siRNA knockdown found to increase pAKT in PTEN− cells only (Figure 3C). Mechanistically, PLX4720 increased IGF-I signaling in the PTEN− cells, with the IGFR1 inhibitor NVP-ADW-742 (21) being found to abrogate the PLX4720-mediated increase in pAKT signaling (Supplemental Figure 7).
Figure 3. Loss of PTEN is associated with PI3K/AKT signaling following BRAF inhibition.
(A) PTEN+ (WM35, WM164, WM983A) and PTEN− (M233, WM9, WM793, 1205Lu) cells were treated with PLX4720 (24 hrs: 0.03-3 μM) and probed for phospho-PDK1 (pPDK1), total PDK1, phospho-AKT (pAKT), total AKT (tAKT), phospho-S6 (pS6) and total S6. Numbers indicate relative intensity of pPDK1 normalized to PDK1 and pAKT normalized to tAKT. (B) PLX4720 increases pAKT following PTEN knockdown. WM35 cells were incubated with non-targeting siRNA (NT) or two different PTEN specific siRNA’s (PTEN) before treatment with either vehicle or PLX4720 (3 μM). (C) siRNA knockdown of BRAF increases pAKT in melanoma cell lines that are PTEN−. WM164, (PTEN+) and WM793 (PTEN−) cells were incubated with lipofectamine alone (L), non-targeting siRNA (NT) or BRAF-specific siRNA (BRAF). Protein was extracted, resolved and probed for BRAF, pAKT, total AKT and GAPDH.
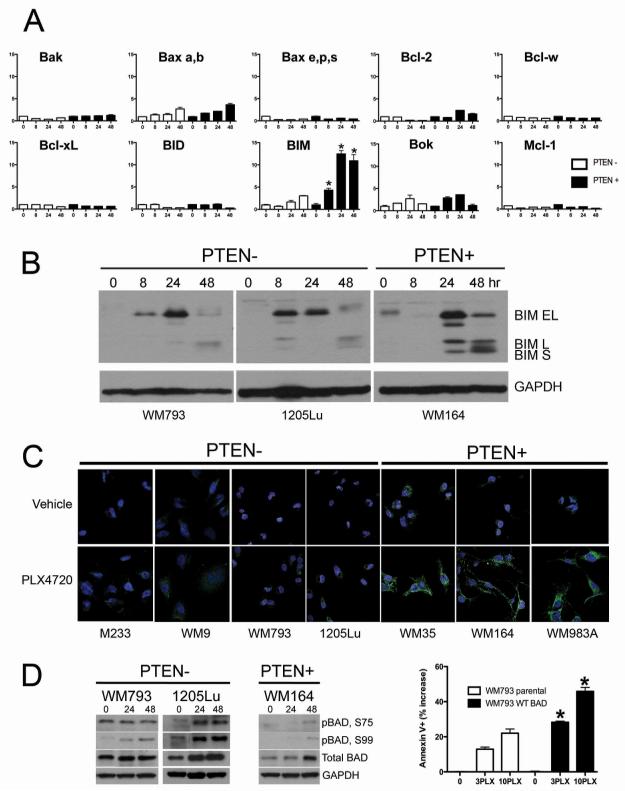
PLX4720 treatment differentially regulates BIM in PTEN+ and PTEN− cells
We next used LC-MRM to quantify the PLX4720-induced changes in the expression of 17 members of the Bcl-2 protein family (Figure 4A shows results for 9 proteins). The only pro-apoptotic protein to demonstrate significant differences between the PTEN− and PTEN+ cell lines was BIM (Figure 4A) (14-fold and 4-fold increases in the PTEN+ and PTEN− cell lines, respectively). Western blots and immunofluorescence staining confirmed the LC-MRM data and showed a greater degree of PLX4720-induced (3 and 10μM) BIM expression in the PTEN+ cell lines compared to PTEN− cell lines (Figure 4B,C and Supplemental Figure 8). In parallel, we observed that PLX4720 also increased the inactivation of BAD (as shown by increased phospho-BAD) in the PTEN− cells (Figure 4D) and that overexpression of BAD (not shown) in the PTEN− cells enhanced PLX4720-mediated apoptosis (Figure 4D). PLX4720 treatment also increased total BAD expression in both the PTEN+ and PTEN− cell lines. Little PLX4720-induced changes in Mcl-1 expression were observed in the PTEN+ and PTEN− cell lines (Supplemental Figure 9).
