Abstract
Survivin is a universal tumor antigen that is being currently targeted in vaccine app roaches against cancer. Our study here examined the immunogenicity of a novel variant of an HLA-A0201-binding decamer peptide from region 95-104 of Survivin (ELMLGEFLKL) with a T→M modification at position 3 in the peptide. We found that this new modified 10-mer peptide had enhanced HLA-A0201 binding and induced a stronger T-cell response over its wild type counterpart peptide (ELTLGEFLKL) in select HLA-A0201+ normal donors. In addition, when compared to the previously characterized altered 96-104 peptide (LMLGEFLKL) from the same region of Survivin currently used in vaccine trials, we found that both peptides had similar immunogenicity, but donor T cells preferentially reacted strongly to either one or the other, but not strongly to both. These results suggest that these two closely related Survivin peptides yield distinct T-cell responses and that most individuals dominantly respond to one or the other altered peptide. We also found a novel association between positive reactivity to the new altered decamer Survivin peptide in some individuals and their expression of the HLA-C0701 allele along with HLA-A0201. Thus, vaccinating with both the 10-mer and 9-mer peptides would be required to immunize a maximum number of individuals in the HLA-A0201+ population and could lead to more consistent T-cell responses against this region of Survivin.
Keywords: Tumor antigen, Survivin, HLA-A0201, epitope, T cell
INTRODUCTION
Cancer evades the body’s defense system because it is an “altered self” and not perceived as a foreign threat thus rendering it very poorly immunogenic. However the subtle differences between normal and cancerous cells can still be noticed by the immune system. Tumor-associated antigens (TAA) are self antigens that are over-expressed and/or mutated in the cancerous cell while being absent or expressed at very low levels in normal cells. This aberrant expression generates new epitopes that are displayed on HLA class I and II molecules on the surface of the cancer cell that can be cross-presented by antigen-presenting cells (APC). It should signal the expansion of specific T cells able to kill the malignant cell. Indeed tumor-specific T cells do infiltrate tumors and there is an inverse correlation between number of infiltrating T cells and tumor growth or metastasis potential [1]. However, numerous tumor escape mechanisms can prevent activation or effector function of T cells. Proper activation of the immune system toward tumors requires the “unveiling” of the cancer neo-epitopes. Much effort in the field of tumor immunotherapy has been focused on the identification of specific cytotoxic CD8+ T cell (CTL) epitopes derived from different TAA for vaccination purposes.
It is now becoming clear that in order to avoid tumor escape it is imperative to target a tumor antigen that is essential to the tumor survival. One of these antigens is Survivin (Sur). It is expressed at high levels in virtually all malignancies and commonly referred to as a universal tumor antigen, while having no or very low expression in normal tissues. Studies have identified immunogenic epitopes restricted by different HLA class I molecules in the Sur sequence [2-6].
Results from phase I clinical trials using Sur-derived peptides have been published. The peptides used were restricted by HLA-A0201 or HLA-A24 which are respectively the most prevalent HLA subtypes in Caucasian and Asian populations. Both vaccinations have proven to be safe in a Phase I clinical trials [7-10]. The efficacy reported with the HLA-A24 peptide vaccination was limited to minor clinical response even if immune response to the peptide was induced. A Phase I clinical trial in HLA-A0201+ patients with a peptide from the 96-104 region of Sur with a T→M substitution at position 2 (LMLGEFLKL) was immunogenic in a subset of patients and induced an infiltration of peptide-specific CD8+ T-cells into tumors. Another case report showed that vaccination with this same peptide in a pancreatic cancer patient induced a complete remission of a metastatic lesion in the liver [11]. These initial clinical studies revealed that Sur is immunogenic and that immune response directed at this protein could lead to tumor regression. However, the reports indicated that only a fraction of the patients develop an immune response to the studied peptide and even among these groups, a limited number show a clinical response. These pitfalls could potentially be overcome by improving the immunogenicity of Sur peptides used as well as by optimization of the vaccine adjuvant and patient selection.
The altered HLA-A0201 (HLA-A0201) Sur 96-104 nonamer peptide (96-104m) that has been used in the clinic has increased HLA-A0201 binding and boosts CD8+ T-cell responses while retaining the capacity of the expanded T cells to recognize the wild-type peptide [2]. Evidence suggests that the 96-104 Sur peptide is naturally processed and presented by cancer cells, induces an immune response and is able to elicit positive clinical response. This illustrates the immunogenicity of this segment of Sur. The closely related decamer 95-104 peptide (95-104wt) has also been shown to be naturally processed and presented by cancer cells and has been tested also in clinical trials using peptide-pulsed dendritic cells (DC) in prostate cancer patients [12]. However, its binding to HLA-A0201 is weaker than 96-104m and may limit its immunoreactivity. Modifications of the 95-104wt peptide have not been tested; these may trigger enhanced HLA-A0201-binding and enhanced reactivity.
In this paper, we designed and tested an altered 95-104 Sur peptide with a T→M change at position 3 (referred to as 95-104m) to determine its HLA-A0201-binding activity and immunogenicity. We found that this new altered decamer Sur peptide bound HLA-A0201 with higher affinity than its wild-type counterpart and expanded antigen-specific CD8+ T-cell responses in normal donors much more that 95-104wt and with a comparable magnitude or stronger than 96-104m. T-cells activated with 95-104m recognized Sur-over-expressing tumor cells. Interestingly, during our studies driving out normal donor CD8+ T-cell lines with the altered nonamer or the new altered decamer, we found a dichotomy in the immunogenicity of the two peptides. This was manifested in the observation that certain individuals could raise a T cell line by repeated stimulations with 95-104m, but remarkably much weaker activation of T cells was obtained in a parallel culture using 96-104m peptide, or vice versa. Interestingly, when using a mixture of both peptides in repeated stimulations the resulting expanded T cells were dramatically less reactive to either peptide. To summarize, reactivity to a mixture of both peptides was poor, although when used separately over 80% of the HLA-A0201+ normal donors were able to mount an immune response to at least one of the peptides, and in each case, reactivity to one peptide or the other dominated the response. These results suggest 95-104m peptide may increase the immunogenicity of the 95-104 Sur peptide used in clinical trials and that both 96-104m as well as 95-104m should be used in peptide vaccine clinical trials to improve immunization levels in the HLA-A0201+ patient population against this Sur 95-104 region.
MATERIAL AND METHODS
Cell lines and reagents
T2 and MDA-MB 453 breast cancer cell lines were obtained from ATCC. All basal media used to grow cell lines contained 1 mM Glutamax™ (Invitrogen, Carlsbad, CA), a stable form of glutamine and were supplemented with 10% fetal bovine serum (FBS), Penicillin/Streptomycin, and 50 mM 2-mercaptoethanol. T2 cells were maintained in RPMI 1640. MDA-MB-453 and CF-Pac-1 cells were grown in DMEM (Invitrogen) supplemented as above. Melanoma cell line, Mel 2320, was maintained in RPMI 1640 supplemented as above. MDA-MB 453 cells expressing HLA-A0201 were generated by transducing the cell line with HLA-A0201-expressing lentiviral construct, as previously described [13]. Peptides were obtained from SynBioSci (California) or RS Synthesis LLC (Louisville, KY) and were HPLC purified to at least 95% purity. The following HLA-A0201-binding Sur peptides were used: LMLGEFLKL, LTLGEFLKL, ELMLGEFLKL, and ELTLGEFLKL. The HLA-A0201-binding peptides, ILKEPVHGV or SLYNTVATL, from HIV rev and gag, respectively, were used as negative controls. Functional grade (azide free) anti-HLA-Class I antibody clone W6/32 was from eBioscience (San Diego, CA).
