Abstract
Helicobacter pylori is a very successful human-specific bacterium worldwide. Infections of the stomach with this pathogen can induce pathologies including chronic gastritis, peptic ulcers and even gastric cancer. Highly virulent H. pylori strains encode the cag (cytotoxin-associated genes) pathogenicity island which expresses a type IV secretion system (T4SS). This T4SS forms a syringe-like pilus structure for the injection of virulence factors such as the CagA effector protein into host target cells. This is achieved by a number of T4SS proteins including CagI, CagL, CagY and CagA which by itself bind the host cell integrin member β1 followed by delivery of CagA across the host cell membrane. A role of CagA interaction with phosphatidylserine has also been shown to be important for the injection process. After delivery, CagA becomes phosphorylated by oncogenic tyrosine kinases and mimics a host cell factor for the activation or inactivation of some specific intracellular signaling pathways. Here we review recent progress in characterizing CagA-dependent and CagA-independent signalling capabilities of the T4SS which include the induction of membrane dynamics, disruption of cell-to-cell junctions, actin-cytoskeletal rearrangements as well as pro-inflammatory, cell cycle-related and anti-apoptotic transcriptional responses. The contribution of these signalling pathways to pathogenesis during H. pylori infections is discussed.
Keywords: Helicobacter pylori, signalling, type IV secretion, VirB5, VirB10
Introduction
Helicobacter pylori colonizes the surface area of the gastric mucosa in the human stomach and is one of the most adapted microbial pathogens. About 50% of the world's population carries this bacterium, causing chronic, often asymptomatic gastritis in all infected humans, and more severe gastric diseases in up to 10–15% of infected people depending on the geographical region [1-3]. Infections commonly occur in early childhood and, if not treated by antimicrobial therapy, H. pylori can persist lifelong. Although H. pylori infections are often diagnosed with a pronounced cellular inflammation status, which is triggered by the host innate and adaptive immune systems, the bacteria are commonly not eliminated. Numerous mechanisms of immune evasion have been reported and H. pylori became a paradigm for chronic infections. Studies of H. pylori have revealed not only its ability to colonize individual hosts for many decades, but also that this bacterium has co-existed with humans for a very long period through history. Genetic studies indicate that H. pylori spread during human migrations from east Africa more than 60,000 years ago [4]. Based on this long time of co-evolution, there are some indications that colonization by H. pylori could have been beneficial for their human carriers and this probably provided selective advantages [3]. In the modern world, however, H. pylori infections can cause a heavy burden of morbidity and mortality as a consequence of peptic ulcer disease, mucosa-associated lymphoid tissue (MALT) lymphoma and, the most dangerous complication, gastric adenocarcinoma [1-3]. Gastric adenocarcinoma is the second leading cause of cancer-related death in the world and approximately 649,000 people die from this malignancy each year [1].
The clinical outcome of H. pylori infections is determined by highly complex host-pathogen interactions. Disease progression is constrained by various parameters such as the bacterial genotype, environmental determinants and genetic predisposition of the host. For example, specific polymorphisms in human genes with crucial functions in immunoregulatory and pro-inflammatory signaling such as interleukin-1β (IL-1β), Nod (nucleotide oligomerization domain), tumour necrosis factor alpha (TNF-α) or interleukin-8 (IL-8) have been associated with an increased risk of developing disease including gastric cancer as summarised in excellent review articles [1-3,5; for more details see Glossary file Doc. S1 in the Supplementary Material]. During the last two decades, the cellular and molecular mechanisms acquired by H. pylori to undermine host defences have been investigated intensively (Fig. 1). H. pylori isolates are surprisingly diverse both in their genome sequences and pathogenicity. Dozens of bacterial factors have been identified to influence the pathogenesis of H. pylori. There are two classical secreted virulence factors present in H. pylori, the vacuolating cytotoxin (VacA) and the CagA protein encoded by the cag (cytotoxin-associated genes) pathogenicity island (cagPAI). VacA interacts with numerous host surface receptor molecules and can trigger various responses including pore insertion into the cell membrane, modification of endo-lysosomal functions, cell vacuolation, apoptosis and immune inhibition [1-3]. Much research interest worldwide is also focused on CagA which is encoded by more virulent strains but is typically missing in less virulent H. pylori isolates. Thus, the protein has been recognized as a marker for the cagPAI locus [6,7]. Other well-known pathogenicity-associated processes include flagella-driven H. pylori motility, urease-triggered neutralization of pH, several adhesins (BabA/B, SabA, AlpA/B, HopZ, OipA and others) mediating binding of H. pylori to gastric epithelial cells, glycosylation of cholesterol by HP0421, cleavage of E-Cadherin-triggered opening of cell-to-cell junctions by the protease HtrA, downregulation of antimicrobial nitric oxide production by arginase RocF as well as γ-glutamyl transpeptidase (GGT) which inhibits T-cell proliferation and others as summarised in Fig. 1 [1-3,5,8]. In addition, H. pylori induces a pronounced pro-inflammatory phenotype in infected gastric epithelial cells by multiple signalling activities that stimulate the transcription factors NF-κB and/or AP-1 [5,9]. There are also numerous other reported marker genes for H. pylori-induced disease development (e.g. iceA and dupA), but their biological function is widely unclear. Here we review the various cagPAI functions and multiple host cell signalling pathways with emphasis on their potential role in the pathogenesis of H. pylori.
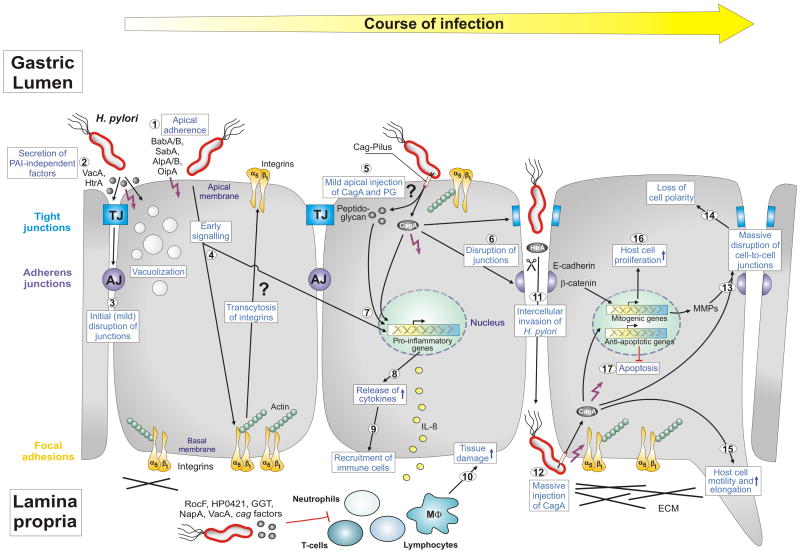
Figure 1.