Figure 4. LC-MRM identifies differential regulation of BIM in PTEN+ and PTEN− cell lines following PLX4720 treatment.
(A) Representative LC-MRM data showing the fold-changes in the expression of Bak, Bax, Bcl-2, Bcl-w, Bcl-xL, BID, BIM, Bok and Mcl-1 over internal standard in the WM164 (PTEN+) and 1205Lu (PTEN−) cell lines following treatment with PLX4720 (10μM, 0-48 hrs). Statistical analysis of BIM fold change in PTEN− versus PTEN+, *p<0.05. (B) Western blot showing BIM expression following PLX4720 treatment (10μM, 0-48 hrs) in PTEN− (WM793, 1205Lu) and WM164 cell lines (PTEN+). (C) Immunofluorescence staining, showing expression of BIM (green) and DAPI (blue) staining of PTEN− (M233, WM9, WM793, 1205Lu) and PTEN+ (WM35, WM164, WM983A) cells following PLX4720 treatment (3μM, 48 hrs). (D) Western blot showing BAD phosphorylation following treatment with PLX4720 (0-48 hrs) in PTEN− (WM793, 1205Lu) and PTEN+ WM164. Annexin V binding following treatment with 3 or 10μM PLX4720 (48 hrs) showing increased apoptosis in WM793 stably overexpressing WT BAD.
PTEN is required for efficient BIM upregulation following BRAF inhibition
We next explored the link between PTEN expression status and PLX4720-mediated induction of BIM. siRNA knockdown of PTEN using two siRNA sequences led to the inhibition of PLX4720-induced BIM expression in PTEN+ cells (Figure 5A). We next determined whether re-introduction of wild-type PTEN (PTEN-wt) or lipid phosphatase mutated PTEN (PTEN-G129E) into a PTEN− cell line enhanced BIM expression when BRAF was inhibited. In these studies we used an isogenic pair of WM793 melanoma cell lines that expressed either doxycycline inducible PTEN-wt or PTEN-G129E mutant. Control studies showed that doxycyline (100 ng/ml, 48 hr) increased expression of PTEN in both cell lines (Figure 5B). The impaired lipid phosphatase function of the G129E mutant was confirmed by the fact that only the induction of PTEN-wt suppressed pAKT activation (Figure 5B). The role of PTEN in the PLX4720-mediated induction of BIM was confirmed by the enhanced expression of BIM seen when PTEN-wt was induced compared to when PTEN-G129E was induced (Figure 5B) and was paralleled by a significant increase in PLX4720-mediated apoptosis (P<0.05) (not shown). Interestingly, the addition of PLX4720 decreased the expression of PTEN through mechanisms that are not currently clear. The effects of PI3K/AKT signaling upon the suppression of BIM were mostly mediated through AKT3, with siRNA knockdown of AKT3 found to increase BIM expression when BRAF was inhibited (Supplemental Figure 10). As a final test of the relevance of BIM induction in the PLX4720-induced apoptotic response we showed that siRNA knockdown of BIM led to an impairment of PLX4720 (3 and 10μM) induced apoptosis (Figure 5D).
Figure 5. PTEN is required for efficient upregulation of BIM following BRAF inhibition.
(A) WM164 cells (PTEN+) were incubated with non-targeting siRNA (NT) or transfected with two PTEN specific siRNA’s (I and II) before treatment with PLX4720 (3 or 10μM, 48hrs). Protein was resolved and probed for expression of BIM, GAPDH and PTEN. (B) Induction of PTEN-wt but not PTEN-G129E in WM793 (PTEN−) cells was sufficient to increase BIM expression when BRAF was inhibited. Left panel: Western blot shows induction of PTEN− wt and PTEN−G129E following doxycycline treatment. Right panel: Induction of PTEN-wt + PLX4720 induces BIM more efficiently than PTEN-G129E + PLX4720. (C) BIM is required for PLX4720-induced apoptosis in PTEN+ cells. WM164 and WM983A cells were incubated with non-targeting siRNA (NT) or transfected with two BIM specific siRNA’s (BIM I and BIM II) prior to treatment with PLX4720 (3 or 10μM, 48 hrs). Flow cytometry studies showed a significant reduction in TMRM loss and Annexin V binding when cells were transfected with BIM siRNA compared to non-targeting control siRNA (*P<0.05).