Peptide binding assay using T2 cells
T2 cells (HLA-A0201+ and TAP2-deficient) were washed and cultured for 5 h in serum free AIM-V medium. Cells were then further washed, plated at 250,000 cells per well in a 48-well plate and incubated overnight with the different peptides at various concentrations. The culture was supplemented with 5 μg/ml β2-microglobulin in a final volume of 600 μl of serum free AIM-V medium. The next day the cells were stained for HLA-A2 expression using purified monoclonal anti-HLA-A2 antibody (clone BB7.2, purified in house from the hybridoma obtained from ATCC) followed by incubation with Alexa Fluor 488 labeled goat anti-mouse IgG (Invitrogen, Carlsbad, CA). Dead cells were excluded from the analysis by 7AAD staining. Samples were read on BD FACSCanto II flow cytometry analyzer using FACSDiva and analyzed with FlowJo software.
PBMC, HLA typing, and selection of HLA-A0201+ samples
Buffy coats from normal donors were obtained from Gulf Coast Regional Blood Bank (Houston, TX) and processed the same day the sample was drawn. PBMC were isolated from buffy coats by density separation (HistoPaque1077, Sigma-Aldrich) and cryopreserved. An aliquot of cells was sent to the MD Anderson Cancer Center’s HLA-typing lab for high resolution molecular typing. Only PBMC from donors confirmed to be HLA-A0201 by high resolution typing were used in subsequent experiments.
Generation of peptide-specific T cell lines
T cell lines were raised using either HLA-A0201+ PBMC enriched for T-cells by prior depletion of monocytes using a plastic-adherence step (90 min at 37°C), or enriched CD8+ T cells obtained by subjecting total PBMC to negative selection of CD8+ T cells using magnetic beads (Miltenyi). Peptide-specific T cell lines were generated using repeated stimulations with peptide-pulsed autologous antigen-presenting cells (APC) in T-cell culture medium (TC-CM) consisting of Iscove’s Modified Dulbecco’s Medium containing Glutamax supplemented with 10% human AB serum (Sigma), 20μg/ml gentamycin, 1mM pyruvate, and 50μM β-mercaptoethanol. The primary stimulation was done using irradiated (2,000 Rads) peptide-loaded autologous mature dendritic cells (DC), at a DC:T-cell ratio 1:10, and subsequently using irradiated (2,000 Rads) peptide-pulsed autologous CD40-ligand-activated B cells for subsequent rounds of stimulation (at a B cells:T cell ratio of 1:1). For each stimulation, the APC were pulsed with 20 μg/ml of each indicated peptide for 2-3 h at 37°C in media supplemented with 2-2.5 μg/ml human β2-microglobulin (Sigma-Aldrich) and washed once before being used to stimulate T cells. The pulsed APC were mixed with PBMC enriched for T cells (6 × 106 per well) or purified CD8+ T cells (2 × 106 per well). Each round of stimulation lasted for a period of 10-12 days. Cells were fed with cytokines every 3-4 days (20 U/ml IL-2 initially and 5 ng/ml IL-7 after day 7). DC were differentiated from plastic-adherent monocytes isolated from donor PBMC cultured with 1,000 ng/ml GM-CSF and 1,000 ng/ml IL-4 and matured using 20ng/ml TNF-α and 100 U/ml IFN-α as previously described [14]. Mature DC expressed high levels of CD86 and HLA class II, intermediate levels of CD80 and CD83 and were positive for CD11c and negative for CD14 (data not shown). Activated B cells were generated using a previously published methodology [15]. Briefly, PBMC were cultured over a bed of irradiated (10,000 Rads) CD40L-expressing 3T3 fibroblasts with 100 U/ml recombinant human IL-4, 5μg/ml recombinant human insulin and 0.6 μM cyclosporin A. The cells were re-stimulated every 4-5 days with 3T3 CD40L cells in fresh media containing the same additives. B cells were used as APC for the second to fifth round of stimulation.
IFN-γ ELISPOT assay
T cell lines were assessed for reactivity to the stimulating peptide using IFN-γ ELISPOT after the third round of stimulation with peptide-pulsed APC. Briefly 96 well ELISPOT plates (Millipore) were coated over night with 5 μg/ml anti-IFN-γ capture antibody (clone Mab 1-D1K, Mabtech) at 4°C in carbonate-bicarbonate buffer pH 9.6. The plates were washed 3 times with PBS and blocked with PBS, 2% BSA for 45 min at room temperature. After three washes in PBS the T cells and peptide were added to the wells in 200 μl final volume (200,000 T cells per well with 20 μg/ml final peptide concentration in TC-CM) and incubated for 16-18 h at 37°C. The plates were further washed with water and ELISPOT wash buffer (PBS, 0.05% Tween-20). A biotinylated anti-IFN-γ detection antibody (clone Mab 7-B6-1, Mabtech) was then added at 1 μg/ml (100μl/well) in ELISPOT wash buffer containing 1% BSA (blocking buffer) and the plates incubated at 37°C for 1 h and then washed 3 times in water and ELISPOT buffer. The plates were further incubated with Extravidin-alkaline phosphatase (Sigma) diluted 1/5000 in blocking buffer (100μl/well) for 60-90 min at room temperature. The washed plates were developed with BCIP/NBT substrate (Sigma) for few minutes or until the spots are visible. The reaction was stopped by rinsing with water. The plates were allowed to dry and the spots were quantified using an ELISPOT reader (C.T.L. Technologies, Minneapolis, MN).
CTL assays
A 51Cr release assay was used to evaluate CTL-mediated killing of tumor cell lines. Target cells (2-4 × 106) were labeled with 100 μCi of 51Cr for 30-60min at 37°C. Labeled target cells were subsequently washed with serum-containing culture media and plated at 5000 target cells/well in a 96 well round bottom plate. Effector T cells were mixed at different ratios with the target cells in a total volume of 200 μl and incubated for 4-6h at 37°C. For experiments using peptide-pulsed targets, the labeled targets were pre-incubated at 1 × 106/ml with 20 μg/ml of peptide for 1 h and washed before mixing with the effector cells. In every experiment target cells alone (without effector cells) was used to determine the spontaneous release and target cells plus 2% Triton X-100 was used to obtain maximal release. CTL activity was calculated as percent specific 51Cr release (% release), where % release = (Experimental release-minimum release/Maximum release-minimum release) × 100. CTL assays were also performed using a flow cytometry caspase 3-cleavage assay in target cells as described previously [16]. Briefly, the target cells were labeled with DDAO-SE (Molecular Probes-Invitrogen) and washed. The labeled targets cells were incubated with T cells from the peptide-activated T cell lines at different effector:target ratios for 4 h and then fixed, permeabilized, and stained with a PE-conjugated anti-cleaved caspase 3 rabbit monoclonal antibody. Stained cells were acquired in a FACScanto II flow cytometer with data analysis using FlowJo software.
Molecular modeling of peptides interaction with HLA-A0201 molecule
The predicted structure of Sur peptides bound in the HLA-A0201 binding groove was generated by adapting available crystallographic data from HIV gag peptide bound to HLA-A0201 (pdb2C7U). The HIV peptide was removed and replaced with Sur peptides. Structure analysis was conducted using PyMol software to measure the distance between specific atoms to determine the distance between the ‘floor’ of the HLA-A0201 groove and the highest point reached by the peptide.