Model of Helicobacter pylori-induced epithelial-barrier disruption and pathogenesis. The interplay between polarized gastric epithelial cells and a variety of bacterial pathogenicity factors modulates multiple host responses during the course of infection, as indicated. H. pylori expresses several adhesins such as BabA, BabB, SabA, AlpA and AlpB as well as OipA which mediate apical binding of the bacteria (1). Attached H. pylori or those in the mucus secrete virulence factors into the medium (HtrA protease, VacA cytotoxin and others) (2), which could trigger mild opening of tight junctions (TJs) and adherens junctions (AJs) at early time points of infection (3). While internalized VacA causes cellular vacuolization, a hallmark of the ulceration process, HtrA cleaves the junctional protein E-cadherin [8]. Another intriguing possibility of junction disruption could be the transcytosis of basal integrins to the apical surface by early, but unknown, cagPAI-independent signaling (4). Apical exposure of some integrin molecules such as integrin α5β1 could stimulate the T4SS pilus to inject CagA and peptidoglycan into cells (5). Injected CagA can then be targeted to TJs and AJs followed by further disruption of these junctions (6). Injected CagA and peptidoglycan (5) in addition to OipA (1) can trigger nuclear factor-kB (NF-κB) activation (7) and the release of proinflammatory cytokines such as IL-8 (8). These cytokines can alter the secretion of mucus and induce changes in gastric-acid secretion and homeostasis. They also attract immune cells to infiltrate from the blood stream into the gastric mucosa (9), where they cause substantial tissue damage at the site of infection (10). H. pylori also express numerous factors to suppress immune cell functions as indicated. The result of the above described processes is local epithelial disruption enabling some H. pylori of entering the intercellular space between adjacent cells and reaching the basal membranes (11). In this manner, the bacteria could access integrins and inject CagA (12). Injected CagA can then induce the massive disruption of cell junctions (13) and loss of cell polarity (14). The induction of metalloproteases (MMPs) might enhance this effect in addition to HtrA. Finally, CagA can induce multiple pathways to trigger host-cell motility and elongation (15) and the onset of mitogenic genes and cell proliferation (16), and it can inhibit apoptosis (17). The interplay of each of these pathways could result in substantial deregulation and oncogenic transformation of gastric epithelial cells in vivo and, thus, are important for H. pylori pathogenesis. Specific steps labeled with question marks are untested or speculative aspects of the model. Other specific abbreviations used: α5β1 (chains of the integrin receptor), cag (cytotoxin-associated genes), ECM (extracellular matrix), IL-8 (interleukin 8), HP0421 (cholesterol-alpha-glucosyltransferase), MΦ (macrophage), NapA (neutrophil-activating protein A), PAI (pathogenicity island), PG (peptidoglycan), RocF (arginase enzyme). For more details see text and glossary in the Supplementary Material (Doc. S1). This model was updated from Wessler and Backert [15] with kind permission from Blackwell Publishing.
The cag pathogenicity island encodes a type IV secretion system: two pilus assembly models
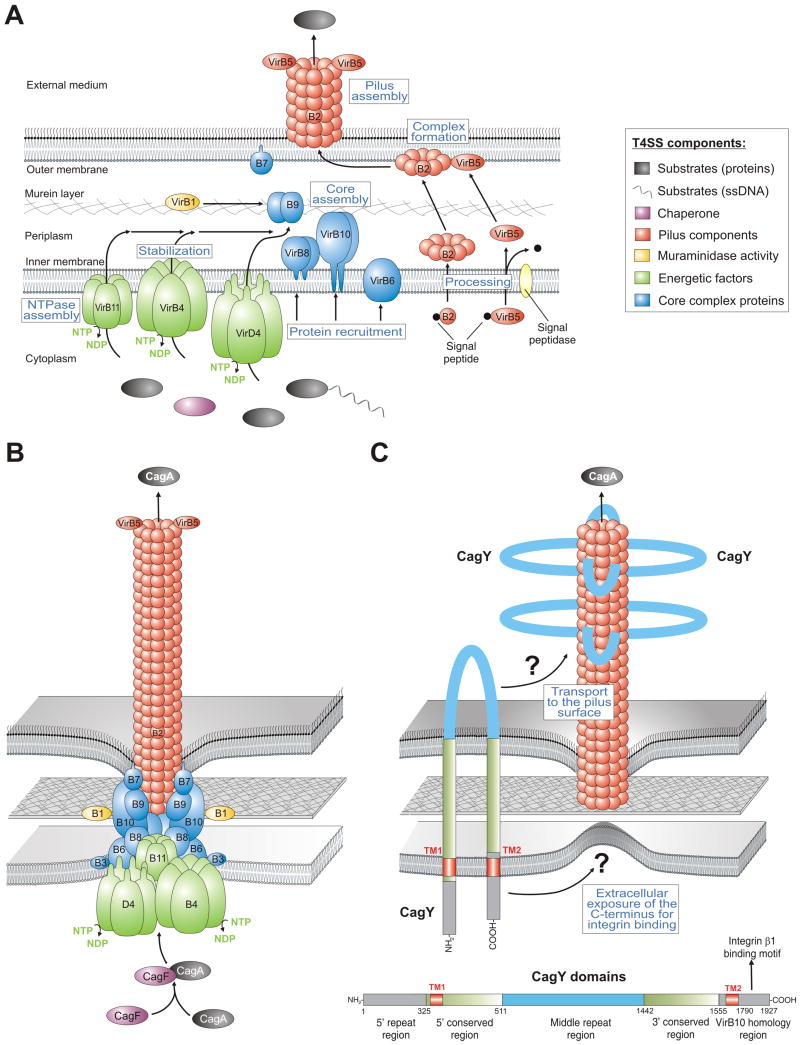
Intensive research in recent years has demonstrated that the cagPAI encodes functional components of a type IV secretion system (T4SS). This T4SS represents a needle-like structure (also called T4SS pilus) protruding from the bacterial surface and is induced by host cell contact to inject virulence factors [10,11]. T4SS transporters are commonly found in many Gram-negative bacteria and are evolutionary related to DNA conjugation machines [6]. The group of T4SSs is diverse both with respect to delivered substrates (DNA-protein complexes or proteins) and recipients, which can be either a bacterium of the same or other species, or organisms from a different kingdom (e.g. plants, fungi or mammalian cells). In addition to H. pylori, T4SSs have been found in Agrobacterium, Bordetella, Bartonella, Legionella, Anaplasma and other pathogens. T4SS transporters typically consist of 11 VirB proteins (encoded by virB1-virB11 genes) and the so-called coupling protein (the NTPase VirD4). The prototypic and best characterised T4SS is the VirB or T-DNA transfer system of Agrobacterium tumefaciens. The agrobacterial VirB proteins can be grouped into three categories: (i) the core components or putative channel (VirB6-10), (ii) the pilus-associated components (VirB2, and possibly VirB3 and VirB5) and (iii) the energetic components (the NTPases: VirB4 and VirB11). VirB1 is an enzyme with muraminidase activity possibly enabling localized lysis of murein to achieve T4SS assembling at a given location [6]. In Agrobacterium, signal peptidase-I removes signal peptides from precursors of the main pilus component VirB2 and the minor pilus component VirB5, followed by cyclization of VirB2 by an unknown factor. Processed VirB2 and VirB5 subsequently associate with the membranes as stabilised by VirB4 and VirB8. Stabilized and properly oriented VirB5 then forms a complex with VirB2, which is a key step in the formation of the T4SS pilus assembly subcomplex. A model for individual steps in the assembly of the agrobacterial T4SS is summarised in Fig. 2A. When looking at the H. pylori T4SS, all orthologs of the 11 VirB proteins and VirD4 have been identified to be encoded in the cagPAI as well as some accessory factors [4,6] leading to a T4SS model similar to that of Agrobacterium (Fig. 2B). In line with these conclusions, immunogold-labelling studies indicated that the tips of the T4SS pilus are decorated with CagL [11], a proposed VirB5 ortholog [6]. In a second model (Fig. 2C), it was proposed that the T4SS appendages in H. pylori are covered locally or completely by CagY (VirB10) [10] and the model includes all identified VirB components, except VirB5 [12]. Interestingly, CagY is a very large protein (about 250 kDa) that contains two transmembrane domains with the mid domain (also called the repeat domain) exposed to the extracellular space [10]. Thus, it is still not fully clear if the H. pylori T4SS pilus is more similar to that of Agrobacterium which is mainly composed of VirB2 subunits and VirB5 or if it is composed of CagY as major pilus subunit or if it is a mix of both (Fig. 2B/C).