Dual BRAF/PI3K inhibition enhances BIM expression and apoptosis in PTEN− cells
One of the major effects of PTEN is to limit PIP3 levels through its lipid phosphatase activity. We next treated PTEN− cell lines with a PI3K inhibitor (GDC-0941, 3μM or LY294002 10μM), PLX4720 (3 and 10μM), or the two drugs in combination, and showed that combined PI3K and BRAF inhibition increased the level of BIM expression in both Western blot and immunofluorescence studies (Figure 6A). Both the MAPK and PI3K/AKT pathways are known to regulate BIM RNA expression levels through the transcription factor FOXO3a (22-25). In agreement with this, PLX4720 treatment increased the nuclear accumulation of FOXO3a in the PTEN+ but not PTEN− melanoma cells (Figure 6B and not shown). Consistent with a role for increased AKT signaling suppressing BIM expression in PTEN− cells, dual BRAF and PI3K inhibition increased nuclear FOXO3a localization in the PTEN− cell lines (Figure 6B) and enhanced the level of BIM mRNA (Figure 6B). siRNA knockdown of FOXO3a was further found to block PLX4720-mediated upregulation of BIM in PTEN+ cells (Supplemental Figure 11). The observation that PLX4720 treatment led to increased PI3K/AKT signaling in PTEN− melanoma cell lines suggested that dual BRAF/PI3K inhibition could be one strategy to overcome intrinsic resistance. In agreement with this the combination of PLX4720 with the PI3K inhibitor GDC-0941 significantly enhanced the levels of apoptosis observed in PTEN− melanoma cell lines compared to either the BRAF or PI3K inhibitor alone (Figure 6C). Similar results were also observed in a 3D spheroid assay, where combined PLX4720 (3μM) and LY294002 (10μM) treatment prevented the recovery of cell growth observed when melanoma spheroids were treated with either drug alone (Figure 6D). The proposed mechanism for BIM regulation following BRAF inhibition in PTEN+ and PTEN− melanoma cell lines is shown in Supplemental Figure 12.
Figure 6. Dual PI3K/BRAF inhibition upregulates BIM and enhances apoptosis in PTEN− cells.
(A) (left) Western blot of 1205Lu cells treated with PLX4720 (3μM, 48 hrs), the PI3K inhibitor GDC-0941 (3μM, 48 hrs) or both drugs in combination (P+G). Right panel: Immunofluorescence staining of BIM (green) and DAPI (blue) in PTEN− cells following PLX4720 treatment (3μM, 48 hrs) the PI3K inhibitor LY294002 (10μM, 48 hrs) or both drugs in combination (PLX+LY). (B) Left panel: Immunofluorescence staining of PTEN−1205Lu following combined inhibition (3μM PLX4720 + 10μM LY294002, 48 hrs) increases nuclear localization of FOXO3a (green). DAPI is shown in blue. Magnification:X40. Right panel: Combined inhibition (3μM PLX4720 + 10μM LY294002, 48 hrs) increases PTEN− WM793 BIM mRNA levels to those observed with single BRAF inhibition (3μM PLX4720, 48 hrs) in the PTEN+ WM35. (C) PTEN− cells were treated with PLX4720 (3μM, 48 hrs), GDC-0941 (3μM, 48 hrs) or a combination of the two drugs (3P+3G) before Annexin-V staining was analyzed by flow cytometry (*indicates P<0.05 between the drug combination and each inhibitor alone). (D) Combined BRAF/PI3K inhibitor treatment blocks the escape of 1205Lu cells (PTEN−) from therapy. Spheroids of 1205Lu cells were treated with either PLX4720 alone (3 and 10μM: data shows 3μM), LY294002 (10μM) alone or a combination of the two drugs for 72hrs. In other studies, spheroids were treated with drugs for 72hrs and then allowed to recover for 120hrs. Micrograph shows viability staining (green=live cells, red=dead cells). Magnification x10.