RESULTS
Sur 95-104m and 96-104m bind to HLA-A0201 with similar affinity
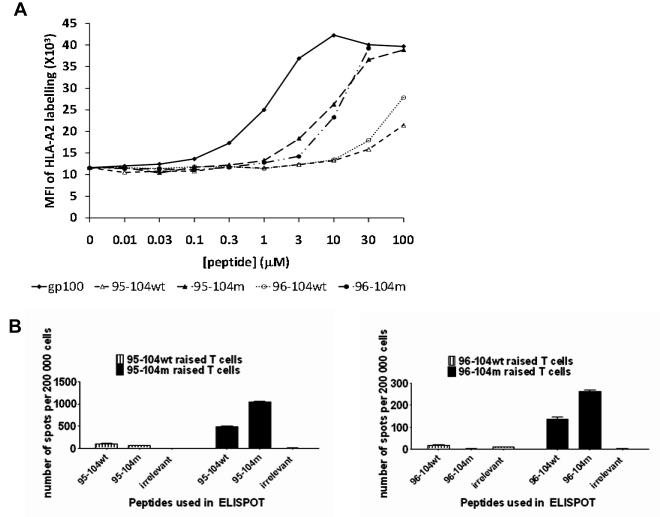
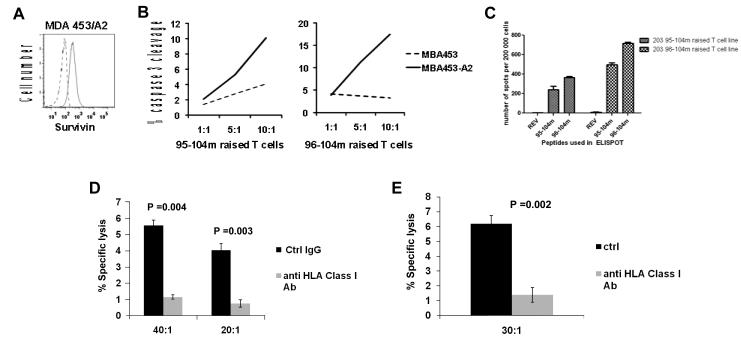
We first examined the binding Sur 96-104m and the new 95-104m peptides to HLA-A0201 molecules compared to their respective wild-type peptides using a classical T2 binding assay. Due to a defect in TAP molecules expression, HLA class I molecules produced in T2 cells are not loaded with endogenous peptides and therefore are not stably expressed at the surface of T2 cells unless an exogenous peptide is bound. Thus, this assay measures the stabilization of cell surface HLA-A0201 expression (as an increased MFI) caused by the binding of the test peptide to unstable empty HLA molecules in the T2 cells [17]. As shown in Fig. 1A, 95-104m induced a marked increase in HLA-A0201 MFI at the different concentrations over its wild-type counterpart and induced a similar dose-dependent increase in HLA-A0201 expression as 96-104m. The modified gp100 peptide 209-217 (210M) [18] was included as a known good binder and positive control (Fig. 1A). In both cases, the wild-type peptide did not show saturation in the concentration range shown. Thus, the new altered 95-104m peptide has a much greater affinity for HLA-A0201 than its wild type counterpart. The respective affinity of 95-104m and 96-104m peptides for HLA-A0201 molecule was found to be comparable.
Fig. 1.
A. Binding of Survivin-derived peptides from the 95-104 amino acid sequence to the HLA-A0201 molecule. Wild type (wt) or modified (m) versions of the peptides were compared. 95-104wt (ELTLGEFLKL), 95-104m (ELMLGEFLKL), 96-104wt (LTLGEFLKL) or 96-104m (LMLGEFLKL) have been tested in a classical T2 binding assay for their capacity to stabilize HLA-A0201 expression on the surface of T2 cells (see methods). Peptide concentrations of 0.01-100uM have been tested. Results of one out two experiments with similar results are shown. B. Altered 95-104 and 96-104 peptides had increased immunogenicity in comparison to their wild type counterpart. Enumeration of cells secreting IFN-γ by ELISPOT after 3 rounds of stimulation using 95-104wt or 95-104m (left panel), 96-104wt or 96-104m (right panel). Data from left and right panel are derived from one donor. Similar data has been obtained with two additional donors.
Sur 95-104m and 96-104m are both immunogenic although they selectively activate different populations of normal HLA-A0201+ donors
We then proceeded to test the immunogenicity of Sur 95-104m peptide in comparison to the 96-104m peptide in T cells from normal HLA-A0201+ donors. Peptide-specific T cell lines were raised by repeatedly activating the cells with peptide-pulsed autologous APC. The APC were pulsed with 20 μM of peptide, a concentration that resulted in a near saturation of HLA-A0201 molecules on T2 cells for both peptides (Fig. 1A). After three rounds of stimulation, the T cells were assayed for peptide specificity by measuring IFN-γ release in an ELISPOT assay. The T-cell lines raised against any of the two peptides consisted of >80% CD8+ T cells after three stimulations (data not shown). We first tested whether 95-104m peptide was immunogenic and induced better HLA-A0201-restricted T-cell responses compared to the wild-type peptide. As shown in Fig. 1B, similar to 96-104m peptide, the new 95-104m peptide induced T-cells were capable of peptide-specific IFN-γ production at markedly higher levels than its wild-type counterpart. Thus, as predicted by the T2 binding assays, both altered peptides induced increased antigen-specific T-cell responses.
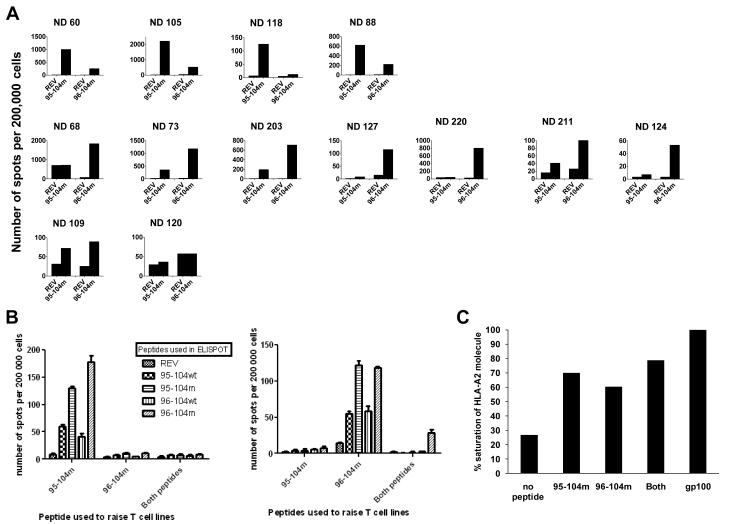
We then compared the immunogenicity of 95-104m versus 96-104m using IFN-γ ELISPOT in T cell cultures raised from 13 HLA-A0201+ normal donors. As before, we performed three repeated stimulations of T-cells from PBMC using autologous peptide-pulsed APC after which IFN-γ ELISPOT was performed. Both the 95-104m and 96-104m induced peptide-specific T-cell responses of variable intensity in 11 out of 13 different normal HLA-A0201+ donors analyzed (Fig. 2). Interestingly there was a dichotomy in the intensity of the respective peptide-specific T-cell responses. Indeed for any of the 11 responsive donors, one of the two altered peptide would elicit a response far superior to the other altered peptide by a factor of at least 3 when enumerating the IFN-γ secreting cells by ELISPOT (Table 1, Fig. 2A). When T-cells were repeatedly stimulated with APC simultaneously pulsed with both altered Sur peptides at the same concentration (20 μM) IFN-γ ELISPOT responses against both peptides either alone or in combination were poor in comparison to T cell lines generated from the same donors in the same set of experiments but using single peptide pulsed APC (Fig. 2B). We investigated if a change in the cytokine secretion profile occurred in the cells stimulated with both peptides and looked for the appearance of other cytokines in the supernatants collected 48h after peptide stimulation of T-cell lines generated with single peptides or against both mixed peptides. Supplementary Figure 1 presents results of a multiplex cytokine detection run on supernatants harvested after 48h of co-incubation of the peptide specific T cell line with cognate peptide. Aside from IFN-γ, no other cytokines were detected at significant levels therefore arguing that the lack of IFN-γ secretion in the T cell lines generated with both peptides is not the result of a switch to a Th2 cytokine secretion profile. An intracellular IL-17 staining was also negative (data not shown). The combination of both peptides pulsed at 20 μM each (40 μM total peptide concentration) however was still able to stabilize HLA-A0201 expression on T2 cells to the same saturation level as the use of 20 μM of single peptides, although not reaching 100% saturation as may have been expected (Fig. 2C) suggesting that the poor antigen-specific IFN-γ response elicited was not due a cross-inhibition of peptide binding to HLA-A0201.
Fig. 2.