Figure 2.
Models for the assembly and assembled structure of T4SSs in Agrobacterium tumefaciens and Helicobacter pylori. (A) Proposed assembly of the prototypical Agrobacterium VirB/VirD4 T4SS machinery is shown. The T4SS is a multi-component protein complex spanning the inner and outer membranes of Agrobacterium and other Gram-negative bacteria. Current knowledge of T4SS assembly, cellular localization of its components is shown in a simplified manner. The coupling protein VirD4 and structural components (VirB1-VirB11) are typically required for secretion and are depicted according to their proposed functions. A model for T-pilus assembly in Agrobacterium showing the proposed VirB4-VirB8-VirB5-VirB2 interaction sequence leading to the formation of VirB2-VirB5 complexes followed by T-pilus assembly. The assembled T4SS then triggers the secretion of substrates from the bacterial cytoplasm directly into the cytoplasm of infected host cells or into the extracellular milieu. (B) Model-1 for the assembled T4SS machinery in H. pylori assuming that all VirB1-11 proteins are encoded by the cagPAI and assemble in a similar fashion as proposed for A. tumefaciens [6]. The reported substrates for this T4SS are CagA and peptidoglycan. (C) Model-2 proposes that the T4SS requires basically the same VirB proteins as in panel B with one major difference. The pilus surface is proposed to be covered with CagY molecules. In contrast to VirB10 in many T4SSs, H. pylori VirB10 (CagY) is a very large protein (ca. 250 kDa, domain structure and amino acid positions shown for CagY of strain 26695, accession number NP_207323.1) carrying two transmembrane domains (TM1 and TM2) to form a hairpin-loop structure in the membranes as depicted [10]. Immunogold labelling against the loop region in CagY indicated that this part of the protein is exposed to the extracellular space and is transported to the pilus surface by a yet unknown mechanism [10]. However, recent data demonstrated that the very carboxy-terminus of CagY can bind to the host receptor integrin β1 [13]. How the latter domain can be exposed to the extracellular space in order to make contact with integrin β1 is not yet clear and must be clarified in future studies.
However, the only reported effector molecules injected by the H. pylori T4SS are peptidoglycan and CagA. Immunogold-stainings indicated the presence of CagA at the tips of T4SS pili, thus providing the first direct evidence that CagA might be delivered through these surface appendages, an observation which has not yet been reported for any other known T4SS effector protein in the bacterial world [11]. Investigation of the injection mechanism has shown that delivery of CagA requires a host cell receptor, the integrin member β1 [11,13] and phosphatidylserine [14]. Numerous structural T4SS components have been demonstrated to bind to integrin β1 including CagL [11], or CagA, CagI and CagY but excluding CagL [13]. However, while very little is known about CagA and CagI in the above context, CagL has been investigated in more detail. Like the human extracellular matrix protein fibronectin, CagL carries a RGD-motif shown to be important for interaction with integrin β1 in vitro and on host cells, as well as downstream signaling to activate the kinases FAK and Src [11], but mutation of the RGD-motif in CagL had no defect in T4SS functions such as phosphorylation of injected CagA during infection in another study [13]. These reports indicate that there is a controversy in the literature about the importance of the CagL RGD-motif in T4SS functions and host cell signaling. Another unsolved question is the proposed structure of CagY with respect to which domain is exposed to the extracellular space. The repeat domain in the middle of CagY has been shown to be accessible to labelling by antibodies made specifically against this subdomain [10]; however, yeast-two hybrid screens and other in vitro binding studies indicated that the very carboxy-terminus interacted with integrin β1 [13] which has been proposed to be cytoplasmic [10] as shown in Fig. 2C. Thus, the role of the CagL RGD-motif and CagY topology for injection of CagA is not yet solved. It is also unclear why the effector protein CagA itself can bind to the extracellular domain of integrin β1, because one would expect that such high binding affinity inhibits the injection process. However, these studies clearly showed that H. pylori targets integrin β1 as a receptor for the T4SS, and deletion of cagI, cagL and cagY genes disrupt T4SS functions almost completely [11,13]. Thus, each of these factors exhibits important functional roles but their concerted interaction activities are unknown.
However, since a receptor is involved in host recognition by the T4SS it can be proposed that CagA is not injected into target cells at random positions but rather in a tightly controlled manner [15]. Importantly, integrins are normally not found at the apical membrane but at the basal membrane of polarized gastric epithelial cells (Fig. 1). This suggests the existence of a sophisticated control mechanism by which H. pylori injects CagA [11]. Basically, there are two major injection models which can be considered. First, H. pylori could inject its T4SS effectors across the basolateral membrane (Fig. 1). A possible scenario is that early exposed cagPAI-independent factors such as the H. pylori adhesins as well as HtrA, VacA, OipA and others may loosen intercellular epithelial junctions at locally restricted areas before a limited number of bacteria may gain access to integrins and inject CagA. The basal injection model of CagA can also explain why H. pylori does not cause more gastric damage in infected individuals and may only inject virulence proteins into target cells under specific conditions in vivo. Second, cagPAI-independent signaling events might stimulate the transcytosis of integrin molecules from the basal to the apical side of the cells, a process that has been suggested for integrin β1 [15]. Indeed, disruption of host-cell polarity by another pathogen (enteropathogenic Escherichia coli) enabled basal membrane proteins to migrate apically. Transcytosis of integrins would therefore enable H. pylori an intriguing possibility to target the integrin β1 receptor at apical membranes (Fig. 1). The latter scenario would explain how H. pylori T4SS substrates might be injected apically, possibly in part, to further disrupt intercellular junctions or activate early signaling events leading to the induction of pro-inflammatory genes, respectively.