Discussion
The current study has focused upon the mechanisms underlying the intrinsic resistance observed in melanoma patients recently treated in the phase-I trial of PLX4032 (4). Melanomas are known to have constitutive activity in many signaling pathways whose outputs converge to regulate cell cycle entry and survival. Of these, melanoma initiation and progression is known to be dependent upon both the Ras/Raf/MEK/ERK and PI3K/AKT pathways (12, 26-28). The mechanisms underlying this signaling activity differ according to the initiating oncogenic event. Thus melanomas with activating NRAS mutations rarely harbor concurrent alterations in either BRAF or PTEN/AKT as Ras stimulates both the Raf/MEK/ERK and PI3K/AKT pathways (29, 30). In contrast, melanomas with BRAF mutations require other mechanisms to activate their PI3K/AKT signaling and frequently show inactivation/deletion of PTEN or increased expression of AKT3 (31-34).
We began by investigating PTEN expression across a large sample of melanocytic lesions and found that PTEN was lost in 10-27% of melanomas. Although PTEN loss overlapped with the level of pAKT staining it was not always well correlated, agreeing with previous observations that other mechanisms may underlie the increased AKT activation associated with melanoma progression (35). Our results agree with other published studies on smaller numbers of melanoma samples (n=16-39), and confirm that reduced PTEN expression is a significant oncogenic event for a restricted subgroup of melanomas (31, 32, 36). Although PTEN was retained in non-atypical nevi, a significant number (23%) of atypical nevi lacked expression, suggesting this to be an early event in melanoma development. This idea is supported by recent mouse modeling studies showing that the conditional expression of the BRAF V600E mutation leads to melanoma development only when PTEN is suppressed (28).
Although lack of PTEN expression did not predict for sensitivity of BRAF V600E mutated melanoma cell lines to the growth inhibitory effects of PLX4720, there were significant differences in PLX4720-mediated apoptosis between PTEN+ and PTEN− melanoma cell lines. Initially, we hypothesized that PTEN− melanoma cell lines would show higher levels of AKT activity and that this would mediate resistance to PLX4720. Instead, we observed that drug treatment increased AKT signaling in the PTEN−cell lines. The effects upon AKT signaling were PTEN dependent, and could be recapitulated in PTEN+ melanoma cell lines when PTEN was knocked down using siRNA. The increase in AKT signaling observed in the PTEN− cell line panel was associated with PDK1 phosphorylation and increased expression of IGF-I. These effects were reversed following pre- treatment with the IGF1R inhibitor NVD-ADW-742 (21) suggesting a link between BRAF inhibition and enhanced IGF1R-mediated PI3K signaling. Similar findings, linking BRAF/MEK inhibition to increased IGF signaling, have been recently reported by two other groups (37, 38).
AKT plays a critical role in cancer development through its ability to regulate cell survival through the direct phosphorylation of BAD, the stimulation of ribosomal S6 kinase signaling, the inhibition of FOXO signaling and the inhibition of glycogen synthase 3-kinase (35). To determine the mechanism of PLX4720-induced apoptosis induction in the PTEN+ melanoma cell lines, LC-MRM analysis was used to quantify the relative expression of members of the Bcl-2 protein family (20). For the majority of proteins examined, PLX4720 treatment was associated with very similar dynamics in both the PTEN+ and PTEN− cell lines. These findings agree with previous studies and show that BRAF inhibition leads to an increase in the expression in the pro-apoptotic protein BIM (18, 39, 40). In contrast to these studies, which did not distinguish between PTEN+ and PTEN− cell lines, the LC-MRM analysis allowed us to identify significant PTEN− dependent differences in the level of PLX4720-induced BIM expression. BIM is a pro-apoptotic BH3-only member of the Bcl-2 protein family that exists in three major splice forms; extra long (BIM-EL), long (BIM-L) and short (BIM-S) (41, 42). It exerts its cytotoxic activity by binding to and antagonizing the anti-apoptotic proteins Bcl-2, Bcl-w, Bcl-XL and Mcl-1 (41, 42). Expression of BIM is regulated both transcriptionally and post-transcriptionally by a number of signaling pathways, including BRAF/MEK/ERK, JNK, p38 MAPK and PI3K/AKT (39, 41, 43, 44). In melanoma, the BRAF V600E mutation regulates BIM expression through the MEK/ERK pathway-mediated phosphorylation of the extra-long form of BIM (BIM-EL) at Serine 69, leading to its subsequent degradation by the proteasome (39, 45). Our study is the first to demonstrate that the level of BIM expression following BRAF inhibition is also determined by PTEN status and that the differing levels of BIM induction can determine the extent of apoptosis induction when BRAF is inhibited. Apoptosis control in melanoma cells is complex and increased AKT signaling is likely to regulate survival at multiple levels. One of the best known pro-survival substrates of AKT is the cell death inducing molecule BAD. AKT inactivates BAD via phosphorylation at Ser99, which prevents it’s binding to Bax and relieves the antagonism of Bax on Bcl-2 and Bcl-XL (18). A role for Bad inactivation in the escape of PTEN− cells from PLX4720-induced apoptosis was suggested by the preferential inactivation of BAD when BRAF was inhibited and the fact that overexpression of BAD sensitized the same cell line to PLX4720-induced apoptosis. Another candidate pro-apoptotic factor upregulated in melanoma cells following BRAF/MEK/ERK inhibition is BMF (Bcl-2 modifying factor) (40, 46). BMF, which is also regulated through the PI3K/AKT pathway, mediates its apoptotic effects through binding to Mcl-1. Although it is possible that BMF may also be differentially regulated in PTEN+/− cells, we, like other groups, were unable to confirm the selectivity of commercially available BMF antibodies (40, 47).
