Abstract
Superficial spreading melanoma (SSM) and nodular melanoma (NM) are believed to represent sequential phases of linear progression from radial to vertical growth. Several lines of clinical, pathological and epidemiologic evidence suggest, however, that SSM and NM might be the result of independent pathways of tumor development. We utilized an integrative genomic approach that combines single nucleotide polymorphism array (SNP 6.0, Affymetrix) with gene expression array (U133A 2.0, Affymetrix) to examine molecular differences between SSM and NM. Pathway analysis of the most differentially expressed genes between SSM and NM (N=114) revealed significant differences related to metabolic processes. We identified 8 genes (DIS3, FGFR1OP, G3BP2, GALNT7, MTAP, SEC23IP, USO1, ZNF668) in which NM/SSM-specific copy number alterations correlated with differential gene expression (P<0.05, Spearman’s rank). SSM-specific genomic deletions in G3BP2, MTAP, and SEC23IP were independently verified in two external data sets. Forced overexpression of metabolism-related gene methylthioadenosine phosphorylase (MTAP) in SSM resulted in reduced cell growth. The differential expression of another metabolic related gene, aldehyde dehydrogenase 7A1 (ALDH7A1), was validated at the protein level using tissue microarrays of human melanoma. In addition, we show that the decreased ALDH7A1 expression in SSM may be the result of epigenetic modifications. Our data reveal recurrent genomic deletions in SSM not present in NM, which challenge the linear model of melanoma progression. Furthermore, our data suggest a role for altered regulation of metabolism-related genes as a possible cause of the different clinical behavior of SSM and NM.
Keywords: melanoma, nodular, genomics, SNP array, DNA copy number
INTRODUCTION
Superficial spreading melanoma (SSM) and nodular melanoma (NM), the two most common histopathologic subtypes (70% and 20% respectively), are characterized by markedly different clinical presentations and natural histories. NM has a higher rate of recurrence (1, 2) and has not demonstrated the same degree of downward stage migration at initial presentation relative to SSM (3). These differences are generally attributed solely to the advanced thickness of NM, with no prognostic relevance assigned to histopathological subtype in melanomas of equivalent thickness. Previous studies by several groups including ours, however, suggest that underlying molecular differences between the two subtypes may also contribute to the disparate outcomes (4–6).
It is generally accepted that SSM and NM develop along a linear pathway of progression that begins with transformation of epidermal melanocytes and differs between subtypes primarily with respect to the speed with which the transformed melanocytes invade the dermis (7, 8). Until the identification of distinct molecular alterations characterizing acral melanoma such as c-KIT mutation and amplification (9–11), all melanoma subtypes were typically viewed as a relatively homogenous biologic entity (8). Advances in genomic technology in the setting of a broader recognition of the biologic heterogeneity of cancer have changed this long-held view, at least for acral melanoma. As such, clinical trials evaluating the efficacy of c-KIT inhibitor imatinib for patients with metastatic melanoma from an acral primary are ongoing (12). Currently, there is no definitive evidence to demonstrate that SSM and NM are molecularly different enough to warrant a unique molecular classification such as the one that has now been assigned to acral melanoma.
It is difficult to reconcile observed differences between SSM and NM, such as the minimal degree of epidermal involvement and the lack of a detectable radial growth phase in NM, with the current linear progression model. To our knowledge, there are no studies that have evaluated the genomic and gene expression alterations that characterize SSM and NM. Using an integrative genomic approach, we examined the hypothesis that SSM and NM follow separate pathways of development from the transformed melanocyte to the invasive primary melanoma that are characterized by subtype specific molecular alterations.
MATERIALS AND METHODS
Melanoma cell lines and human tissues
Cell lines included normal melanocytes cultured from infant foreskin (NHM), immortalized melanocytes (Hermes1 and 2B), primary SSM (WM35, WM1552c, WM1575), primary nodular melanoma (WM39, WM853.2), primary vertical growth phase (VGP) (WM98.1, WM853.2), metastatic nodular melanoma (Lu451, SK-147), and metastatic melanoma of unknown primary histologic subtype (501MEL, A375, SK-MEL-19, -29, -85, -94, -100, -103, -173, -187, -192, -197). Metastatic cell lines were kindly provided by Alan Houghton (Memorial Sloan-Kettering Cancer Center, New York, NY), and primary cell lines (as well as Lu451) were purchased commercially from the Wistar Institute (Philadelphia, PA). Cell lines were not further tested or authenticated (see Suppl. Methods for details on cell lines and tissues).
SNP DNA array
DNA was hybridized to the Affymetrix Genome-Wide Human SNP 6.0 Array as per the manufacturer protocol (Affymetrix Santa Clara, CA). CEL files were generated using the GeneChip Command Console software (Affymetrix), and the Birdseed v2 algorithm (13) was used to make the genotype calls and summarize the probe-set intensity values for ensuing copy number analysis. A single reference genome profile assayed in the same batch was created using values from the 2 normal melanocytes cultured from infant foreskin (NHM).
Gene expression array
RNA was hybridized to the Affymetrix U133A 2.0 array as per manufacturer protocol. Array data were normalized using the robust multichip average (RMA) and filtered for expression values lower than 16. The first analysis undertaken was an exploratory principal component analysis (PCA) which allows for the visualization of relationships between samples in a multidimensional dataset(14). Data were then log transformed to perform an analysis of variance (ANOVA) and a two-sample unpaired t-test with p-values adjusted for multiple comparisons (Bonferroni correction). A single factor, two-level ANOVA design was utilized to examine sources of measured variability between the SSM and NM. A software package D-chip was used to facilitate the analysis. MAIME-compliant data for the Affymetrix SNP6.0 and U133A 2.0 gene expression array have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) [GSE22306 (GSE22301 and GSE22305)].
Integrative genomic analyses
Correlation of copy number data with gene expression was performed using the Partek Genomic Suite (Partek St Louis, MO). Copy number gains and losses were detected using a Hidden Markov Model algorithm in the Partek copy number workflow for unpaired samples. (see Suppl. Methods for detailed analysis).
External validation
Unprocessed data for 13SSM and 5NM human tissues were downloaded from the 2007 publication by Jaeger et al(4). Data were then normalized using RMA and log transformed for a two-sample unpaired t-test with a Bonferroni correction for multiple testing. GenePattern was used for clustering and gene set enrichment analysis. The complete list of 540 transcripts differentially expressed between radial growth phase (RGP) and VGP in the 2009 publication by Scatolini et al. (15) was also used as an external validation.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
qRT-PCR was used to verify the results of gene expression microarray data. Data were exported using SDS 2.3 and analyzed using the methods of Pfaffl (16) assuming 100% efficiency of amplification with normalization to four housekeeping genes: TMPRSS2, PDLIM4, DHX15, and SNF8. These genes were selected based on their uniform expression across the data set (coefficient of variance <10%). Fold change was evaluated relative to melanocyte controls. Primer sequences are summarized in Supplementary Table 1A.
Genomic qPCR
Five genes were evaluated for validation using genomic PCR: DIS3, G3BP2, MTAP, SEC23IP, and USO1. Gene-specific primers are shown in Supplementary Table 1B and housekeeping genes, UBE2E1 and GNS, were used as previously described (17)(see Suppl. Methods for detailed protocol).