Functional studies of H. pylori using animal infection models and transgenic mice
Recent functional studies in animal models have provided compelling evidence for the importance of CagA and cagPAI in H. pylori pathogenesis. Mongolian gerbils (Meriones unguiculatus), several knockout and other mice (e.g. INS-GAS mice) and Rhesus monkeys have been shown to serve as useful in vivo models to study H. pylori-induced pathology. However, each animal model has distinct advantages and disadvantages and can, therefore, be only considered as complementary systems. The most extensively studied model with respect to host inflammatory and physiological responses to H. pylori is the Mongolian gerbil [1,16-18]. Mongolian gerbils have been shown to develop similar H. pylori-induced pathology as compared to humans. H. pylori reproducibly induces gastric inflammation in this system and cagPAI-positive as well as various H. pylori mutants colonize gerbils sufficiently well, which allow the examination of the role of bacterial virulence determinants in gastric injury. For example, gerbils were challenged by the cagPAI-positive strain TN2 and its isogenic mutants of cagE (virB4) or vacA for 62 weeks [16]. The wild-type and vacA mutants induced severe gastritis, whereas cagE mutants induced far milder changes. Gastric ulceration was induced at the highest rate (22/23) by wild-type H. pylori, followed by the vacA mutant (19/28). No ulcers were found in the gerbils infected with the cagE mutant (0/27) or in controls (0/27). Intestinal metaplasia was also found in the gerbils infected with the wild-type (14/23) or vacA mutant (15/28). Gastric cancer developed in one gerbil with wild-type infection and in one with vacA mutant infection [16]. These early data suggested that cagPAI-positive H. pylori can induce gastritis and gastric ulcer in gerbils with an important role of the T4SS. Further studies indicated that H. pylori strain B128 (also harbouring a functional cagPAI) increased plasma gastrin, a factor known to promote gastric epithelial hyperproliferation, but not infection with isogenic mutants lacking either cagA or cagY [17]. Enhanced corpus colonization with the parental wild-type strain was also evident. Virulence factors such as the cagPAI are therefore likely to impact on gastric physiological changes observed with H. pylori infection either directly, via permitting colonization of the corpus mucosa as a consequence of increased acid tolerance or indirectly, via promoting enhanced inflammation. Interestingly, infection of gerbils by H. pylori led to the development of inflammation-induced gastric adenocarcinoma in some but not all studies, highlighting the possible importance of different gerbil lines, diet, genetic differences between H. pylori strains and probably other parameters [1,17,18]. For example, gerbils infected with the cagPAI-positive strain 7.13, a gerbil-adapted derivative of B128, developed gastric dysplasia by only four weeks in 88% of gerbils, which was accompanied by adenocarcinoma in 25% of animals [18]. By eight weeks, gastric adenocarcinomas were present in 75% of infected gerbils that were sacrificed at this time-point and gastric adenocarcinomas were accompanied by severe lymphofollicular gastritis. Importantly, CagA and the T4SS played a crucial role in gastric cancer development of gerbils [18]. Consequently, further efforts have been put into identifying the mechanism of H. pylori -associated carcinogenesis. A first direct causal link between CagA and oncogenesis in vivo was identified by the generation of transgenic C57BL/6J mice expressing CagA in the absence of H. pylori [19]. CagA transgenic mice showed gastric epithelial hyperplasia and some mice developed gastric polyps and adenocarcinomas of the stomach and small intestine. Systemic expression of CagA further induced leukocytosis with IL-3/GM-CSF hypersensitivity and some mice developed myeloid leukemias and B cell lymphomas [19]. These results indicate that H. pylori can rapidly induce gastric adenocarcinoma in gerbils in a T4SS-dependent manner and that expression of CagA alone in transgenic mice is sufficient to induce severe malignant lesions. Therefore, it is clear that CagA and the T4SS play a central role in the pathogenesis of H. pylori in vivo.
H. pylori in vitro infection models: T4SS-dependent but CagA-independent cellular signalling
In addition to the above discussed in vivo models, the use of several in vitro cell culture systems has been very efficient for studying signalling cascades which are of relevance to H. pylori disease development. In particular, gastric epithelial and colonic cell lines (e.g. AGS, AZ-521, Caco2, HEp-2, KATO-III, MKN-28, MKN-45, NCI-N87 and others), primary gastric epithelial cells and professional phagocytes, including human polymorphonuclear leucocytes (PMNs) and human or murine macrophage cell lines (e.g. J774A.1, JoskM, RAW264.7, THP-1, U937 and others) have been utilized. In the following sections we will highlight some of these in vitro studies and begin with the T4SS-dependent but CagA-independent signaling pathways as summarised in Fig. 3A.
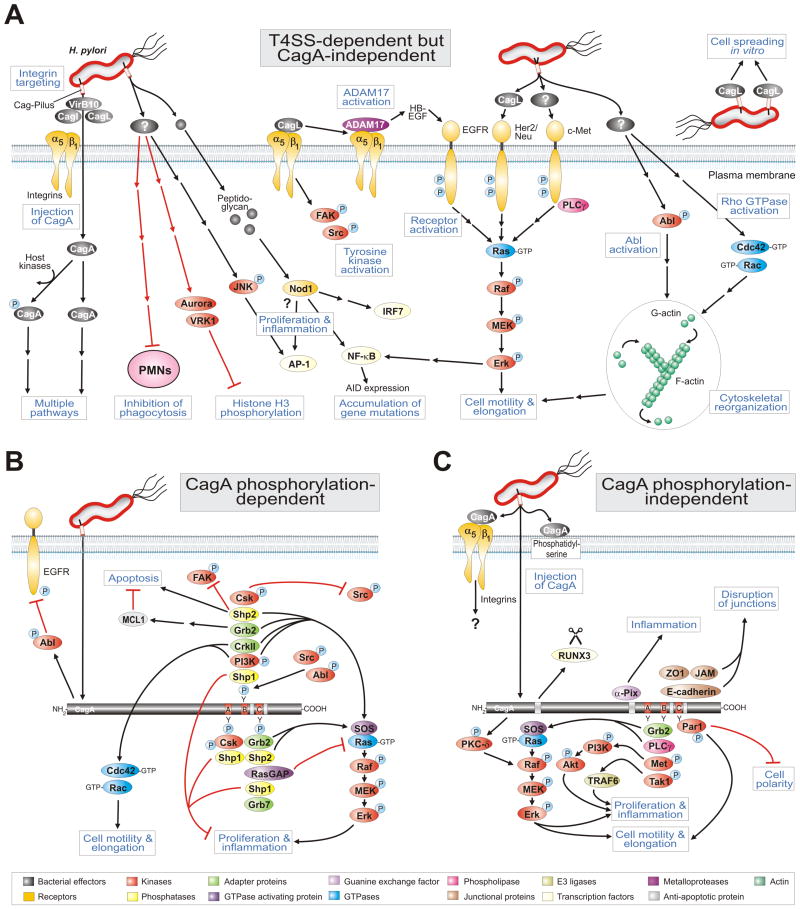
Figure 3.