Response to 95-104 and 96-104 altered peptides in normal donors. A. Thirteen HLA-A0201 normal donor PBMCs were used to raise T cell lines against 95-104m or 96-104m Survivin peptides and reactivity to the cognate peptide was assessed after 3 rounds of stimulation by IFN-γ ELISPOT. Results are expressed in number of spots per 200,000 cells. HIV REV peptide (ILKEPVHGV) was used to control for non-specific IFN-γ secretion. For each donor, the two first columns on the left are testing the ability of the 95-104m raised T cell line to recognize 95-104m peptide (or irrelevant REV peptide). The two last columns on the right side of the graph represent the ability of the 96-104m raised T cell line to recognize 96-104m peptide (or irrelevant REV peptide). The donors have been classified according to the preference of the mounted response as 10-mer responders (top row), 9-mer responders (middle row) or no preference/very weak response (bottom row). B. Pulsing APCs with the two altered peptides simultaneously to raise T cell lines abrogates the response to any of the single peptide. Cell lines were raised against individual altered peptide or a mixture of both for 3 rounds of stimulation after which response to individual peptide or to the mixture of the two was assessed by IFN-γ ELISPOT. Left panel represents results from a 95-104m responder, and the right panel represents the results from a 96-104m responder. These experiments represent the results of two 95-104m responders and two 96-104m responders tested. C. Binding of either individual or mixed altered peptides to HLA-A0201 is equivalent. T2 binding assay using 20μm of either peptide or a mixture of both (20μM of each) was performed and % saturation of HLA-A0201 molecule was calculated relative to gp100(209-217)210M mean fluorescence intensity (MFI) at saturation.
Table 1.
Summary of survivin m95-104 and m96-104 peptide responses in relation to full HLA-A , B, and C sub-types of donor T cells.
| 95-104m raised T cells | 96-104m raised T cells | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Normal donor # |
HLA-A | HLA-A | HLA-B | HLA-B | HLA-C | HLA-C | # specific dots for 95-104m per 106 cells |
# specific dots for 96-104m per 106 cells |
|
| HLA-A0201 homozygous | ND 73 | 0201 | 0201 | 35EAZA | 44EAPK | 04xx | 05xx | 1350 | 5500 |
| ND 105 | 0201 | 0201 | 27ADDM | 40EADJ | 01EAJA | 03EASA | 11365 | 2495 | |
| ND 203 | 0201 | 0201 | 3906 | 4405 | 0702 | 0202 | 1175 | 3530 | |
|
HLA-A0201 heterozygous
HLA-Cw7+ |
ND 60 | 0101 | 020101 | 08DYSE | 18EKCS | 0701 | 0701 | 5180 | 1307 |
| ND 88 | 010101g | 020101 | 07EJZK | 08EAAY | 0702 | 0701 | 3282 | 948 | |
| ND 118 | 0201 | 29xx | 18EKRW | 44ENCM | 0701 | 16AJ | 605 | 55 | |
| ND 109 | 0201 | 01xx | 08EANH | 15EATX | 03xx | 0701 | 214 | 267 | |
| ND 211 | 010101g | 020101 | 08XKT | 44FCXH | 0701 | 05AC | 84 | 277 | |
| ND 220 | 0201 | 03xx | 08XKT | 1801 | 0701 | 1203 | 105 | 3850 | |
| ND 68 | 0201 | 6802 | 07ENHH | 14GES | 0702 | 08AEY | 1855 | 8625 | |
|
HLA-A0201 heterozygous
HLA-Cw7− |
ND 124 | 01xx | 0201 | 5001 | 57EJTZ | 06xx | 06xx | 15 | 215 |
| ND 127 | 0201 | 32xx | 40EJTW | 44DYSF | 02xx | 16MR | 10 | 580 | |
| ND 120 | 0201 | 68xx | 14BH | 15EAUH | 03xx | 08WN | 30 | 30 |
These results also precluded that the reactivity of the decamer 95-104m peptide was due to a truncation (e.g., due to serum exonuclease activity) of the decamer into a nonamer during peptide pulsing of the APC because pulsing both peptides did not enhance the response and, rather, inhibited the response. In addition, all donors tested would have reacted to the nonamer 96-104m peptide if the responses against the 95-104m were due to conversion into the nonamer.
Thus, the new Sur 95-104m peptide is immunogenic and induced enhanced T-cell responses over its wild-type counterpart. However, interestingly both the 95-105m and 96-104m elicited a selective CD8+ T-cell response in some HLA-A0201+ normal donor T cells, but not in others. This intriguing result suggests a dichotomy in the human population in reacting to those two closely related peptides. The observation that some donors react to the modified decamer but not to the modified nonamer implies that the reactivity against the decamer was not due to truncation into the nonamer form of the peptide. Interestingly, the response against the mixture of both peptides was weak even though stimulations with single peptides could elicit considerable response to one peptide or the other with the same HLA-A0201+ donor T cells.
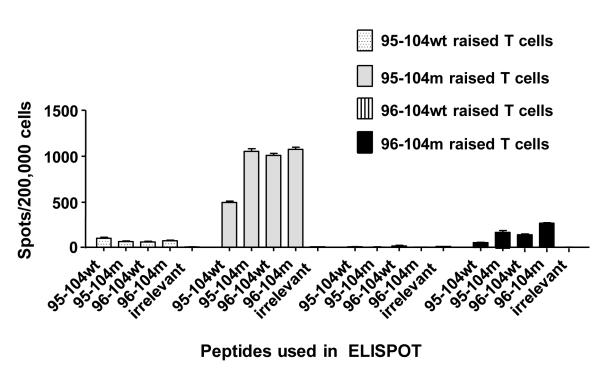
Sur 95-104m peptide induces T-cells cross-reactive against the native 95-104 and 96-104 peptide
In order for the new Sur 95-104m peptide to be useful in anti-cancer vaccination it must cross-react with the native 95-104 peptide and also induce T-cells capable of recognizing and killing tumor cells presenting processed and presented 95-104 from the wild type Sur protein (95-104wt). We first examined the cross-reactivity of the decamer Sur 95-104m peptide to its wild-type counterpart as well as the nonamer Sur 96-104m and its wild-type counterpart. T-cells from HLA-A0201+ normal donors (previously found to be reactive to 95-104m) were stimulated with Sur peptides 95-104m or wild-type 95-104 over a number of rounds of stimulation and expansion, as before. After 3 stimulations, the lines were tested for reactivity against the stimulating peptide (95-104m) or against wild-type peptide in an IFN-γ ELISPOT. Reactivity against the wild-type and altered nonamer peptides was also evaluated. T cell lines reacting preferentially to the 96-104m and wild-type 96-104 peptides were also tested in a similar fashion. T cell lines raised using the wild-type 95-104 and 96-104 peptides induced only a low response against the native peptide used to raise them and all the Sur decamer and nonamer peptides tested, as expected due to the low HLA-A0201 binding activity of the native peptides (Fig. 3). In contrast, the 95-104m induced a strong response and expanded T cells that cross-reacted with the wild-type 95-104 as well as both the altered and wild-type 96-104 peptides equally well (Fig. 3). The donor presented in Fig. 3 mainly reacted to 95-104m peptide, but a similar pattern of cross-reactivity to the mutant or wild-type 96-104 or 95-104 was observed in donors reacting preferentially to 96-104m (data not shown). Thus, although each altered peptide in this Sur region activated and expanded T cells from different and not the same individuals, the expanded T cells showed effector cell activity that was cross-reactive against altered and wild-type versions of both peptides.
Fig. 3.
Recognition of the wild type Survivin peptides by T cell lines raised with altered Survivin peptides and plasticity of recognition between the 9-mer and 10-mer Survivin peptides. Results from one normal donor where 4 T cell lines were raised (one for each peptide) and after the third round of stimulation the cell lines were tested in IFN-γ ELISPOT for recognition of the four test peptides including the one they have been raised with. Results are representative of 8 normal donors assessed in separate experiments.