Western blot
Membranes were probed with anti-MTAP polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA); anti-ALDH7A1 polyclonal antibody (Epitomics Burlingame, CA); and anti-EPB41L3 (Abnova, Taiwan) at dilutions of 1:200, 1:1000, and 1:500, respectively (see Suppl Methods for detailed protocol).
Lentiviral infection and growth curve
MTAP coding sequence was amplified from pBS/MTAP (MHS1010-7295727, open Biosystems) by PCR and then cloned into the PstI and NdeI restriction site of pWPI lentiviral vector (gift of Dr. Hernando), downstream of the EF1 promoter (see Suppl. Methods for detailed protocol).
Immunohistochemistry (IHC) on TMAs
IHC was performed using a tissue microarray of 20NM and 20SSM cases. Tissue cores were evaluated for expression of ALDH7A1 (rabbit polyclonal antibody, Abcam Inc., Catalog # ab80187) diluted 1:500, EPB41L3 (purified MaxPab mouse polyclonal antibody, Abnova Corporation., Cat. # H00023136-B01P) at 1:500, and MTAP (monoclonal antibody (M01) clone 2G4 purified mouse immunoglobulin, Abnova, Cat. # H00004507-M01) at 1:2000 (see Suppl. Methods for detailed protocol).
An attending pathologist (FD) scored the expression of ALDH7A1, MTAP, and EPB41L3 in each core on a scale from 0–2. The association between staining intensity and histologic subtype was assessed by the t-test or chi-square test for each marker. The chi-square test was also used to compare the number of cores with a specific intensity score between the NM and SSM groups. A multivariate general linear model was used to compare the adjusted mean expression of ALDH7A1 between the NM and SSM case groups, after controlling for thickness and ulceration. All p-values are two-sided with statistical significance evaluated at the 0.05 alpha level. All analyses were performed in SPSS Version 18.0 (SPSS Inc., Chicago, IL).
Analysis of genetic and epigenetic alterations of ALDH7A1
ALDH7A1 exons 2, 12, 14 and 15, and introns 5 and 16, were sequenced (Supplementary Methods, primers shown in Suppl. Table 1C). To analyze the methylation and acetylation status of ALDH7A1 promoter, WM1552c cells were treated with 1uM of methyltransferase inhibitor 5-azacytidine (aza-CR), followed by an additional 24h incubation with 1uM 5-azacytidine (aza-CR) plus 200ng/ml of histone deacetylase inhibitor Trichostatin A (TSA). ALDH7A1 mRNA expression level was analyzed by real-time PCR (primers shown in Suppl. Table 1D) (see Suppl. Methods for detailed protocol).
RESULTS
SNP array identifies recurrent NM/SSM-specific copy number alterations
In order to investigate the genomic alterations characteristic of SSM vs. NM, we performed high resolution SNP array on a panel of 22 melanoma cell lines inclusive of normal melanocytes, primary SSM (WM1552c, WM35), primary NM (WM39, WM278), metastatic NM (Lu451, SK-MEL-147) and metastatic melanoma. The list of significantly altered genomic segments across all the cell lines was then filtered to identify those specific to SSM and NM. SNP array identified 408 SSM-specific regions with significant genomic alteration (212 gains, 196 losses), and 543 NM-specific genomic alterations (295 gains, 248 losses) (Suppl. Table 2).
Gene expression profiling reveals 114 genes differentially expressed between SSM and NM
Principal component analysis (PCA) showed that the melanocytes formed a tight and distinct cluster, while there was a high degree of variance among the melanoma cell lines. SSM cell lines clustered together and appeared to be closely related to a larger group of metastatic cell lines. On the contrary, the NM cell lines, particularly the NM primaries, did not cluster together and also did not fall within the larger cluster of metastatic and SSM cell lines. All but one of the 4 NMs fell outside 2 standard deviations of the mean expression value for all other melanomas (including SSM and metastatic). Both of the NM primaries demonstrated distinct expression profiles relative to the other melanoma cell lines (data not shown). These results, although exploratory in nature, suggested that there was enough difference with regard to the gene expression profiles between NM and SSM to justify further investigation of the sources of these differences.
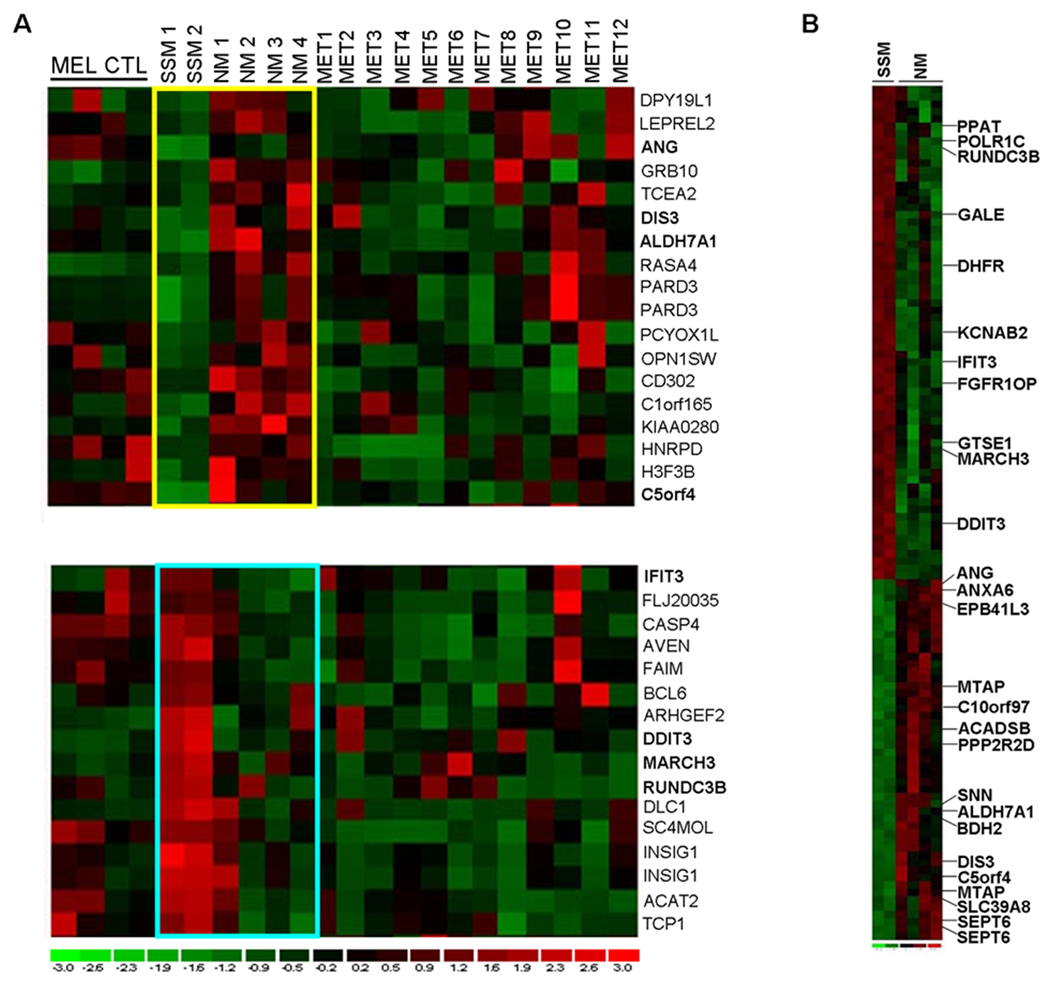
Two statistical analyses were then performed to identify differentially expressed genes between SSM and NM. The first analysis (ANOVA) revealed a list of 79 gene probe IDs differentially expressed between SSM and NM relative to each other and relative to normal melanocytes and metastatic melanoma cells (P<0.05, Bonferroni correction) (Figure 1A). The second statistical analysis (t-test, SSM vs. NM) identified 114 differentially expressed genes (116 probe IDs) (P<0.05, Bonferroni correction; Figure 1B and Suppl. Table 3).