Model for the role of H. pylori T4SS in host cell signaling processes which may effect pathogenesis. (A) The H. pylori T4SS pili are induced upon contact with host cells and can inject effector molecules such as the CagA protein and peptidoglycan in a manner dependent on integrin β1. Injected CagA can then induce cascades as depicted in the panels below. Panel A highlights a multitude of known T4SS-dependent but CagA-independent pathways involved in the activation of receptor and non-receptor tyrosine kinases, pro-inflammatory signalling, Rho GTPase activation, scattering and motility of gastric epithelial cells as well as suppression of histone phosphorylation and H. pylori phagocytosis by immune cells. Two particular T4SS factors have been reported to be involved in some but not all of these responses. The known signaling functions for injected peptidoglycan as well as pilus-exposed or recombinant CagL are shown. For numerous other pathways the actual T4SS factor is yet unknown as also indicated. (B) CagA phosphorylation-dependent and (C) phosphorylation-independent signal transduction events. CagA is injected into the host cell membrane of infected gastric epithelial cells which also requires phosphatidylserine. The tyrosine kinases Src and Abl phosphorylate injected CagA. CagA can then modulate various signaling cascades associated with cell polarity, cell proliferation, actin-cytoskeletal rearrangements, cell elongation, disruption of tight and adherens junctions, pro-inflammatory responses and suppression of apoptosis, as depicted. Black arrows indicate activated sigalling pathways and red arrows correspond to inactivated cascades. Panels B and C were updated from Backert et al. [26] with kind permission from WILEY. For specific abbreviations and terms used in this figure, please see text and glossary in the Supplementary Material (Doc. S1).
Very early experiments have shown that H. pylori can actively block its own uptake by professional phagocytes [20]. Vital H. pylori are necessary to block the phagocytic uptake, and H. pylori also abrogated the phagocytes' ability to engulf latex beads or adherent Neisseria gonorrhoeae bacteria as control. This antiphagocytic phenotype was dependent on a functional T4SS because isogenic virB7 and virB11 mutants abrogated this effect [20]. Interestingly, the actual factor involved was not CagA because isogenic cagA mutants also blocked phagocytosis. These data indicate that H. pylori expresses a yet unknown T4SS factor exhibiting antiphagocytic activity which may play an essential role in the immune escape of this persistent pathogen (Fig. 3A). However, the majority of studies were performed on the interaction of H. pylori with cultured gastric epithelial cells. For example, H. pylori was reported to change histone H3 phosphorylation by a T4SS-dependent but CagA-independent process (Fig. 3A). Infection with cagPAI-poitive H. pylori strains decreases H3 phosphorylation levels both at serine residue 10 and threonine residue 3 [21]. These observations were based on mitotic histone H3 kinases such as vaccinia-related kinase 1 (VRK1) and Aurora B which were not fully activated in infected cells, resulting in a transient H. pylori -induced pre-mitotic arrest [21]. Together, these results show that H. pylori subverts key cellular processes such as cell cycle progression by a yet unknown T4SS factor. In addition, the results of numerous reports indicated that structural components of the T4SS but not CagA itself were required for the induction of pro-inflammatory signaling including the activation of NF-κB and AP-1 (Fig. 3A). This suggested that the T4SS might inject factors in addition to CagA or that the T4SS itself triggers the effect. Despite intensive efforts including a systematic mutagenesis of all cagPAI genes, the hypothetical additional effector remained unknown for many years. Another possible candidate was H. pylori peptidoglycan which can be recognized by Nod1, an intracellular pathogen-recognition molecule [5]. These observations suggested that T4SS-dependent delivery of peptidoglycan is responsible for activation of Nod1→NF-κB-dependent pro-inflammatory responses such as secretion of IL-8 [5]. Interestingly, cagPAI-positive H. pylori can induce the NF-κB-dependent expression of AID (a DNA-editing enzyme) in host target cells, which resulted in the accumulation of mutations in the tumour suppressor protein TP53 [22]. Thus, induction of AID might be a mechanism whereby gene mutations could emerge during H. pylori-associated gastric carcinogenesis. However, the actual bacterial T4SS factor(s) and pathways which activate both transcription factors, NF-κB and AP-1, are highly controversial in the literature and still not fully clear [9].
Infection of gastric epithelial cells with H. pylori was also reported to profoundly activate numerous receptor tyrosine kinases (RTKs) in a T4SS-dependent fashion including the epidermal growth factor receptor EGFR [23,24], hepatocyte growth factor receptor c-Met [23] and Her2/Neu [23]. Studies on the downstream signalling indicated that each of these RTKs can activate the MAP kinase members MEK and Erk1/2 (Fig. 3A). However, while activation of EGFR has been shown to induce pro-inflammatory responses leading to the secretion of IL-8 [24], activation of c-Met (but not EGFR or Her2/Neu) was involved in cell scattering and motogenic responses of infected gastric epithelial cells [23]. Interestingly, the non-receptor tyrosine kinase c-Abl and the small Rho GTPases Rac1 and Cdc42 are also activated by a T4SS-dependent but CagA-independent mechanism and play a role in triggering the scattering and motility of infected gastric epithelial cells (Fig. 3A). However, the actual T4SS factor involved was also unclear for many years.