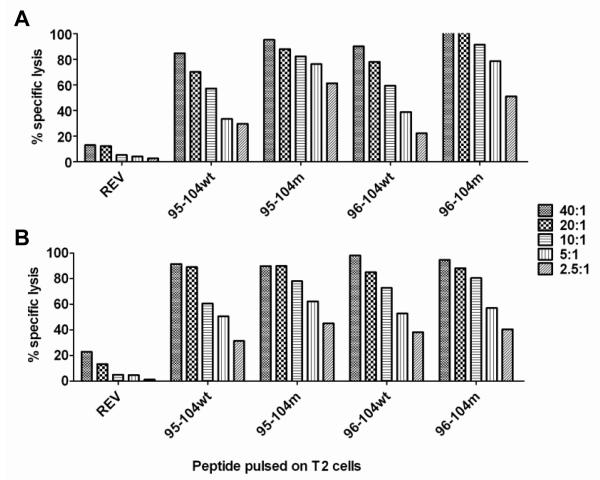
T-cell lines from HLA-A0201+ individuals activated against 95-104m or 96-104m were further tested in CTL assays against peptide-pulsed T2 target cells using 51Cr-release assays. T2 cells were loaded with either one of four peptides (95-104m, wild-type 95-104, 96-104m, or wild-type 96-104). T2 cells pulsed with an HLA-A0201-binding peptide from HIV REV was used as negative control. As shown in Fig. 4A, T-cells raised against 95-104m efficiently killed 95-104m-loaded and wild-type 95-104-loaded target cells as well as target cells loaded with 96-104m and wild-type 96-104 peptide. Similar results were obtained with T-cell lines from preferentially 96-104m-reactive donors (Fig. 4B).
Fig. 4.
T cell lines raised with Survivin altered peptides can kill T2 targets pulsed with the 4 test peptides. T cell lines raised against 95-104m (A) or 96-104m (B) from normal donors were tested in a 51Cr-release CTL assay to evaluate their ability to kill T2 cells pulsed with the 4 test peptides (or HIV REV negative control); the 10-mer 95-104 wild type or modified, the 9-mer 96-104 wild type or modified at the different effector-to-target (E:T) ratios shown on the right. Cytotoxicity assay was performed after the fourth round of stimulation. The two cell lines showed were derived from different donors since we could not have a donor who reacted strongly to both altered peptides. This is representative of 4 separate experiments in 4 different donors.
Thus, T-cells raised by repeated stimulation using the 95-104m peptide cross-react against wild-type versions of both Sur 95-104 and Sur 96-104 peptides, as determined by cytokine secretion (IFN-γ ELISPOT assays) and ability to kill antigen-loaded targets (CTL assays using peptide-pulsed T2 cells). The cross-recognition of both 9-mer and 10-mer wild type peptides, by T cells specific for 95-104m suggests that they would also recognize a tumor cells presenting these naturally processed and presented wild-type Survivin epitopes.
Sur 95-104m-reactive T cells recognize and kill HLA-A0201+ Sur-expressing tumor cells
Next, we asked the question of whether Sur 95-104m peptide primed T cells could recognize and kill HLA-A0201+ tumor cells expressing Sur in order to be considered as an additional peptide vaccine candidate for cancer in addition to Sur 96-104m and other Sur peptides currently being used in the clinic in HLA-A0201+ cancer patients. There is evidence in the literature that 95-104 and 96-104 Survivin peptides may be naturally processed and presented by cancer cells. T cells specific for the wild type Sur 95-104 able to kill autologous HLA-A0201+ tumor cells have been found in the blood of cancer patients [2, 19]. We tested 95-104m-activated T cell lines generated from HLA-A0201+ normal donors against the Survivin-expressing breast cancer cell line MDA-MB-453 (MDA 453) originally from an HLA-A0201− patient that was stably transduced with an HLA-A0201-expressing lentiviral vector. MDA 453 transduced with HLA-A0201 (MDA 453/A2) expressed Survivin, as determined using intracellular staining and flow cytometry analysis (Fig. 5A). Non-HLA-A0201-transduced MDA 453 cells also expressed a similar level of Survivin (data not shown). T-cell lines specific for 95-104m were raised as before and tested after 3-4 rounds of stimulation. We generated peptide-specific T cell lines against Sur 95-104m and Sur 96-104m from one of the rare donors who could raise lines to both peptides (ND 203) and then tested the CTL activity against MDA 453 cells using a sensitive flow cytometry-based assay measuring cleaved caspase 3 in target cells [16]. Fig. 5B shows the percentage of target cells positive for cleaved Caspase 3 (in the process of apoptosis) at different effector to target ratios (E:T). Fig. 5C shows the IFN-γ secretion of the T cell lines used in Fig. 5B when exposed to their cognate peptide overnight in an ELISPOT assay. Both T cell lines show HLA-A0201 restricted killing of targets and the amplitude of the killing was proportional to the frequency of the IFN-γ secreting cells in the culture detected by ELISPOT (Fig.5C), with the 96-104m-reactive T cell line showing higher killing due to the higher frequency of these peptide-specific T cells. Fig. 5D and E show the results of killing by a 95-104m raised T cell line from a different normal donors in a standard 51Cr assay using this time targets that expresses HLA-A0201 naturally. The target chosen in Fig. 5D was a primary melanoma cell line freshly established from an HLA-A0201+ patient (Mel 2320), while the target used in Fig. 5E was a pancreatic cancer cell line (CF-Pac-1). Both target cell lines expressed HLA-A0201 on the cell surface and Suvivin intracellulary, as determined by flow cytometry analysis ([13]; data not shown). Targets and T cells were co-incubated in the presence of an anti-HLA-Class I antibody (clone W6/32) or an isotype control antibody. The sizeable reduction in killing measured when anti-Class I is added suggests the killing was HLA restricted.
Fig. 5.
T cells raised against the new 95-104m peptide can kill unpulsed targets. A. Survivin expression in MDA 453 breast cancer cell line transduced with HLA-A0201 molecule (MDA 453/A2). Cells were fixed, permeabilized, stained with anti-Survivin-FITC or isotype control and assessed by flow cytometry analysis (isotype control in broken line, Survivin expression in black line). B. T cell lines from the same HLA-A0201+ individual raised against the Sur 95-104m or Sur 96-104m peptide by 4 rounds of stimulation with peptide-pulsed APC preferentially kill MDA 453 breast cancer cells expressing HLA-A0201, as measured using a Caspase 3-cleavage CTL assay performed using wild-type MDA 453 (HLA-A0201-negative) or MBA 453/A2 cells as targets. C. INF-γ ELISPOT showing reactivity of T cell lines used in B to Survivin peptides after 3 rounds of stimulation. D-E. 95-104m raised T cell line can kill a target naturally expressing HLA-A0201. D. Killing of a primary HLA-A0201+ melanoma cell line freshly established from a patient enrolled in an adoptive cell therapy trial at MD Anderson was assessed in a standard 51Cr assay. Targets and T cells were incubated with 40 μg/ml of an anti- HLA-Class I antibody (clone W6/32), or with the same concentration of an isotype control antibody at 40:1 or 20:1 effector:target ratio for 5 h at 37°C. E. 95-104m raised T cell line can kill pancreatic cancer cell line CF-Pac-1 (HLA-A0201+). Targets and T cells were incubated with 20 μg/ml of an anti- HLA-Class I antibody (clone W6/32), or with the same concentration of an isotype control antibody at 30:1 effector:target ratio for 5h at 37°C.