In addition to regulating PIP3 levels in the cytoplasm through its lipid phosphatase function, PTEN also localizes to the nucleus where it exerts its tumor suppressor function through lipid phosphatase-independent effects upon the regulation of chromosomal integrity, p53 acetylation and the expression of cyclin D1 (48). As the lipid phosphatase-dependent and -independent functions of PTEN are likely to be very different, we re-expressed either wild-type PTEN or a PTEN mutant with impaired lipid phosphatase function (G129E) in melanoma cells that were PTEN− (49). These studies confirmed the requirement for the lipid phosphatase function of PTEN in the suppression of BIM expression, with PLX4720 treatment inducing only a weak upregulation of BIM protein when PTEN G129E was expressed. The importance of the lipid phosphatase function in the suppression of BIM expression was supported by experiments showing that combined BRAF/PI3K inhibition and siRNA knockdown of AKT3 both enhanced the level of BIM expression and increased the level of apoptosis in the PTEN− cells. In other systems, AKT downregulates BIM expression by phosphorylating and inactivating the transcription factor FOXO3a (22, 24). In agreement with these reports, we confirmed that PLX4720 treatment led to increased phosphorylation of FOXO3a in the PTEN+ cells only and demonstrated that siRNA knockdown of FOXO3a abrogated the increase in BIM expression.
In summary, we have identified an important role for PTEN loss in the intrinsic resistance of BRAF V600E mutated melanoma cells to the BRAF inhibitor PLX4720. These studies further suggest that increased BIM expression may be a useful biomarker in predicting clinical response to BRAF inhibition and demonstrates that LC-MRM is a useful method for monitoring BIM expression that could be translated to patient assessment. This work also provides a rationale for dual BRAF/PI3K inhibitor treatment in the management of melanomas that are BRAFV600E/PTEN−.
Supplementary Material
Acknowledgements
We thank Gideon Bollag (Plexxikon) for the PLX4720. KSMS was supported by The Melanoma Research Foundation, The Bankhead-Coley Research Program (09BN-14), The American Cancer Society (#93-032-13) and the NIH/National Cancer Institute (U54 CA143970-01). The Moffitt Proteomics Facility is supported by the US Army Medical Research and Materiel Command (DAMD17-02-2-0051), the NIH/NCI (P30-CA076292) and the Bankhead-Coley Research program (06BS-02-9614).