FIGURE 1.
A) ANOVA of 22 cell lines identifies 79 transcripts differentially expressed between normal melanocytes, SSM, NM and metastatic lines (P<0.05, Bonferroni correction). Smaller distinct subsets of genes are overexpressed in NM relative to SSM (yellow box) and overexpressed in SSM compared to NM (blue box). B) Direct comparison of gene expression between NM and SSM identifies 114 differentially expressed transcripts (P<0.05, t-test; Bonferroni correction). Genes indicated on the heat map (B) are those that are differentially expressed in both statistical analyses (ANOVA and t-test; N=25 total genes). MEL CTL–melanocyte control, SSM-superficial spreading melanoma, NM-nodular melanoma, MET-metastatic melanoma. SSM 1, 2 (WM1552c, WM35) and NM 1, 2 (WM278, WM39) are primary melanomas. NM 3 and 4 (Lu451, SK-MEL-147) are metastases from known nodular primaries. MET 1–12 are metastases of unknown primary histologic subtype.
Functional annotation shows that differentially expressed transcripts between SSM and NM are related to metabolic pathways
Gene ontology (GO) analysis of differentially expressed genes between SSM and NM revealed enrichment for GO biologic process (BP) terms related to primary, cellular, and nucleic acid metabolic processes (Suppl. Table 4). The most enriched term with regard to number of genes was “metabolic process” (total gene count 65, 55.1%; P=0.04) which included genes such as methylthioadenosine phosphorylase (MTAP), aldehyde dehydrogenase 7A1 (ALDH7A1), angiogenin (ANG), and dihydrofolate reductase (DHFR) (Suppl. Table 4). The two most statistically significant GO BP terms were “nucleic acid metabolic process” (total gene count 38, 32.2%, P=0.003) and “cellular component organization/biogenesis” (total gene count 27, 22.9%; P=0.001).
To prioritize the 114 differentially expressed NM/SSM, we focused on the 25 genes with differential expression in both analyses (the ANOVA and t-test; Suppl. Table 5). The gene showing the highest and most significant differential expression between SSM and NM was MTAP (Fold change = −21.5; P=0.0003) in which expression was lower in SSM. The gene with the second highest fold change between NM and SSM expression was ALDH7A1 (Fold change = −18.0; P=0.03), and the second most significant difference was erythrocyte membrane protein band 4.1 (EPB41L3, FC = −6.2; P=0.001)(Suppl. Table 5). MTAP, ALDH7A1 and EPB41L3 were underexpressed in SSM relative to NM, and all genes are related to cellular metabolism, consistent with the results of the pathway analysis (Suppl. Table 4).
Integrative genomic workflow yields 8 genes in which SSM/NM specific copy number changes are correlated with NM/SSM differential expression
NM/SSM-specific copy number gains and losses that mapped to regions coding for genes found to be differentially expressed between NM and SSM in our initial analysis (N=114) were evaluated for statistical correlation between copy number and mRNA expression level. The integrative analysis revealed a list of 8 genes (DIS3, FGFR1OP, G3BP2, GALNT7, MTAP, SEC23IP, USO1, ZNF668) that met the following criteria: 1) NM/SSM-specific copy number gain or loss; 2) differential gene expression between NM and SSM; and 3) copy number significantly correlated with gene expression (Table 1). Included in this list of 8 genes were MTAP and FGFR1OP, the two genes with the highest degree of fold change between NM and SSM in opposite directions (MTAP lower in SSM, FGFR1OP higher in SSM), suggesting that these genes may hold increased biologic relevance.
TABLE 1.
List of genes that are differentially expressed between NM and SSM, have NM/SSM-specific copy
| Gene Symbol | Gene Expression | p-value* | Copy Number Status | Correlation† | p-value§ |
|---|---|---|---|---|---|
| FGFR1OP | NM<SSM | 0.001 | DEL NM | 0.94 | 0.005 |
| SEC23IP | SSM<NM | 0.01 | DEL SSM | 0.94 | 0.005 |
| GALNT7 | SSM>NM | 0.009 | AMP SSM | 0.94 | 0.005 |
| G3BP2 | SSM<NM | 0.02 | DEL SSM | 0.94 | 0.005 |
| MTAP | SSM<NM | 0.0003 | DEL SSM | 0.89 | 0.02 |
| DIS3 | SSM<NM | 0.03 | DEL SSM | 0.83 | 0.04 |
| ZNF668 | NM<SSM | 0.02 | DEL NM | 0.83 | 0.04 |
| USO1 | SSM<NM | 0.04 | DEL SSM | 0.83 | 0.04 |
number alterations, and in which mRNA expression is significantly correlated with DNA copy number.
T-test, with Bonferroni correction for multiple comparisons.
Spearman’s rank correlation coefficient.
By Spearman’s rank.