Recent in vitro studies showed a profound role of recombinant CagL in activation of EGFR, Her3/ErbB3, Src and Fak kinases in an RGD-dependent manner [25]. Investigation of the molecular mechanism how CagL can activate EGFR has demonstrated to involve ADAM17, a metalloprotease involved in catalyzing ectodomain shedding of receptor tyrosine kinase ligands. In non-stimulated cells, ADAM17 is normally in complex with the integrin member α5β1 and inactive. During acute H. pylori infection, however, it was shown that CagL dissociates ADAM17 from the integrins and activates ADAM17 (Fig. 3A). This was confirmed by infection with a ΔcagL deletion mutant, which is entirely defective in the latter response and by genetic complementation with the wild-type cagL gene or biochemical complementation by addition of extracellular CagL restoring this function. In addition, during integrin binding studies using intact host cells and immobilised CagL on petri dishes it was found that CagL mimics some important functions of human fibronectin [25]. Fibronectin is a 250-kDa eukaryotic protein containing an RGD-motif which plays crucial roles in promoting cell adhesion, migration, and intracellular signaling. It was shown that purified CagL alone can directly trigger intracellular signaling pathways upon contact with mammalian cells and can even complement the spreading defect of fibronectin-/- knock-out cells in vitro [25]. During interaction with various human and mouse cell lines, CagL mimics fibronectin in triggering cell spreading and focal adhesion formation. CagL-mediated activation of the above mentioned kinases was essential in these processes. Interestingly, fibronectin activates a similar range of tyrosine kinases but not Her3/ErbB3 [25]. These findings suggest that the VirB5 ortholog CagL does not only exhibit functional mimicry with fibronectin but is also capable of activating fibronectin-independent signaling events. Interestingly, when the purified repeat region 2 or the carboxy-terminus of CagY was immobilized on petri dishes, neither of these fragments could induce efficient cell spreading [25]. Remarkably however, when CagL was mixed with CagY, the repeat region 2 but not the integrin β1-interacting carboxy-terminus [13] enhanced the CagL effect [25]. This finding suggests that the internal repeat region of CagY and CagL may act cooperatively, and that the carboxy-terminal interaction of CagY with integrin β1 has a different function, further confirming that the observed cell spreading effect is specific for CagL. If other H. pylori factors such as extracellularly added CagY, CagI or CagA can also trigger similar and/or other intracellular signalling pathways, and whether CagL-mediated activation of EGFR, Her3/ErbB3, Src and Fak contributes to the injection of CagA during H. pylori infection was not yet investigated and needs to be addressed in future studies.
Phosphorylation-dependent cell signaling of injected CagA
An important reason why CagA was actually discovered as an injected effector protein is the very early observation that it undergoes tyrosine-phosphorylation (CagAPY) by the host cell kinases Src and Abl [15]. Site-directed mutagenesis and mass spectrometry of CagA in H. pylori or transfected CagA identified numerous phosphorylation sites as the Glu-Pro-Ile-Tyr-Ala (EPIYA)-motifs A, B, C and/or D [7,26]. In infected or transfected epithelial host cells, CagAPY has then been shown to interact with the Src homology 2 (SH2) domains of numerous eukaryotic signalling proteins. The first detected binding partner of CagAPY was the tyrosine phosphatase Shp2 (Fig. 3B). Since then, nine other host cell factors were also reported to interact with CagA in a phosphorylation-dependent fashion: the tyrosine phosphatases Shp1, the adaptor proteins Grb2, Grb7 and Crk, phosphoinositide-3-kinase (PI3K), Ras GTPase activating protein RasGAP, and as well as the tyrosine kinases Csk, Src and Abl [26]. Thus, CagAPY appears to mimick a tyrosine-phosphorylated host cell protein and therefore seems to act as a kind of masterkey or picklock to highjack crucial signaling pathways in the host. The various CagAPY-SH2 domain interactions play complex roles in H. pylori-induced actin-cytoskeletal rearrangements, scattering and elongation of infected host cells in culture as summarised in Fig. 3B.
Gastric epithelial cells infected with H. pylori in vitro start to migrate and acquire a morphology that has been originally described as the “hummingbird phenotype”. This phenotype results from two successive events: the induction of cell scattering and cell elongation. While induction of early cell motility mainly depends on a CagA-independent T4SS factor [23], cell elongation is clearly triggered by CagAPY [6,7]. Transfection experiments demonstrated that the CagAPY-Shp2 interaction stimulates the phosphatase activity of Shp2 which contributes to cell elongation by activating the Rap1→B-Raf→Erk signalling cascade and by direct dephosphorylation and inactivation of focal adhesion kinase, FAK [7]. Further studies have indicated that the CagAPY-induced cell elongation phenotype also involves tyrosine dephosphorylation of cortactin, vinculin and ezrin, three well-known actin-binding proteins [6]. The phosphatases involved in this scenario, however, remained unknown and does not require Shp2. Instead, CagAPY can inhibit Src activity both by direct interaction of both proteins or by binding of CagAPY to Csk, a negative regulator of Src [6,7]. Because Src is the primary kinase phosphorylating CagA in the EPIYA-motifs, inhibition of Src by CagAPY generates a classical negative feedback-loop that appears to control the amount of intracellular CagAPY. Cortactin, ezrin and vinculin are all Src substrates, and Src inactivation causes dephosphorylation of these proteins and is crucial for triggering cell elongation [26]. In addition, interaction of CagAPY with CrkII/Abl and/or PI3K may also activate the small Rho GTPases Cdc42 and Rac1, binding of CagAPY to Grb2 or Shp2 may regulate proliferative and pro-inflammatory signalling via the MAP kinase pathway while interaction of CagAPY with Shp1 may downregulate the latter response (Fig. 3B). Finally, several additional binding partners were identified such as Grb7 and RasGAP (Fig. 3B). The potential function of these two factors in molecular pathogenesis, however, is yet unknown and needs to be investigated in future studies. Taken together, CagAPY interacts with a surprisingly high number of host proteins to induce signalling pathways involved in cell scattering, elongation and probably other phenotypes.
Phosphorylation-independent signalling of CagA
Remarkably, not all interactions of injected or transfected CagA depend on its tyrosine phosphorylation. Together, 12 cellular interaction partners of non-phosphorylated CagA have been identified [26]. These interactions have been reported to induce the disruption of cell-to-cell junctions, loss of cell polarity and induction of pro-inflammatory and mitogenic responses (Fig. 3C). The first detected interaction partner of non-phosphorylated CagA was the adapter protein Grb2, and Grb2 is the only host factor described to interact with both non-phosphorylated and additionally with phosphorylated EPIYA motifs as described above [26]. In particular, non-phosphorylated CagA was shown to interact with Grb2 both in vitro and in vivo, which provides a mechanism by which Grb2-associated Sos (son of sevenless, a guanine-exchange factor of the small GTPase Ras) is recruited to the plasma membrane (Fig. 3C). The CagA/Grb2/Sos complex can promote Ras-GTP formation, which in turn stimulates the Raf→Mek→Erk signalling cascade leading to cell scattering as well as activation of nuclear transcription factors involved in cell proliferation and expression of the anti-apoptotic myeloid cell leukaemia sequence-1 (MCL-1) protein [27]. In line with these findings, CagA was also shown to function as a mimetic of the eukaryotic Grb2-associated binder (Gab) adaptor protein in transgenic Drosophila which can further explain how CagA triggers this signalling cascade [7]. Interestingly, CagA can also interact with RUNX3, a tumor suppressor which is frequently inactivated in gastric cancer, by a novel identified WW domain in the amino-terminal region of CagA [28]. In particular, CagA induces the ubiquitination and degradation of RUNX3, thereby extinguishing its ability to inhibit the transcriptional activation of RUNX3 (Fig. 3C). Additional evidence for a role of CagA in manipulating nuclear responses came from whole-genome microarrays and functional studies investigating host cell gene expression after infection of target cells with wild-type H. pylori, isogenic cagA and cagPAI mutants as well as CagA transfection. For example, it was shown that under certain circumstances CagA can also induce the transcription factor NF-κB influencing the expression of multiple target genes such as IL-8 in a CagA phosphorylation-independent manner as discussed recently [26]. These data suggest the presence of distinct EPIYA-independent domains within CagA which play essential roles in protein targeting and alteration of host-cell transcription signalling pathways.