Molecular modeling reveals differences in the interaction of 95-104m and 96-104m with HLA-A0201
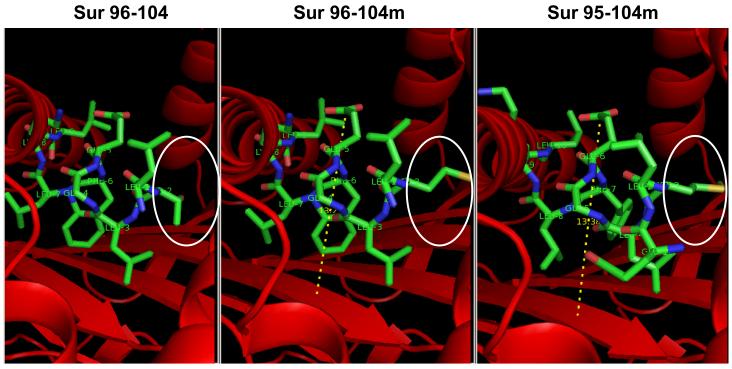
We next examined the modeled interaction of the two altered peptides with HLA-A0201, as well as the wild type 96-104 (Fig. 6). The predicted structure of Survivin peptides into HLA-A0201 binding groove was generated by adapting available crystallographic data from HIV gag peptide bound to HLA-A0201 (pdb2C7U). The HIV peptide was removed and replaced with Survivin peptides. Structure analysis was conducted in PyMol software to measure the distance between specific atoms to determine the distance between the ‘floor’ of the HLA-A0201 groove and the highest point reached by the peptide. That distance was measured to be 13.38 Å for Survivin 95-104m peptide and 13.21Å in the case of the 96-104 peptide. This modeling highlights a difference in the way the two modified peptides interact with HLA molecule. The distance between the “floor” of the MHC molecule and the higher point reached by the peptide is slightly longer in the case of 95-104m. Therefore this longer peptide protrudes from the HLA peptide-binding groove and we hypothesize that this difference could substantially alter the interaction with the TCR.
Fig. 6.
The predicted structure of Survivin peptides (green) into HLA-A0201 binding groove (red) was generated by adapting available crystallographic data from HIV gag peptide bound to HLA-A2*0201 (pdb2C7U). The HIV peptide was removed and replaced with Survivin peptides. From left to right are presented the peptide/MHC structure of the 96-104 in its wild type form (LTLGEFLKL), 96-104 modified (LMLGEFLKL) and finally our novel 95-104 modified (ELMLGEFLKL). The area circled in white highlights the difference in structure imposed by the mutation T→M at position 97, affecting both 96-104m and 95-104m peptides. The dotted line symbolizes the distance between the ‘floor’ of the HLA molecule and the highest point reached by the peptide.
Full HLA class I typing at the A, B, and C locus reveals possible contribution of HLA-C alleles to Sur 95-10m versus 96-104m reactivity
We went on and performed a full HLA class I typing on the A, B, and C loci from the 13 donors tested in this study. This was done to try to better understand why the Sur 95-104m and 96-104m peptides showed selectivity in terms of their ability to generate peptide-specific T cells from PBMC, where most donors responded against one or the other peptide and only some generated T cell lines reactive against both peptides. Among the 13 donors, those who were homozygous for HLA-A0201 generated T cell lines reactive against both peptide, as tested in IFN-γ ELISPOT assays following the third round of stimulation (Table 1, top section). Typing for different B alleles did not uncover any trend in terms of Sur 95-104m versus 96-104m selectivity. However, typing for the HLA class I C allele sub-type found that all individuals (7/7) that were heterozygous for HLA-A0201 but had at least one HLA-C0701 allele generated a T cell line reactive to 95-104m (Table 1, middle section), whereas none of the donors heterozygous for HLA-A0201 and negative for HLA-C0701 could raise a 95-104m specific T cell line (Table 1, bottom section). In contrast, response to 96-104m peptide did not seem to be influenced by the presence of HLA-C0701. Thus, more detailed typing of all HLA class I loci revealed that individuals heterozygous for HLA-A0201 carrying an HLA-C0701 allele were able to generate T cell lines to the new Sur 95-104m peptide.
We went on to search a number of HLA class I peptide binding algorithms available on-line (e.g., SYFPEITHI, BIMAS, WAPP, Rankpep, ProPred-I, nHLAPred, MAPPP), but, while these available sites have an algorithm predicting peptide binding to HLA-C0702, no predictive algorithm exists for the HLA-C0701 allele, especially for decamers. We looked at HLA-C0702 nevertheless using the BIMAS site, but no appreciable binding of either Sur 95-104m or Sur 96-104m was predicted to HLA-C0702 (data not shown). Using BIMAS, HLA class I molecules that are predicted to bind to Sur 95-104m are B7 and Cw3 (0301), while 96-104m is predicted to bind to Cw3 (0301) and B2705. Notably, in our panel of normal donors used only two were positive for HLA-B7 (ND 68 and 88) and they both were highly reactive to 95-104m, whereas ND 105 which is Cw3+ was the most reactive to the 95-104m peptide.
DISCUSSION
Initial work characterizing Survivin as a tumor-associated antigen has uncovered few immunogenic epitopes restricted by HLA-A0201 along the 142 amino acids sequence of the protein. Reactivity to peptides 95-104 and 96-104 has been most consistently observed in patients. The 9-mer peptide 96-104 has been modified in position 2 (T97M) to better anchor the HLA molecule which enhances the immunogenicity of the peptide. This modified peptide, along with the 95-104wt, is one of a number of HLA-A0201 restricted Survivin epitope that are undergoing clinical testing [7, 11, 12, 20]. Although modifications at anchor residues in 9-mer HLA-A0201-binding peptides (position 2 and 9) have been tested for enhance binding and HLA-A0201-restricted immunoreactivity, little is known about whether modifications in 10-mer HLA-A0201-restricted peptides behave comparably. Alterations in 10-mer HLA-A0201 peptides have not been considered so far.
We hypothesized that introducing the T97M alteration at position 3 in the 10-mer 95-104 peptide could stabilize binding to HLA-A0201 and could potentially lead to a more immunogenic Sur peptide in a similar fashion as this amino acid change does at position 2 in the 9-mer 96-104 Sur peptide. We found that the use of the 10-mer peptide encompassing the region 95-104 and where T97 was modified to M was actually more immunogenic for a significant proportion of our normal donors (4 out of 13 or 30%). To our knowledge, this is the first description of the use of peptide 95-104 where the threonine 97 is changed to a methionine. By augmenting the affinity of the 95-104 peptide, we could better study its immunogenicity. Very interestingly, this report shows that a methionine at position 3 of a 10-mer peptide increases binding to HLA-A0201 molecule, in a similar fashion than it does for the 9-mer peptide (methionine at position 2 in this case). It is well known that M in position 2 of a 9-mer peptide serves as anchoring residue for HLA-A2 but is much less appreciated that it can also play this role in position 3 of a 10-mer. Although it has been proposed by Rupppert and Sette that an aliphatic residue in position 3 of a 10-mer is associated with high binders, their model says that E (glutamic acid) at position 1 would be very detrimental to the interaction [21]. Our molecular modeling data shows that both peptides bind in a similar way to HLA-A0201, but that the 10-mer extends out of the HLA-binding groove. We hypothesize that this might alter the contact with the TCR.
A great majority (11/13 or 85%) of the normal donors tested responded to at least one of the tested modified Sur peptides but not with the same potency. A dichotomy was observed where the response to one peptide was always much stronger than the other. This disparity was not anticipated since both altered peptides bind with similar affinity to HLA-A0201 according to the T2 binding assay. Interestingly, the original report testing the modified Sur 96-104 peptide also found a dichotomy in the immunogenicity of the 96-104 modified peptide compared to 95-104 (wild type) in PBMC samples from cancer patients. Peripheral blood lymphocytes from melanoma, breast cancer and CLL patients were found to contain effector cells that reacted with 96-104m or 95-104wt peptide (in IFN-γ ELISPOT) but rarely to both [2]. Although modifying the second amino acid to a methionine increased the proportion of the screened patients responding to 96-104 peptide, 40-50% of the melanoma and breast cancer patients were still not responding to the peptide. Based on our data, it is possible that a good proportion of these non-responding patients, if re-evaluated, could turn out to be reactive for the 95-104m peptide.
It is unclear at the moment what regulates the dichotomy in the ability to generate T-cell lines in response to 96-104m and 95-104m peptides. Both peptides are clearly able to bind HLA-A0201 molecule with about the same affinity (Fig. 1). Interestingly, all three donors homozygous for HLA-A0201 (73, 105 and 203) could respond to both peptides with at least 1,150 IFN-γ producing T cells per million cells after three rounds of stimulation, although not all of them responded better to the same peptide (Table 1). However, among normal donors heterozygous for HLA-A0201, only donors co-expressing HLA-C0701 (7/7 donors) showed a strong tendency to be able to respond to the altered 10-mer (response meaning in this case a minimum of at least 84 IFN-γ producing T cells per million cells). The small number of donors analyzed does not allow to correlate any other co-expressed HLA class I molecule to improved response to the altered 9-mer.