Array results verified using genomic qPCR and qRT-PCR
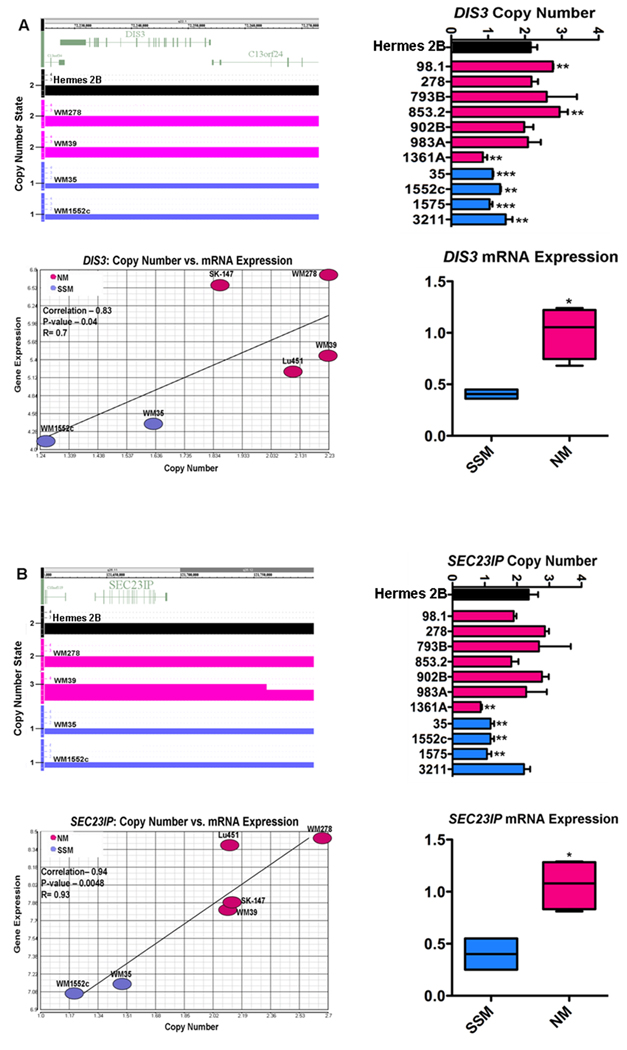
The copy number status of USO1, G3BP2, DIS3, and SEC23IP was evaluated in an expanded panel of NM/vertical growth phase (VGP) cell lines (N=7) and SSM/radial growth phase (RGP) cell lines (N=4). Genomic PCR verified the presence of significant deletions in 100% (P<0.05, N=4) of SSM cell lines relative to melanocytes in the regions of USO1, G3BP2, and DIS3 (DIS3, Figure 2A). In the region of SEC23IP, genomic PCR verified significant deletions in 75% (P<0.01, N=4) of the SSM cell lines (Figure 2B). Of note, mitotic control homolog DIS3 was significantly deleted in all 4 RGP-like SSM cell lines assessed by genomic PCR, whereas 4 of the 7 VGP-like NM cell lines were diploid and 2 had significant (P<0.01) amplification (Figure 2A). The same 5 genes were also evaluated for mRNA expression by qRT-PCR using the melanoma cell lines from the array. qRT-PCR verified the pattern of differential gene expression observed in the array for all genes (ie, down in SSM relative to NM), with 3 genes (MTAP, DIS3, SEC23IP) showing a statistically significant (P<0.05, t-test) difference in expression between NM and SSM (DIS3 and SEC23IP, Figure 2; MTAP, Figure 4C).
FIGURE 2.
A) Clock-wise from top left: copy number alterations identified in SSM (blue) and NM (pink) cell lines in the region of DIS3 using SNP array; genomic qPCR verification of SSM/NM-specific copy number alterations in an expanded panel of cell lines; qRT-PCR verification of differential mRNA expression between NM (WM278, WM39, Lu451, SK-MEL-147) and SSM (WM1552c, WM35) cell lines in DIS3; and significant correlation between DIS3 copy number and DIS3 gene expression (Spearman’s Rank correlation). B) The same analysis of SEC23IP in the same NM and SSM cell lines. *P<0.05, **P<0.01, ***P<0.001
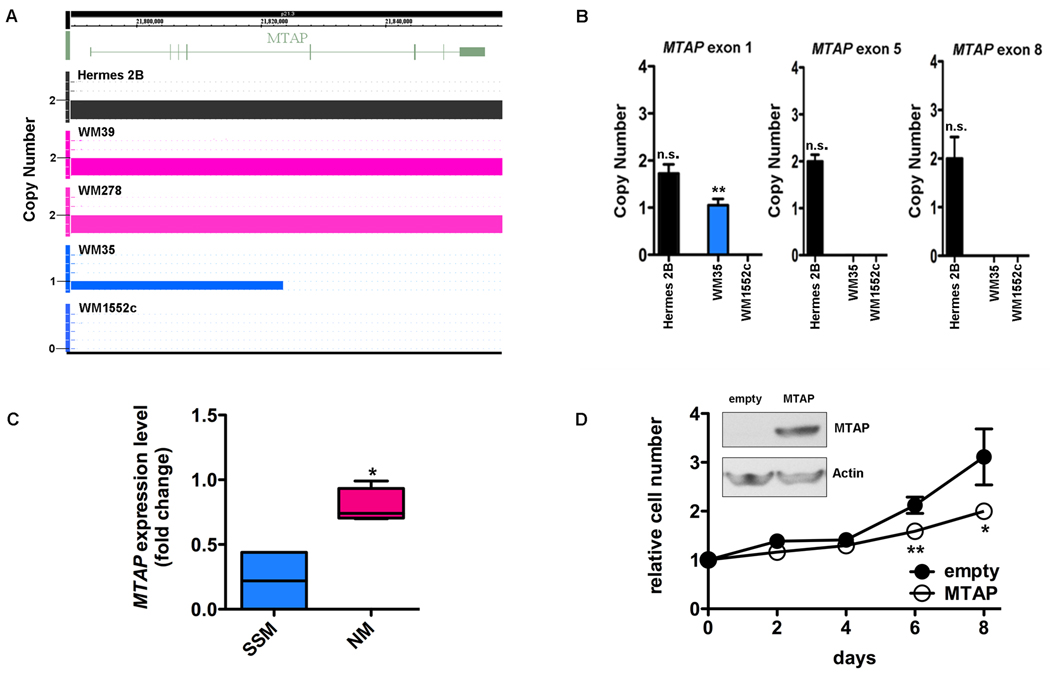
FIGURE 4.
A) SNP array in the region of MTAP shows that primary immortalized melanocyte Hermes 2B (control, black) and primary nodular melanoma cell lines (pink) have 2 copies of MTAP. There is a heterozygous deletion in SSM cell line WM35 (blue) spanning exons 1–4 and a homozygous deletion in exons 5–8. The deletion in WM1552c is homozygous and encompasses all exons. B) Genomic PCR of the MTAP gene in SSM cell line WM1552c confirms genomic losses in exons 1, 5, and 8, consistent with the large area of genomic deletion noted on SNP array. In SSM cell line WM35 (blue), only exons 5 and 8 are homozygously deleted which verifies the focal genomic loss detected using the SNP array. C) qRT-PCR verification of differential mRNA expression between NM (WM278, WM39, Lu451, SK-MEL-147) and SSM (WM1552c, WM35) cell lines in MTAP. D) Ectopic expression of MTAP in SSM cell line WM1552c by lentiviral infection (inset) results in decreased growth relative to control MTAP null cells. Mean ± SD, N=3. *P<0.05, **P<0.01, n.s.-not significant (relative to normal human genomic DNA in B).