References
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Dhomen N, Marais R. BRAF signaling and targeted therapies in melanoma. Hematology/oncology clinics of North America. 2009;23:529–45. ix. doi: 10.1016/j.hoc.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Smalley KS, Nathanson KL, Flaherty KT. Genetic subgrouping of melanoma reveals new opportunities for targeted therapy. Cancer Res. 2009;69:3241–4. doi: 10.1158/0008-5472.CAN-08-4305. [DOI] [PubMed] [Google Scholar]
- 4.Flaherty KT, Puzanov I, Kim KB, Ribas A, MacArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010 doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King AJ, Patrick DR, Batorsky RS, Ho ML, Do HT, Zhang SY, et al. Demonstration of a genetic therapeutic index for tumors expressing oncogenic BRAF by the kinase inhibitor SB-590885. Cancer Res. 2006;66:11100–5. doi: 10.1158/0008-5472.CAN-06-2554. [DOI] [PubMed] [Google Scholar]
- 8.Paraiso KH, Fedorenko IV, Cantini LP, Munko AC, Hall M, Sondak VK, et al. Recovery of phospho-ERK activity allows melanoma cells to escape from BRAF inhibitor therapy. Br J Cancer. 2010;102:1724–30. doi: 10.1038/sj.bjc.6605714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smalley KSM, Sondak VK. Melanoma - an unlikely poster child for targeted therapy. N Engl J Med. 2010;363:876–8. doi: 10.1056/NEJMe1005370. [DOI] [PubMed] [Google Scholar]
- 10.Smalley KS, Lioni M, Palma MD, Xiao M, Desai B, Egyhazi S, et al. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas. Mol Cancer Ther. 2008;7:2876–83. doi: 10.1158/1535-7163.MCT-08-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazar V, Ecsedi S, Szollosi AG, Toth R, Vizkeleti L, Rakosy Z, et al. Characterization of candidate gene copy number alterations in the 11q13 region along with BRAF and NRAS mutations in human melanoma. Mod Pathol. 2009;22:1367–78. doi: 10.1038/modpathol.2009.109. [DOI] [PubMed] [Google Scholar]
- 12.Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006;5:1136–44. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 13.Tran MA, Gowda R, Sharma A, Park EJ, Adair J, Kester M, et al. Targeting V600EB-Raf and Akt3 using nanoliposomal-small interfering RNA inhibits cutaneous melanocytic lesion development. Cancer Res. 2008;68:7638–49. doi: 10.1158/0008-5472.CAN-07-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smalley KS, Contractor R, Haass NK, Lee JT, Nathanson KL, Medina CA, et al. Ki67 expression levels are a better marker of reduced melanoma growth following MEK inhibitor treatment than phospho-ERK levels. Br J Cancer. 2007;96:445–9. doi: 10.1038/sj.bjc.6603596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sondergaard JN, Nazarian R, Wang Q, Guo D, Hsueh T, Mok S, et al. Differential sensitivity of melanoma cell lines with BRAFV600E mutation to the specific Raf inhibitor PLX4032. J Transl Med. 2010;8:39. doi: 10.1186/1479-5876-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaswamy S, Nakamura N, Vazquez F, Batt DB, Perera S, Roberts TM, et al. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci USA. 1999;96:2110–5. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boisvert-Adamo K, Aplin AE. Mutant B-RAF mediates resistance to anoikis via Bad and Bim. Oncogene. 2008;27:3301–12. doi: 10.1038/sj.onc.1211003. [DOI] [PubMed] [Google Scholar]
- 19.Smalley KS, Contractor R, Haass NK, Kulp AN, Atilla-Gokcumen GE, Williams DS, et al. An organometallic protein kinase inhibitor pharmacologically activates p53 and induces apoptosis in human melanoma cells. Cancer Res. 2007;67:209–17. doi: 10.1158/0008-5472.CAN-06-1538. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Gruidl M, Remily-Wood E, Liu RZ, Eschrich S, Lloyd M, et al. Quantification of beta-catenin signaling components in colon cancer cell lines, tissue sections, and microdissected tumor cells using reaction monitoring mass spectrometry. J Proteome Res. 2010;9:4215–27. doi: 10.1021/pr1005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Akiyama M, et al. Inhibition of the insulin-like growth factor receptor-1 tyrosine kinase activity as a therapeutic strategy for multiple myeloma, other hematologic malignancies, and solid tumors. Cancer Cell. 2004;5:221–30. doi: 10.1016/s1535-6108(04)00050-9. [DOI] [PubMed] [Google Scholar]
- 22.Yang JY, Chang CJ, Xia W, Wang Y, Wong KK, Engelman JA, et al. Activation of FOXO3a is sufficient to reverse mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitor chemoresistance in human cancer. Cancer Res. 2010;70:4709–18. doi: 10.1158/0008-5472.CAN-09-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunters A, Madureira PA, Pomeranz KM, Aubert M, Brosens JJ, Cook SJ, et al. Paclitaxel-induced nuclear translocation of FOXO3a in breast cancer cells is mediated by c-Jun NH2-terminal kinase and Akt. Cancer Res. 2006;66:212–20. doi: 10.1158/0008-5472.CAN-05-1997. [DOI] [PubMed] [Google Scholar]