Another important consequence of phosphorylation-independent CagA interactions in polarised epithelial cells is the disruption of cell-to-cell junctions (Fig. 3C). In particular, tight and adherens junctions are essential for the integrity of the gastric epithelium [15]. CagA interferes with these intercellular junctions via several pathways. Injected CagA associates with the epithelial tight-junction scaffolding protein ZO-1 (zona occludens-1) and the transmembrane protein JAM (junctional adhesion molecule), causing an ectopic assembly of tight-junction components at sites of bacterial attachment [2]. Non-phosphorylated CagA was also reported to interact with the transmembrane cell-cell junction protein E-cadherin [7]. Later it was found that CagA forms a complex with c-Met recruiting E-cadherin and the Armadillo-domain protein p120 catenin indicating that the interaction between CagA and E-cadherin is not direct [29]. However, whether the 135 kDa c-Met receptor is phosphorylated and activated upon H. pylori infection is a matter of debate [26]. Thus, the role of c-Met in H. pylori–induced signalling is not fully clear and needs to be investigated more thoroughly in future. Controversy also exists whether CagA can induce disruption of the E-cadherin complex followed by the release of β-catenin, which has been proposed for transfected CagA or H. pylori-infected AGS cells [7,18]. There is some doubt if vector-based overexpression of CagA induces cellular effects which are really comparable to that of CagA injected by H. pylori. In addition, AGS cells do not express E-cadherin and show abnormal β-catenin distribution making them unsuited for the investigation of β-catenin signaling [29]. Emphasizing this aspect, using MDCK cells which express E-cadherin and β-catenin without any mutations, it was demonstrated that CagA is not involved in H. pylori-induced β-catenin signal transduction pathways during infection [30]. These data were also supported by several other research groups as recently shown.
However, the role of CagA in inducing the loss of cell polarity is much clearer. The kinase Par1b/MARK2 (partitioning-defective 1/microtubule affinity-regulating kinases), a central regulator of cell polarity, was found to play a role in H. pylori-induced signalling. Non-phosphorylated CagA can directly bind Par1b and inhibits its kinase activity to trigger the loss of cell polarity [7]. In addition, more recent studies also showed that CagA not only binds to Par1b but also to other members of this kinase family (including Par1a, Par1c and Par1d), and that these interactions contribute to the H. pylori-induced elongation phenotype of AGS cells. Taken together, these findings demonstrate that injected or transfected CagA can interfere with Par1, c-Met and E-cadherin signalling and may also activate NF-κB, thereby contributing to H. pylori-induced pro-inflammatory responses [9,26]. However, the downstream pathways of CagA appear very diverse and possible crosstalk among those and other bacterial factors needs to be dissected in more detail. Finally, there are two additionally reported binding partners of CagA, α-Pix and integrin β1 as mentioend above (Fig. 3C), but the functional importance of these interactions is yet unclear and also needs to be investigated in future studies.
Concluding remarks
H. pylori represents one of the most successful human pathogens, which induces severe clinical symptoms only in a small subset of individuals mirroring a highly balanced degree of co-evolution of H. pylori and humans. Studies of host-bacterial interactions and virulence factors CagA and the T4SS have provided us with many fundamental insights in processes leading to H. pylori pathogenesis. The number of more than twenty known cellular interaction partners of CagA is very astonishing and remarkable for a bacterial effector protein. The current hypothesis implies a model with translocated CagA as an ‘eukaryotic’ signaling mimetic molecule either present in a large multiprotein complex or simultaneously in separated locations of infected target cells, which may have important impact on the multi-step pathogenesis of H. pylori. The high number of binding partners also reflects the integrated network of complex signal transduction pathways in host cells, which is coordinated through CagA itself or initiated by the T4SS-host interaction underlining their overall importance as observed in numerous in vitro and in vivo studies. In future, it will be important to search for additional injected proteins because it seems rather unlikely that the entire cagPAI was aquired during evolution to only inject one effector protein (CagA) and peptidoglycan. Interestingly, recent computational predictions suggested that proteins encoded by HP0522 and HP0535 maybe novel T4SS effectors [4]. Whether HP0522 and HP0535 are indeed translocated by the T4SS is only one open question among many others that need to be answered in future studies in order to uncover the complex mechanisms how H. pylori interacts with host cells.
Supplementary Material
Acknowledgments
We apologize to all H. pylori researchers whose original work could not be cited due to length restriction of this article and only 30 references being allowed. The work of S.W. was supported by the Deutsche Forschungsgemeinschaft (We2843/2–1) and BGAG-Stiftung Walter Hesselbach. The work of S.B. is supported through grants from the Deutsche Forschungsgemeinschaft DFG grant (Ba1671/8-1), from the National Institute Of Diabetes And Digestive And Kidney Diseases (R56DK064371), and from University College Dublin (R11408).
Footnotes
Supporting information: The following supplementary material is available:
Doc. S1. Glossary.