Array results verified by in silico analysis of external datasets
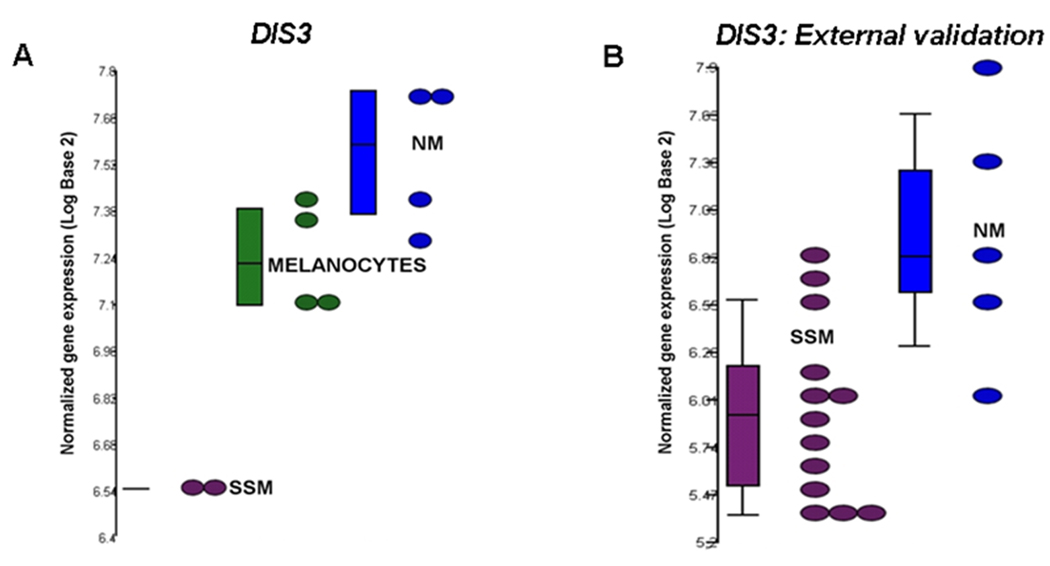
Re-analysis of a publically available gene expression data set by Jaeger et al (4) yielded a list of 291 transcripts differentially expressed between NM and SSM (not shown). Also used as an external validation was a data set from the 2009 study by Scatolini el al (15). All 5 genes with NM/SSM-specific copy number and gene expression changes validated using qPCR and qRT-PCR (USO1, G3BP2, DIS3, SEC23IP, MTAP) were validated as having the same trend of expression (ie, down in SSM relative to NM) in the external data set by Jaeger (DIS3, Figure 3 and not shown). Only 3 of these genes (MTAP, SEC23IP, and G3BP2) were included in the Scatolini data set, but all were validated as having the same trend of expression between subtypes, with G3BP2 also showing statistically significant lower expression in SSM relative to NM in the external data set (15).
FIGURE 3.
In silico analysis of an external validation data set (Jaeger, Clin Cancer Res; 2007). DIS3 in melanoma cell lines (A) shows lower expression in SSM (WM1552c, WM35) relative to both normal (green) and NM (WM278, WM39, Lu451, SK-MEL-147) which is validated in the external data set of human melanoma tissues (B).
Overexpression of MTAP in SSM reduces cell growth
SNP array identified two SSM-specific deletions in the region of MTAP (9p21.3) that were significantly correlated with gene expression (Figure 4A). The deletion in SSM cell line WM1552c is large and encompasses the entire gene (Figure 4A), while the deletion in SSM cell line WM35 is more focal, affecting only the last 4 of 8 exons (Figure 4A). Consistent with the array results, genomic PCR of the MTAP gene for cell line WM1552c demonstrated complete loss of DNA in all 3 exons assessed (exons 1, 5, and 8; Figure 4B). Genomic PCR of MTAP in SSM cell line WM35 showed retained DNA in exon1 but loss in exons 5 and 8 which is consistent with the position of the copy number loss identified by SNP array (Figure 4B). Furthermore, ectopic expression of MTAP by lentiviral infection caused a significant decrease in growth of SSM cell line WM1552c, supporting an oncosuppressor role (Figure 4D).
Validation of array results using western blot
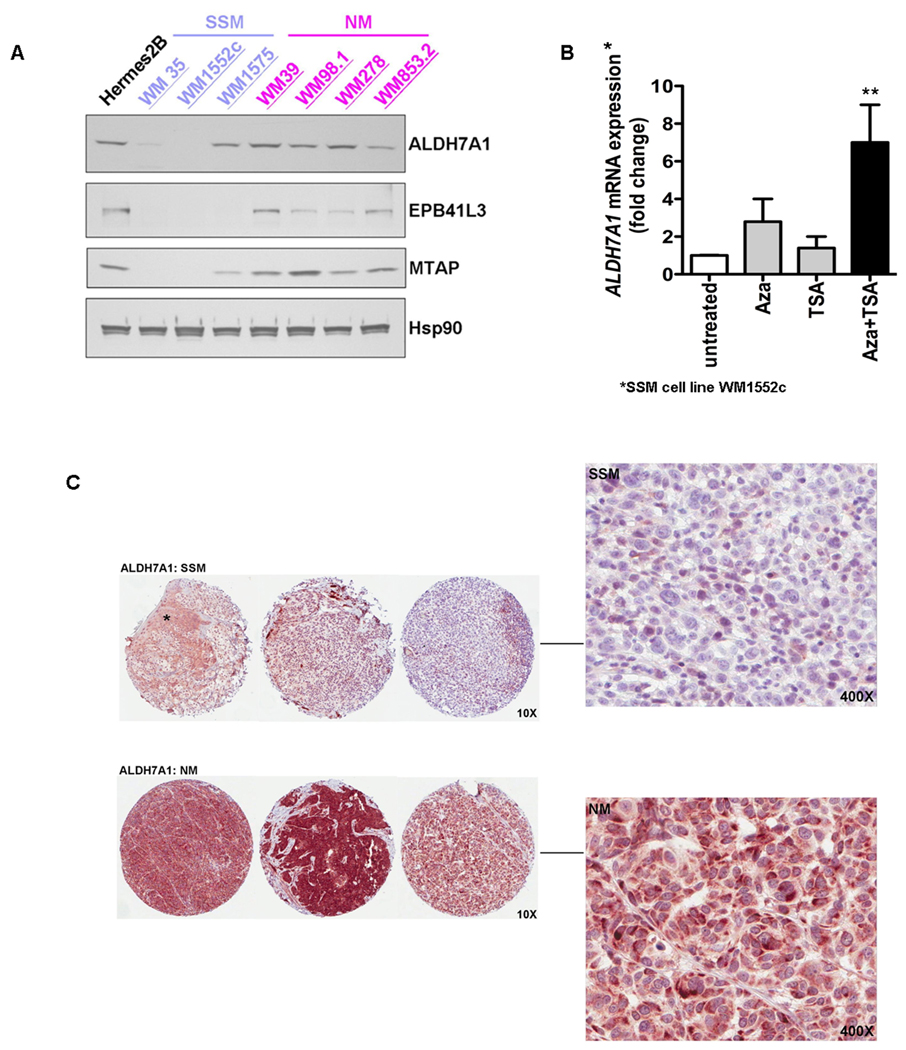
ALDH7A1 and EPB41L3 were also prioritized for protein validation studies because, second only to MTAP, they had the highest (ALDH7A1) and most significant (EPB41L3) fold changes noted in the gene expression array (Suppl. Table 5). Expression of ALDH7A1, EPB41L3, and MTAP was assessed by western blot in the same primary melanoma cell lines used in the array as well as an additional SSM cell line (WM 1575) and 2 additional RGP-like cell lines (WM98.1 and WM 853.2). Consistent with the array results, ALDH7A1, a gene involved in the detoxification of aldehydes generated by lipid peroxidation, showed complete loss of expression in 2 of the 3 SSM cell lines relative to NM and melanocyte controls (Figure 5A). Also consistent with the array results, EPB41L3 showed complete loss of expression in all SSM cell lines relative to melanocytes. EPB41L3 showed higher expression in NM cell lines relative to SSM but lower expression in NM relative to normal which also verified the array results (Figure 5A). MTAP expression was completely lost in the 2 SSM cell lines from the array (WM35 and WM 1552c) which was consistent with the genomic loss, and showed low expression in the additional SSM cell line (WM1575) relative to both NM and normal melanocytes.