The mechanism behind the possible contribution of the HLA-C0701 allele in helping drive out T cells responding to Sur 95-104m is unclear at present, but may involve a new paradigm involving cooperativity between two different HLA class I alleles in expanding low-frequency T-cell responses. It has been shown in mice that the frequency of the precursor T cells in the blood and MHC binding affinity both shape the immunodominance of different epitopes [22, 23]. Since the binding to HLA-A0201 is comparable for both peptides, the precursor frequency might have been the key factor in the response measured. Differences in the respective T cell repertoire of the donors analyzed might account for the dichotomy observed. The only HLA molecule shared by all the donors was HLA-A0201, but the relative contribution of other HLA class I molecules expressed to the final constitution of the repertoire has to be considered. T cells can actually recognize multiple peptides [24, 25], or one peptide in the context of two different class I molecules [26]. In this respect, contributions of HLA-C alleles have generally not been considered due to the dominance of HLA-A allele expression and the fact that much less is known about the nature and immunogenicity of peptides binding to the HLA C alleles. It is possible that a certain degree of additional binding of Sur 95-104m to HLA-C0701 may have led to the ability to generate a minimal frequency of CD8+ T cells responding to this peptide over a critical threshold that then expanded to the levels observed in our experiments. However, once expanded to high enough frequencies, these T cells also cross-reacted with the nonamer 96-104m peptide as well as wild-type counterparts of both the nonamer and decamer presented on HLA-A0201 with or without some contribution of HLA-C0701. This raises an interesting new paradigm regarding how perhaps a cooperativity between HLA class I alleles binding similar sets of peptides may activate responding T cells found at low frequencies to expand to a detectable threshold at which reactivity of the peptide presented by the dominant HLA class I allele can be detected. Unfortunately, at present we could not easily test whether HLA-C0701 is predicted to bind our new Sur 95-104m peptide since such peptide-binding algorithms in silico do not exist at present. Confirmation of this peptide binding will require the development of either surface plasmon resonance peptide binding assays to recombinant HLA-C0701 protein complexes or using TAP-deficient cell lines expressing only HLA-C0701 in which the stabilization of HLA expression can be measured specifically to definitively determine peptide binding to this C allele.
In addition to helping understand why certain individuals generated T cell lines preferentially against the new Sur 95-104m peptide and not the current Sur 96-104 being tested in the clinic via possible HLA-C allele contributions, our HLA typing data in Table 1 also sheds light on why certain individuals can have poor responses to either modified Sur peptide. There have been studies showing that the co-expression of HLA-B7 negatively affects HLA-A0201 restricted responses [27, 28]. Two different groups studying immune response against CMV or EBV found that CD8+ T cell responses associated with HLA-A0201 epitopes were abolished or significantly reduced in individuals co-expressing HLA-A0201 and HLA-B7. Those individuals had a productive response to B7 restricted peptides but not to A0201-restricted peptides. The responses to 5 different HLA-A0201-restricted CMV peptides were down-regulated in two different studies by seemingly the mere presence of B7 [27, 28]. Hence it is also possible that the co-expression of some other HLA class I molecules could regulate the nature of HLA-A0201 presented peptides. Two of the donors analyzed (#109 and #120) expressed the common ancestral Scandinavian HLA haplotype 62.1 (HLA-A0201, B15, Cw3) which has been associated with autoimmunity and rapid progression of melanoma after apparition of metastasis [29, 30]. Of all the normal donors studied, these two donors (#109 and #120) having expressing HLA-A0201 together B15 and Cw3 were the only ones who did not have preferential response to either Sur 95-104m or 96-104m. Moreover, of these two individuals the one who did not have HLA-C0701 (#120) exhibited a very poor generation of T cells reactive to either peptide (Table 1). This further supports a possible role for HLA-C0701 is promoting responses against the new 95-104m peptide.
At this point association between co-expression of various HLA-molecules and modulation of HLA-A2 restricted responses to Survivin peptides is only speculative and requires further examination. We can only suggest that HLA-A0201 restricted responses could be affected by co-expression of certain defined HLA class I molecules. It would be very informative to look at the full HLA-typing (A, B and C) of all the patients enrolled in cancer vaccination trials involving Survivin peptides (or any tumor antigen for that matter) and determine if any correlation between the degree of peptide-specific CD8+ T-cell responses and the presence of specific HLA-C and B allele combinations with HLA-A0201 can be made.
In summary, the data presented in this manuscript shows that a new altered decamer peptide of Survivin has enhanced HLA-A0201 binding and immunoreactivity. Our data also reveals a novel dichotomy in the response of HLA-A0201+ donors to two closely related Survivin peptides previously assumed to derive from the same epitope. Our results support the idea that Survivin 95-104 and 96-104 peptides are recognized by different T cells and therefore represent different epitopes. Clarification of the underlying causes of this immunodominance will need further examination. In this regard, our full HLA class I typing data revealed a possible positive contribution of HLA-C alleles (HLA-C0701 in particular) in regulating this immunodominance. Lastly, our findings argue for the inclusion in cancer immunotherapy vaccination protocols of both the currently used 96-104m peptide and our newly defined 95-104m peptide as a potential approach to significantly increase the frequency of HLA-A0201+ patients potentially reactive to Survivin vaccination. Moreover, our T-cell line generation data showing decreased T-cell responses when both peptides are used simultaneously on APC suggest that measures to reduce interference should be included in the design of future vaccination studies. In order to get priming of T cells with only one peptide at a time a two-site vaccination protocol where vaccination with the different peptides occurs at different times may be optimal. However, at this point we do not know whether the decreased T-cell responses seen when both peptides were used to stimulate the T-cell lines were due to some artifact of the cell culture process. Peptide vaccination experiments in HLA-A0201 transgenic mice will be needed to confirm this.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported in part by an NCI/NIH grant R03CA123426-01 (LR), a matching grant from the Lockton Foundation for Pancreatic Cancer Research (LR), and IRG grant from MD Anderson Cancer Center (LR and DZC), and by the Topfer Family Fund for Pancreatic Cancer Research (DZC). We are grateful to Dr. Sijie Lu (Dept. of Immunology, MD Anderson Cancer Center) for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Oble DA, Loewe R, Yu P, Mihm MC., Jr. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human melanoma. Cancer Immun. 2009;9:3. [PMC free article] [PubMed] [Google Scholar]
- [2].Andersen MH, Pedersen LO, Capeller B, Brocker E-B, Becker JC, thor Straten P. Spontaneous Cytotoxic T-Cell Responses against Survivin-derived MHC Class I-restricted T-Cell Epitopes in Situ As Well As ex Vivo in Cancer Patients. Cancer Res. 2001 Aug 1;61(16):5964–8. 2001. [PubMed] [Google Scholar]