- 24.Sunters A, Fernandez de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003;278:49795–805. doi: 10.1074/jbc.M309523200. [DOI] [PubMed] [Google Scholar]
- 25.Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–48. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smalley KS. Understanding melanoma signaling networks as the basis for molecular targeted therapy. J Invest Dermatol. 2010;130:28–37. doi: 10.1038/jid.2009.177. [DOI] [PubMed] [Google Scholar]
- 27.Cheung M, Sharma A, Madhunapantula SV, Robertson GP. Akt3 and mutant V600E B-Raf cooperate to promote early melanoma development. Cancer Res. 2008;68:3429–39. doi: 10.1158/0008-5472.CAN-07-5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009 doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Research. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 30.Cully M, Downward J. SnapShot: Ras Signaling. Cell. 2008;133:1292–e1. doi: 10.1016/j.cell.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 31.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004;122:337–41. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsao H, Zhang X, Fowlkes K, Haluska FG. Relative reciprocity of NRAS and PTEN/MMAC1 alterations in cutaneous melanoma cell lines. Cancer Res. 2000;60:1800–4. [PubMed] [Google Scholar]
- 33.Davies MA, Stemke-Hale K, Tellez C, Calderone TL, Deng W, Prieto VG, et al. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer. 2008;99:1265–8. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64:7002–10. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 35.Madhunapantula SV, Robertson GP. The PTEN-AKT3 signaling cascade as a therapeutic target in melanoma. Pigment Cell Melanoma Res. 2009;22:400–19. doi: 10.1111/j.1755-148X.2009.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsao H, Mihm MC, Jr., Sheehan C. PTEN expression in normal skin, acquired melanocytic nevi, and cutaneous melanoma. J Am Acad Dermatol. 2003;49:865–72. doi: 10.1016/s0190-9622(03)02473-3. [DOI] [PubMed] [Google Scholar]
- 37.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gopal YN, Deng W, Woodman SE, Komurov K, Ram P, Smith PD, et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res. 2010;70:8736–47. doi: 10.1158/0008-5472.CAN-10-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cartlidge RA, Thomas GR, Cagnol S, Jong KA, Molton SA, Finch AJ, et al. Oncogenic BRAF(V600E) inhibits BIM expression to promote melanoma cell survival. Pigment Cell Melanoma Res. 2008;21:534–44. doi: 10.1111/j.1755-148X.2008.00491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao Y, Aplin AE. Akt3-mediated resistance to apoptosis in B-RAF-targeted melanoma cells. Cancer Res. 2010;70:6670–81. doi: 10.1158/0008-5472.CAN-09-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Connor L, Strasser A, O’Reilly LA, Hausmann G, Adams JM, Cory S, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. Embo J. 1998;17:384–95. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu SY, Lin P, Hsueh AJ. BOD (Bcl-2-related ovarian death gene) is an ovarian BH3 domain-containing proapoptotic Bcl-2 protein capable of dimerization with diverse antiapoptotic Bcl-2 members. Molecular endocrinology (Baltimore, Md. 1998;12:1432–40. doi: 10.1210/mend.12.9.0166. [DOI] [PubMed] [Google Scholar]
- 43.Cai B, Chang SH, Becker EB, Bonni A, Xia Z. p38 MAP kinase mediates apoptosis through phosphorylation of BimEL at Ser-65. J Biol Chem. 2006;281:25215–22. doi: 10.1074/jbc.M512627200. [DOI] [PubMed] [Google Scholar]
- 44.Ley R, Ewings KE, Hadfield K, Cook SJ. Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ. 2005;12:1008–14. doi: 10.1038/sj.cdd.4401688. [DOI] [PubMed] [Google Scholar]
- 45.Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein. Bim. J Biol Chem. 2003;278:18811–6. doi: 10.1074/jbc.M301010200. [DOI] [PubMed] [Google Scholar]
- 46.VanBrocklin MW, Verhaegen M, Soengas MS, Holmen SL. Mitogen-activated protein kinase inhibition induces translocation of Bmf to promote apoptosis in melanoma. Cancer Res. 2009;69:1985–94. doi: 10.1158/0008-5472.CAN-08-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmelzle T, Mailleux AA, Overholtzer M, Carroll JS, Solimini NL, Lightcap ES, et al. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proc Natl Acad Sci U S A. 2007;104:3787–92. doi: 10.1073/pnas.0700115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–14. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 49.Ramaswamy S, Nakamura N, Vazquez F, Batt DB, Perera S, Roberts TM, et al. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci U S A. 1999;96:2110–5. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.