FIGURE 5.
A) Western blot analysis of ALDH7A1, EPB41L3, and MTAP in an expanded panel of SSM (purple) and NM (pink) primary melanoma cell lines. Levels of Hsp90 protein served as loading control. B) There is a significant (**P<0.01) increase in ALDH7A1 mRNA in SSM WM1552c cells after treatment with 1uM azacytidine (Aza) and 200ng/ml Trichostatin A (TSA). C) Nodular melanoma tissue cores (bottom row) show higher expression of ALDH7A1 relative to SSM tissue cores. Cores were scored based on intensity (0–2). Normal skin (asterisk, SSM) served as an internal control (intensity score 1). On high-power (400X), tumor cells in SSM cores show very low level of expression with only scattered, rare cytoplasmic blush whereas the tumor cells in NM cores demonstrate diffuse, intense cytoplasmic staining with a distinctive perinuclear "dot-like” intensification (Golgi pattern).
Regulation of ALDH7A1 expression in SSM
We first performed mutational analysis of ALDH7A1 (exons 2, 12, 14; introns 5, 16) in 7 vertical growth phase (VGP) and 4 radial growth phase (RGP) cell lines that revealed no evidence of mutation (Suppl. Table 6). We then explored whether the lower expression of ALDH7A1 in SSM is due to the epigenetic mechanisms. We found a significant (P<0.01) increase in ALDH7A1 mRNA levels in SSM WM1552c cells after treatment with 1uM azacytidine (aza-CR) and 200ng/ml Trichostatin A (TSA) (Figure 5B). These data suggest that a combination of hypermethylation and deacetylation may be responsible for the lower expression of ALDH7A1 in SSM.
Validation of array results using immunohistochemistry (IHC)
Expression of ALDH7A1, EPB41L3, and MTAP was further evaluated using TMAs of 20 NM and 20 SSM human primary melanoma tissues. Baseline demographic and clinicopathologic data for these cases are presented in Table 2. ALDH7A1 expression was significantly higher in NM cores relative to SSM. The mean intensity score (on a scale from 0–2) of NM cores (N=20) was 1.62 (SD 0.43) compared to 1.03 (SD 0.47) in the SSM cores (N=20) (P<0.0001, t-test). In a multivariate general linear model controlling for tumor thickness and ulceration, the adjusted mean staining intensity score remained significantly higher in NM (1.67, SE 0.11, 95% CI = 1.45–1.88) relative to SSM (0.97, SE 0.11, 95% CI = 0.76–1.19) (P<0.0001). The ALDH7A1 positive cores demonstrated a distinctive, perinuclear "dot-like” intensification consistent with a Golgi pattern of staining (Figure 5C). Almost all cores that were strongly positive (score 2) for EPB41L3 demonstrated a membranous pattern of staining which was more common in SSM than NM [46% (N=18) and 11% (N=5), respectively; P=0.001] (data not shown). There was no significant difference noted in the intensity of MTAP staining between NM and SSM.
TABLE 2.
Baseline demographics and clinicopathologic variables of the 40 primary melanoma cases included on the SSM and NM tissue microarrays.
| SSM (N=20) | NM (N=20) | |
|---|---|---|
| Gender (N,%) | ||
| Male | 16 (80) | 13 (65) |
| Female | 4 (20) | 7 (35) |
| Age at Diagnosis, years | ||
| (median, range) | 62 (29–86) | 71.5 (35–90) |
| Stage (N, %) | ||
| Stage I | 11 (55) | 3 (15) |
| Stage II | 5 (25) | 8 (40) |
| Stage III | 4 (20) | 9 (45) |
| Stage IV | 0 (0) | 0 (0) |
| Thickness, mm | ||
| (median, range) | 1.45 (1–6) | 3.4 (1.24–24) |
| Ulceration (N,%) | ||
| Present | 4 (20) | 12 (60) |
| Absent | 16 (80) | 8 (40) |
| Mitotic index (N,%) | ||
| (per HPF) | ||
| Many | 5 (25) | 11 (55) |
| Moderate | 5 (25) | 7 (35) |
| Few | 10 (50) | 2 (10) |
| Anatomic site | ||
| Extremity | 5 (25) | 8 (40) |
| Axial | 13 (65) | 9 (45) |
| Head and neck | 2 (10) | 3 (15) |
| SLN Biopsy Positive (N, %) | ||
| Yes | 4 (20) | 5 (25) |
| No | 16 (80) | 15 (75) |
| Recurrence (N, %) | ||
| Yes | 5 (25) | 6 (30) |
| No | 15 (75) | 14 (70) |
| Status at last follow-up | ||
| Alive | 16 (80) | 14 (70) |
| Dead | 4 (20) | 6 (30) |
Abbreviations: SSM, superficial spreading melanoma; NM, nodular melanoma; HPF, high power field; SLN, sentinel lymph node biopsy
DISCUSSION
Our results suggest that SSM and NM are not sequential phases of linear progression but rather two different biological entities that are characterized by distinct molecular alterations. Genomic alterations that demonstrated a significant correlation with gene expression were predominantly SSM-specific deletions. Although there are many post-transcriptional and post-translational events that can affect levels of gene expression, the identification of recurrent SSM-deletions that are retained or even amplified in NM cannot be reconciled with the current paradigm of linear progression.
Gene ontology (GO) analysis of differentially expressed genes between SSM and NM showed enrichment for genes related to metabolic processes. Aldehyde dehydrogenase 7A1 (ALDH7A1) demonstrated higher expression in human NM tissues relative to SSM that remained significant in a multivariate model controlling for thickness and ulceration. Recent studies have demonstrated that expression of aldehyde dehydrogenases can identify the presence of cancer stem cell populations in hepatocellular carcinoma (HCC) and breast cancer (18, 19). Overexpression of ALDH7A1 in NM versus SSM may reflect differences in the percentage of tumor initiating cells that could explain NM’s rapid vertical growth. Not only was expression of ALDH7A1 higher in NM relative to SSM and normal skin but SSM expression was lower in SSM relative to normal skin. Our data suggest that, in the absence of genomic losses and premature STOP codons, the lower expression of ALDH7A1 in SSM may be due to an epigenetic mechanism. ALDH7A1 is highly conserved throughout evolution and is homologous to ALDH7B1 in plants where overexpression of the enzyme is believed to indicate a global upregulation of the organism’s response to environmental stress (20). Similarly, ALDH7 and other ALDH family members are highly expressed in the lens of the murine eye where they have a protective effect against ultraviolet (UV)-induced DNA damage (21). Thus, it is possible that the loss of ALDH7A1 observed in SSM makes the subtype more susceptible to and/or more likely to be the result of carcinogenic exposures such as UV light.