This supplementary material can be found in the online version of this article.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- 1.Peek RM, Jr, Crabtree JE. Helicobacter infection and gastric neoplasia. J Pathol. 2006;208:233–48. doi: 10.1002/path.1868. [DOI] [PubMed] [Google Scholar]
- 2.Amieva MR, El-Omar EM. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008;134:306–323. doi: 10.1053/j.gastro.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Atherton JC, Blaser MJ. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest. 2009;119:2475–2487. doi: 10.1172/JCI38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olbermann P, Josenhans C, Moodley Y, Uhr M, Stamer C, Vauterin M, Suerbaum S, Achtman M, Linz B. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6:e1001069. doi: 10.1371/journal.pgen.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Mémet S, Huerre MR, Coyle AJ, DiStefano PS, Sansonetti PJ, Labigne A, Bertin J, Philpott DJ, Ferrero RL. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 6.Backert S, Selbach M. Role of type IV secretion in Helicobacter pylori pathogenesis. Cell Microbiol. 2008;10:1573–1581. doi: 10.1111/j.1462-5822.2008.01156.x. [DOI] [PubMed] [Google Scholar]
- 7.Hatakeyama M. SagA of CagA in Helicobacter pylori pathogenesis. Curr Opin Microbiol. 2008;11:30–37. doi: 10.1016/j.mib.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Hoy B, Löwer M, Weydig C, Carra G, Tegtmeyer N, Geppert T, Schröder P, Sewald N, Backert S, Schneider G, Wessler S. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 2010;11:798–804. doi: 10.1038/embor.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Backert S, Naumann M. What a disorder: proinflammatory signaling pathways induced by Helicobacter pylori. Trends Microbiol. 2010 doi: 10.1016/j.tim.2010.08.003. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 10.Rohde M, Püls J, Buhrdorf R, Fischer W, Haas R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol. 2003;49:219–234. doi: 10.1046/j.1365-2958.2003.03549.x. [DOI] [PubMed] [Google Scholar]
- 11.Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, König W, Backert S. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862–866. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- 12.Kutter S, Buhrdorf R, Haas J, Schneider-Brachert W, Haas R, Fischer W. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J Bacteriol. 2008;190:2161–71. doi: 10.1128/JB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiménez-Soto LF, Kutter S, Sewald X, Ertl C, Weiss E, Kapp U, Rohde M, Pirch T, Jung K, Retta SF, Terradot L, Fischer W, Haas R. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog. 2009;5:e1000684. doi: 10.1371/journal.ppat.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murata-Kamiya N, Kikuchi K, Hayashi T, Higashi H, Hatakeyama M. Helicobacter pylori exploits host membrane phosphatidylserine for delivery, localization, and pathophysiological action of the CagA oncoprotein. Cell Host Microbe. 2010;7:399–411. doi: 10.1016/j.chom.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Wessler S, Backert S. Molecular mechanisms of epithelial-barrier disruption by Helicobacter pylori. Trends Microbiol. 2008;16:397–405. doi: 10.1016/j.tim.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Ogura K, Maeda S, Nakao M, Watanabe T, Tada M, Kyutoku T, Yoshida H, Shiratori Y, Omata M. Virulence factors of Helicobacter pylori responsible for gastric diseases in Mongolian gerbil. J Exp Med. 2000;192:1601–1610. doi: 10.1084/jem.192.11.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rieder G, Merchant J, Haas R. Helicobacter pylori cag-type IV secretion system facilitates corpus colonization to induce precancerous conditions in Mongolian gerbils. Gastroenterology. 2005;128:1229–1242. doi: 10.1053/j.gastro.2005.02.064. [DOI] [PubMed] [Google Scholar]
- 18.Franco AT, Israel DA, Washington MK, Krishna U, Fox JG, Rogers AB, Neish AS, Collier-Hyams L, Perez-Perez GI, Hatakeyama M, Whitehead R, Gaus K, O'Brien DP, Romero-Gallo J, Peek RM., Jr Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci U S A. 2005;102:10646–10651. doi: 10.1073/pnas.0504927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohnishi N, Yuasa H, Tanaka S, Sawa H, Miura M, Matsui A, Higashi H, Musashi M, Iwabuchi K, Suzuki M, Yamada G, Azuma T, Hatakeyama M. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci USA. 2008;105:1003–1008. doi: 10.1073/pnas.0711183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramarao N, Gray-Owen SD, Backert S, Meyer TF. Helicobacter pylori inhibits phagocytosis by professional phagocytes involving type IV secretion components. Mol Microbiol. 2000;37:1389–1404. doi: 10.1046/j.1365-2958.2000.02089.x. [DOI] [PubMed] [Google Scholar]
- 21.Fehri LF, Rechner C, Janssen S, Mak TN, Holland C, Bartfeld S, Brüggemann H, Meyer TF. Helicobacter pylori-induced modification of the histone H3 phosphorylation status in gastric epithelial cells reflects its impact on cell cycle regulation. Epigenetics. 2009;4:577–586. doi: 10.4161/epi.4.8.10217. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto Y, Marusawa H, Kinoshita K, Endo Y, Kou T, Morisawa T, Azuma T, Okazaki IM, Honjo T, Chiba T. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat Med. 2007;13:470–476. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- 23.Churin Y, Al-Ghoul L, Kepp O, Meyer TF, Birchmeier W, Naumann M. Helicobacter pylori CagA protein targets the c-Met receptor and enhances the motogenic response. J Cell Biol. 2003;161:249–255. doi: 10.1083/jcb.200208039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keates S, Sougioultzis S, Keates AC, Zhao D, Peek RM, Jr, Shaw LM, Kelly CP. cag+ Helicobacter pylori induce transactivation of the epidermal growth factor receptor in AGS gastric epithelial cells. J Biol Chem. 2001;276:48127–48134. doi: 10.1074/jbc.M107630200. [DOI] [PubMed] [Google Scholar]
- 25.Tegtmeyer N, Hartig R, Delahay RM, Rohde M, Brandt S, Conradi J, Takahashi S, Smolka AJ, Sewald N, Backert S. A small fibronectin-mimicking protein from bacteria induces cell spreading and focal adhesion formation. J Biol Chem. 2010;285:23515–23526. doi: 10.1074/jbc.M109.096214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: The master key hypothesis. Helicobacter. 2010;15:163–176. doi: 10.1111/j.1523-5378.2010.00759.x. [DOI] [PubMed] [Google Scholar]
- 27.Mimuro H, Suzuki T, Nagai S, Rieder G, Suzuki M, Nagai T, Fujita Y, Nagamatsu K, Ishijima N, Koyasu S, Haas R, Sasakawa C. Helicobacter pylori dampens gut epithelial self-renewal by inhibiting apoptosis, a bacterial strategy to enhance colonization of the stomach. Cell Host Microbe. 2007;2:250–263. doi: 10.1016/j.chom.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Tsang YH, Lamb A, Romero-Gallo J, Huang B, Ito K, Peek RM, Jr, Ito Y, Chen LF. Helicobacter pylori CagA targets gastric tumor suppressor RUNX3 for proteasome-mediated degradation. Oncogene. 2010 doi: 10.1038/onc.2010.304. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira MJ, Costa AM, Costa AC, Ferreira RM, Sampaio P, Machado JC, Seruca R, Mareel M, Figueiredo C. CagA associates with c-Met, E-cadherin, and p120-catenin in a multiproteic complex that suppresses Helicobacter pylori-induced cell-invasive phenotype. J Infect Dis. 2009;200:745–55. doi: 10.1086/604727. [DOI] [PubMed] [Google Scholar]
- 30.Sokolova O, Bozko PM, Naumann M. Helicobacter pylori suppresses glycogen synthase kinase 3beta to promote beta-catenin activity. J Biol Chem. 2008;283:29367–29374. doi: 10.1074/jbc.M801818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.