- [3].Sine Reker, Becker JC, Svane IM, Ralfkiaer E, Straten Pt, Andersen MH. HLA-B35-restricted immune responses against survivin in cancer patients. International Journal of Cancer. 2004;108(6):937–41. doi: 10.1002/ijc.11634. [DOI] [PubMed] [Google Scholar]
- [4].Reker S, Meier A, Holten-Andersen L, Svane IM, Becker JC, thor Straten P, et al. Identification of novel survivin-derived CTL epitopes. Cancer biology & therapy. 2004 Feb;3(2):173–9. doi: 10.4161/cbt.3.2.611. [DOI] [PubMed] [Google Scholar]
- [5].Andersen MH, Pedersen LO, Becker JC, Straten Pt. Identification of a Cytotoxic T Lymphocyte Response to the Apoptosis Inhibitor Protein Survivin in Cancer Patients. Cancer Res 2000. 2000 Feb 1;61(3):869–72. [PubMed] [Google Scholar]
- [6].Hirohashi Y, Torigoe T, Maeda A, Nabeta Y, Kamiguchi K, Sato T, et al. An HLA-A24-restricted cytotoxic T lymphocyte epitope of a tumor-associated protein, survivin. Clin Cancer Res. 2002 Jun;8(6):1731–9. [PubMed] [Google Scholar]
- [7].Otto K, Andersen MH, Eggert A, Keikavoussi P, Pedersen LO, Rath JC, et al. Lack of toxicity of therapy-induced T cell responses against the universal tumour antigen survivin. Vaccine. 2005 Jan 4;23(7):884–9. doi: 10.1016/j.vaccine.2004.08.007. [DOI] [PubMed] [Google Scholar]
- [8].Tsuruma T, Hata F, Torigoe T, Furuhata T, Idenoue S, Kurotaki T, et al. Phase I clinical study of anti-apoptosis protein, survivin-derived peptide vaccine therapy for patients with advanced or recurrent colorectal cancer. J Transl Med. 2004 Jun 13;2(1):19. doi: 10.1186/1479-5876-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Honma I, Kitamura H, Torigoe T, Takahashi A, Tanaka T, Sato E, et al. Phase I clinical study of anti-apoptosis protein survivin-derived peptide vaccination for patients with advanced or recurrent urothelial cancer. Cancer Immunol Immunother. 2009 Mar 18; doi: 10.1007/s00262-009-0691-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tsuruma T, Iwayama Y, Ohmura T, Katsuramaki T, Hata F, Furuhata T, et al. Clinical and immunological evaluation of anti-apoptosis protein, survivin-derived peptide vaccine in phase I clinical study for patients with advanced or recurrent breast cancer. J Transl Med. 2008;6:24. doi: 10.1186/1479-5876-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wobser M, Keikavoussi P, Kunzmann V, Weininger M, Andersen MH, Becker JC. Complete remission of liver metastasis of pancreatic cancer under vaccination with a HLA-A2 restricted peptide derived from the universal tumor antigen survivin. Cancer Immunol Immunother. 2006 Oct;55(10):1294–8. doi: 10.1007/s00262-005-0102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fuessel S, Meye A, Schmitz M, Zastrow S, Linne C, Richter K, et al. Vaccination of hormone-refractory prostate cancer patients with peptide cocktail-loaded dendritic cells: results of a phase I clinical trial. Prostate. 2006 Jun 1;66(8):811–21. doi: 10.1002/pros.20404. [DOI] [PubMed] [Google Scholar]
- [13].Zhu K, Lizee G, Cano P, Fernando-Vina M, Ji B, Abbruzzese JL, et al. HLA-A0201 positive pancreatic cell lines: new findings and discrepancies. Cancer Immunol Immunother. 2007 May;56(5):719–24. doi: 10.1007/s00262-006-0217-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Radvanyi LG, Banerjee A, Weir M, Messner H. Low levels of interferon-alpha induce CD86 (B7.2) expression and accelerates dendritic cell maturation from human peripheral blood mononuclear cells. Scand J Immunol. 1999 Nov;50(5):499–509. doi: 10.1046/j.1365-3083.1999.00625.x. [DOI] [PubMed] [Google Scholar]
- [15].Schultze JL, Michalak S, Seamon MJ, Dranoff G, Jung K, Daley J, et al. CD40-activated human B cells: an alternative source of highly efficient antigen presenting cells to generate autologous antigen-specific T cells for adoptive immunotherapy. The Journal of clinical investigation. 1997 Dec 1;100(11):2757–65. doi: 10.1172/JCI119822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dietrich J, Aagaard C, Leah R, Olsen AW, Stryhn A, Doherty TM, et al. Exchanging ESAT6 with TB10.4 in an Ag85B fusion molecule-based tuberculosis subunit vaccine: efficient protection and ESAT6-based sensitive monitoring of vaccine efficacy. J Immunol. 2005 May 15;174(10):6332–9. doi: 10.4049/jimmunol.174.10.6332. [DOI] [PubMed] [Google Scholar]
- [17].Stuber G, Leder GH, Storkus WT, Lotze MT, Modrow S, Szekely L, et al. Identification of wild-type and mutant p53 peptides binding to HLA-A2 assessed by a peptide loading-deficient cell line assay and a novel major histocompatibility complex class I peptide binding assay. Eur J Immunol. 1994 Mar;24(3):765–8. doi: 10.1002/eji.1830240341. [DOI] [PubMed] [Google Scholar]
- [18].Parkhurst MR, Salgaller ML, Southwood S, Robbins PF, Sette A, Rosenberg SA, et al. Improved induction of melanoma-reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A*0201-binding residues. J Immunol. 1996 Sep 15;157(6):2539–48. [PubMed] [Google Scholar]
- [19].Casati C, Dalerba P, Rivoltini L, Gallino G, Deho P, Rini F, et al. The apoptosis inhibitor protein survivin induces tumor-specific CD8+ and CD4+ T cells in colorectal cancer patients. Cancer Res. 2003 Aug 1;63(15):4507–15. [PubMed] [Google Scholar]
- [20].Andersen MH, Becker JC, Straten P. Regulators of apoptosis: suitable targets for immune therapy of cancer. Nat Rev Drug Discov. 2005 May;4(5):399–409. doi: 10.1038/nrd1717. [DOI] [PubMed] [Google Scholar]
- [21].Ruppert J, Sidney J, Celis E, Kubo RT, Grey HM, Sette A. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell. 1993 Sep 10;74(5):929–37. doi: 10.1016/0092-8674(93)90472-3. [DOI] [PubMed] [Google Scholar]
- [22].Kotturi MF, Scott I, Wolfe T, Peters B, Sidney J, Cheroutre H, et al. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J Immunol. 2008 Aug 1;181(3):2124–33. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Obar JJ, Khanna KM, Lefrancois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008 Jun;28(6):859–69. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].De Silva-Udawatta M, Kumar SR, Greidinger EL, Hoffman RW. Cloned human TCR from patients with autoimmune disease can respond to two structurally distinct autoantigens. J Immunol. 2004 Mar 15;172(6):3940–7. doi: 10.4049/jimmunol.172.6.3940. [DOI] [PubMed] [Google Scholar]
- [25].Mason D. A very high level of crossreactivity is an essential feature of the T -cell receptor. Immunol Today. 1998 Sep;19(9):395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- [26].Lichterfeld M, Williams KL, Mui SK, Shah SS, Mothe BR, Sette A, et al. T cell receptor cross-recognition of an HIV-1 CD8+ T cell epitope presented by closely related alleles from the HLA-A3 superfamily. International immunology. 2006 Jul;18(7):1179–88. doi: 10.1093/intimm/dxl052. [DOI] [PubMed] [Google Scholar]
- [27].Höllsberg P. Contribution of HLA Class I Allele Expression to CD8+ T-Cell Responses against Epstein–Barr Virus. Scandinavian Journal of Immunology. 2002;55(2):189–95. doi: 10.1046/j.0300-9475.2001.01043.x. [DOI] [PubMed] [Google Scholar]
- [28].Lacey SF, Villacres MC, La Rosa C, Wang Z, Longmate J, Martinez J, et al. Relative dominance of HLA-B*07 restricted CD8+ T-Lymphocyte immune responses to human cytomegalovirus pp65 in persons sharing HLA-A*02 and HLA-B*07 alleles. Human Immunology. 2003;64(4):440–52. doi: 10.1016/s0198-8859(03)00028-4. [DOI] [PubMed] [Google Scholar]
- [29].Runge LA, Davey FR, Goldberg J, Boyd PR. The inheritance of Felty’s syndrome in a family with several affected members. The Journal of rheumatology. 1986 Feb;13(1):39–42. [PubMed] [Google Scholar]
- [30].Helgadottir H, Andersson E, Villabona L, Kanter L, van der Zanden H, Haasnoot GW, et al. The common Scandinavian human leucocyte antigen ancestral haplotype 62.1 as prognostic factor in patients with advanced malignant melanoma. Cancer Immunol Immunother. 2009 Feb 13; doi: 10.1007/s00262-009-0669-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.