The utilization of an integrative genomic approach makes it more likely that the alterations identified are biologically relevant. Genes previously identified as differentially expressed between NM and SSM using microarrays have not been further studied at the functional level, thus the underlying mechanisms that contribute to differential gene expression are not understood. Of note, one previous gene expression study of RGP and VGP melanoma revealed the unexpected finding that, within the same patient, the expression profile of the metastatic melanoma was consistent with a RGP-like signature, whereas the nodular primary exhibited a separate, VGP-like expression profile (22). This observation is consistent with our principal component analysis of melanoma cell lines showing that SSM primary cell lines cluster with metastatic lesions, whereas all of the primary NMs fall outside of this cluster. Based on the relatively aggressive clinical behavior of NM relative to SSM, it might be expected that NM would cluster with the metastatic lesions. While we recognize the exploratory nature of the PCA, the findings from these two independent studies showing that gene expression profiles of SSM and metastatic lesions are more closely related than the profiles of nodular primaries supports the hypothesis that NM and SSM represent different biological processes.
An array-based integrative genomic workflow and subsequent verification studies identified SSM-specific deletions in genes that were significantly correlated with gene expression. USO1 and G3BP2 are adjacent genes on chromosome 4q21.1. USO1 has been primarily studied in yeast where it is involved in transport between the endoplasmic reticulum and the Golgi apparatus (23). It is notable that both of our in silico analyses of external data sets confirmed our results showing lower expression of G3BP2 in SSM relative to NM (4, 15). G3BP2 is a relatively recently characterized GTP-ase activating protein that is overexpressed in breast cancer (24) and that plays a role in the regulation of NFkB signaling (25), a pathway known to be altered in melanoma.
DIS3 has been shown to function in the regulation of proper chromosomal segregation during mitosis, and DIS3 mutant yeast cells have increased sensitivity to microtubule destabilizing agents such as thiabendazole (26). In this regard, molecular compound library screening studies by our group showed that mebendazole, a microtubule-destabilizing drug, demonstrated subtype specific efficacy such that SSM cell lines were sensitive, but NM cells were resistant (5). Further supporting the potential biologic relevance of the genes identified by our integrative genomic analysis, two independent gene expression profiling studies of colorectal cell lines and human tissues have identified overexpression of DIS3 as high as 38-fold in primary and metastatic tumors relative to normal colonic mucosa (27, 28). Thus, the overexpression and increased DNA copy number indentified in the region of DIS3 in our NM cell lines may play a role in its rapid vertical growth and aggressive behavior. The same gene, however, also shows genomic deletion and reduced expression in SSM cell lines. Thus, SSM-specific genomic deletions such as the ones noted in the region of DIS3 may play a mechanistic role in the observed increased sensitivity of SSM cells to microtubule destabilizing agents.
A previous gene expression profiling study identified SEC23IP as an important mediator of bone formation and metabolism (29). Interestingly, 3 of the top candidate genes from that study (IFIT3, SEC23IP, and PPP2R2D) were also found to be among the most differentially expressed between NM and SSM in our study. While the link between these two investigations is not immediately apparent, many pathways known to play a role in carcinogenesis, such as interferon signaling and other cytokines, are also important in bone formation and metabolism (30). SEC23IP has also recently been identified as a candidate gene for Waardenberg syndrome, a disorder characterized by craniofacial defects and pigment abnormalities, after it was shown that knockdown of sec23ip in the Xenopus embryo resulted in impaired migration of neural crest cells (31).
Methylthioadenosine phosphorylase (MTAP), maps to 9p21.3 and is commonly co-deleted with tumor suppressor p16 (CDKN2A) (32). MTAP plays a critical role in the salvage pathway of adenine and methionine and has previously been reported to be lost in several tumors including melanoma (33). Our results demonstrate lower expression of MTAP in SSM relative to NM which was significantly correlated with SSM-specific genomic deletions in the region of 9p21.3. MTAP loss in SSM may be particularly relevant given the recent discovery of MTAP variants that are associated with increased number of nevi and with increased risk of melanoma (34, 35). The role of somatic alterations in MTAP with regard to prognosis and tumorigenesis is less clear, but our results suggest that MTAP deletions may be more common in SSM than NM. Additionally, lentiviral-mediated overexpression of MTAP in an SSM cell line with a homozygous deletion resulted in decreased tumor growth compared to MTAP null cells, suggesting that MTAP may function as a tumor suppressor in a subtype-specific fashion. Previous studies have shown that MTAP status may be valuable as a predictor of response to interferon, such that only treatment of patients with MTAP positive tumors confers a survival advantage compared to those untreated (36,37). Thus, if MTAP deletions differentially affect SSM relative to NM, the MTAP status of a tumor may play a role in the development of a subtype-specific treatment strategy. Because MTAP-deficient cells lose the salvage pathway for purine synthesis, they are sensitive to agents that target the de novo purine synthesis pathway. A recent study that evaluated the efficacy of L-alanosine, an inhibitor of de novo adenine synthesis, for the treatment of MTAP-deficient solid malignancies showed no objective responses (38). The authors suggested that improved patient selection based on more standardized, DNA-based techniques for identifying MTAP loss such as fluorescent in situ hybridization (FISH) or genomic PCR might improve results of future studies.
In conclusion, our data support a separate molecular classification of SSM and NM. Further functional studies of genes that are specifically deleted in SSM may better define their roles as potential drivers of subtype specific tumorigenesis.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yutong Zhang of the New York University Cancer Institute and Connie Zhao of the Genomics Resource Center of Rockefeller University for technical assistance in performing the gene and SNP arrays.
Financial Support: This work was supported by NYU Cancer Institute Cancer Center Support Grant (5 P30 CA016087), the Chemotherapy Foundation, the Skin Cancer Foundation, the Elsa Pardee Foundation, and the Marc Jacobs Campaign to support melanoma research (IO). The work was partially supported by the Cornell Clinical Translational Science Center (UL1-RR024996) to PC.
Footnotes
Financial Disclosures: Nothing to disclose.
References
- 1.Corrigan MA, Coffey JC, O'Sullivan MJ, Fogarty KM, Redmond HP. Sentinel lymph node biopsy: is it possible to reduce false negative rates by excluding patients with nodular melanoma? Surgeon. 2006;4:153–157. doi: 10.1016/s1479-666x(06)80085-5. [DOI] [PubMed] [Google Scholar]
- 2.Nowecki ZI, Rutkowski P, Nasierowska-Guttmejer A, Ruka W. Survival analysis and clinicopathological factors associated with false-negative sentinel lymph node biopsy findings in patients with cutaneous melanoma. Ann Surg Oncol. 2006;13:1655–1663. doi: 10.1245/s10434-006-9066-0. [DOI] [PubMed] [Google Scholar]
- 3.Warycha MA, Christos PJ, Mazumdar M, et al. Changes in the presentation of nodular and superficial spreading melanomas over 35 years. Cancer. 2008;113:3341–3348. doi: 10.1002/cncr.23955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaeger J, Koczan D, Thiesen HJ, et al. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin Cancer Res. 2007;13:806–815. doi: 10.1158/1078-0432.CCR-06-1820. [DOI] [PubMed] [Google Scholar]
- 5.Doudican N, Rodriguez A, Pavlick A, Shapiro R, Osman I, Orlow S. Mebendazole induces apoptosis in distinct melanoma subtypes via Bcl-2 dependent mechanism. J Clin Oncol. 2008;26 (May 20 suppl; Abstract #20008. [Google Scholar]
- 6.Ng B, Zakrzewski J, Warycha M, et al. Shedding of distinct cryptic collagen epitope (HU177) in sera of melanoma patients. Clin Cancer Res. 2008;14(19):6253–6258. doi: 10.1158/1078-0432.CCR-07-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnhill RL, Mihm MC., Jr The histopathology of cutaneous malignant melanoma. Semin Diagn Pathol. 1993;10:47–75. [PubMed] [Google Scholar]
- 8.Ackerman AB. Malignant melanoma. A unifying concept. Am J Dermatopathol. 1980;2:309–313. doi: 10.1097/00000372-198000240-00004. [DOI] [PubMed] [Google Scholar]
- 9.Chernoff KA, Bordone L, Horst B, et al. GAB2 amplifications refine molecular classification of melanoma. Clin Cancer Res. 2009;15:4288–4291. doi: 10.1158/1078-0432.CCR-09-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smalley KS, Contractor R, Nguyen TK, et al. Identification of a novel subgroup of melanomas with KIT/cyclin-dependent kinase-4 overexpression. Cancer Res. 2008;68:5743–5752. doi: 10.1158/0008-5472.CAN-08-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 12.Wyman K, Atkins MB, Prieto V, et al. Multicenter Phase II trial of high-dose imatinib mesylate in metastatic melanoma: significant toxicity with no clinical efficacy. Cancer. 2006;106:2005–2011. doi: 10.1002/cncr.21834. [DOI] [PubMed] [Google Scholar]
- 13.Korn JM, Kuruvilla FG, McCarroll SA, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajaram S, Oono Y. NeatMap - non-clustering heat map alternatives in R. BMC Bioinformatics. 11:45. doi: 10.1186/1471-2105-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scatolini M, Grand MM, Grosso E, et al. Altered molecular pathways in melanocytic lesions. Int J Cancer. 2010;126:1869–1881. doi: 10.1002/ijc.24899. [DOI] [PubMed] [Google Scholar]
- 16.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazar V, Ecsedi S, Szollosi AG, et al. Characterization of candidate gene copy number alterations in the 11q13 region along with BRAF and NRAS mutations in human melanoma. Mod Pathol. 2009;22:1367–1378. doi: 10.1038/modpathol.2009.109. [DOI] [PubMed] [Google Scholar]
- 18.Ma S, Chan K, Lee T, et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res. 2008;6:1146–1153. doi: 10.1158/1541-7786.MCR-08-0035. [DOI] [PubMed] [Google Scholar]
- 19.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stroeher VL, Boothe JG, Good AG. Molecular cloning and expression of a turgor-responsive gene in Brassica napus. Plant Mol Biol. 1995;27:541–551. doi: 10.1007/BF00019320. [DOI] [PubMed] [Google Scholar]
- 21.Stagos D, Chen Y, Cantore M, Jester J, Vasiliou V. Corneal aldehyde dehydrogenases: multiple functions and novel nuclear localization. Brain Res Bull. 2010;81:211–218. doi: 10.1016/j.brainresbull.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haqq C, Nosrati M, Sudilovsky D, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noda Y, Yamagishi T, Yoda K. Specific membrane recruitment of Uso1 protein, the essential endoplasmic reticulum-to-Golgi tethering factor in yeast vesicular transport. J Cell Biochem. 2007;101:686–694. doi: 10.1002/jcb.21225. [DOI] [PubMed] [Google Scholar]
- 24.French J, Stirling R, Walsh M, Kennedy HD. The expression of Ras-GTPase activating protein SH3 domain-binding proteins, G3BPs, in human breast cancers. Histochem J. 2002;34:223–231. doi: 10.1023/a:1021737413055. [DOI] [PubMed] [Google Scholar]
- 25.Prigent M, Barlat I, Langen H, Dargemont C. IkappaBalpha and IkappaBalpha /NF-kappa B complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J Biol Chem. 2000;275:36441–36449. doi: 10.1074/jbc.M004751200. [DOI] [PubMed] [Google Scholar]
- 26.Murakami H, Goto D, Toda T, et al. Ribonuclease activity of Dis3 is required for mitotic progression and provides a possible link between heterochromatin and kinetochore function. PloS One. 2007;2 doi: 10.1371/journal.pone.0000317. e317-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang L, Qu L, Ding Y. Protein and mRNA characterization in human colorectal carcinoma cell lines with different metastatic potentials. Cancer Invest. 2007;25:427–434. doi: 10.1080/07357900701512258. [DOI] [PubMed] [Google Scholar]
- 28.Lim J, Kuroki T, Ozaki K, et al. Isolation of murine and human homologues of the fission-yeast dis3+ gene encoding a mitotic-control protein and its overexpression in cancer cells with progressive phenotype. Cancer Res. 1997;57:921–925. [PubMed] [Google Scholar]
- 29.Alam I, Sun Q, Koller D, et al. Differentially expressed genes strongly correlated with femur strength in rats. Genomics. 2009;94:257–262. doi: 10.1016/j.ygeno.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takayanagi H, Kim S, Matsuo K, et al. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature. 2002;416:744–749. doi: 10.1038/416744a. [DOI] [PubMed] [Google Scholar]
- 31.McGary K, Park T, Woods J, Cha H, Wallingford J, Marcotte E. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc Natl Acad Sci U S A. 2010;107:6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conway C, Beswick S, Elliott F, et al. Deletion at chromosome arm 9p in relation to BRAF/NRAS mutations and prognostic significance for primary melanoma. Genes Chromosomes Cancer. 2010;49:425–438. doi: 10.1002/gcc.20753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wild P, Meyer S, Bataille F, et al. Tissue microarray analysis of methylthioadenosine phosphorylase protein expression in melanocytic skin tumors. Arch Dermatol. 2006;142:471–476. doi: 10.1001/archderm.142.4.471. [DOI] [PubMed] [Google Scholar]
- 34.Falchi M, Bataille V, Hayward N, et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat Genet. 2009;41:915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bishop DT, Demenais F, Iles M, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wild PJ, Meyer S, Landthaler M, et al. A potential predictive marker for response to interferon in malignant melanoma. J Dtsch Dermatol Ges. 2007;5(6):456–459. doi: 10.1111/j.1610-0387.2007.06303.x. [DOI] [PubMed] [Google Scholar]
- 37.Meyer S, Wild PJ, Vogt T, et al. Methylthioadenosine phosphorylase represents a predictive marker for response to adjuvant interferon therapy in patients with malignant melanoma. Exp Dermatol. 2010;19(8):e251–e257. doi: 10.1111/j.1600-0625.2010.01072.x. [DOI] [PubMed] [Google Scholar]
- 38.Kindler H, Burris H, Sandler A, Oliff I. A phase II multicenter study of L-alanosine, a potent inhibitor of adenine biosynthesis, in patients with MTAP-deficient cancer. Invest New Drugs. 2009;27:75–81. doi: 10.1007/s10637-008-9160-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.