Abstract
Botulinum neurotoxins (BoNTs) are the most toxic proteins currently known. Current treatments for botulinum poisoning are all protein based with a limited window of opportunity. Inhibition of the BoNT light chain protease (LC) has emerged as a new therapeutic strategy for the treatment of botulism as it may provide an effective postexposure remedy. As such, a small library of 40 betulin derivatives was synthesized and screened against the light chain of BoNT serotype A (LC/A); five positive hits (IC50< 100 μM) were uncovered. Detailed evaluation of inhibition mechanism of three most active compounds revealed a competitive model, with sub-micromolar Ki value for the best inhibitor (7). Unfortunately, an in vitro cell-based assay did not show any protection of rat cerebellar neurons against BoNT/A intoxication by 7.
Keywords: Betulin derivatives, Botulinum neurotoxin, Protease inhibitor
Botulinum neurotoxins (BoNTs), proteins produced by bacteria of the genus Clostridium,1 are responsible for botulism, a disease characterized by peripheral neuromuscular blockade and a characteristic flaccid paralysis of humans. BoNTs are the most poisonous substances known, with serotype A having a lethal dose for a 70 kg human of approximately 0.09-0.15 μg intravenously or intramuscularly, and 0.7-0.9 μg inhalationally.2 Despite their potentially lethal toxicity, BoNTs have emerged as an extremely valuable therapeutic tool for the treatment of a variety of maladies, including strabismus, migraines, and even facial wrinkles.3 However, the potential use of BoNT in a bioterrorist attack remains imminent and the Center for Disease Control (CDC) now classifies this agent as “category A”, placing it among the six highest-priority agents. Current treatments for botulinum poisoning are all antibody based with a limited window of therapeutic effectiveness. A particular drawback with these vaccine approaches is that they cannot reverse the effects after the toxin has reached its target inside the cell.4
Inhibition of the BoNT light chain metalloprotease (LC) has surfaced as a new therapeutic strategy for the treatment of botulism as it may provide an effective post-exposure remedy. BoNTs are synthesized as ~ 150 kDa proteins that are post-translationally activated by proteolytic cleavage to form mature di-chain proteins consisting of a 100 kDa heavy chain (HC) and a 50 kDa light chain (LC) linked by a disulfide bond.5 The HC is responsible for the neurospecific binding, uptake, and translocation of the LC into the cytosol of neuronal cells. The LC is a Zn2+-dependent metalloprotease that cleaves one of three intracellular soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins: syntaxin, vesicle-associated membrane protein (VAMP)/synaptobrevin, or synaptosomal-associated protein of 25 kDa (SNAP-25) depending on the serotype. As a consequence of protein cleavage, release of acetylcholine at the neuromuscular junction is blunted resulting in the loss of neurotransmission. Small molecule inhibitors of the LC may provide an opportunity for development of both pre- and post-exposure therapeutics. In recent years, the LC of serotype A (LC/A) has been a major focus, primarily due to its potency and long duration of paralysis.4 A number of competitive inhibitors of LC/A have been reported the most potent have Ki values of 0.6 – 10 μM,6 several of which coordinate the active site zinc cation required for catalysis (Ki ranging from 0.3 – 12.3 μM).7 More recently, the natural product D-chicoric acid was discovered in our laboratory that putatively binds to an exosite region outside the LC/A active site, displaying noncompetitive partial inhibition.8
Betulin is a naturally occurring, pentacyclic triterpene alcohol belonging to the lupane series of compounds. Betulin is the principle extractive substance of outer birch bark, and it has been extracted from white-barked birches (Betula sp.) in amounts up to 30% dry weight. Betulin 1 can be converted to betulinic acid 18,9 which has plethora of pharmacological properties, such as cytotoxic activity against several tumour cell lines by inducing apoptosis in cells.10 Excitingly, some betulin derivatives have also shown remarkable anti-HIV activity with new mechanisms of action.11 We have previously reported several new bioactivities for these highly interesting compounds, such as anti-leishmanial,12 anti-alphaviral13 and anti-chlamydial activity.14 Finally, structure-activity relationship (SAR) studies and pharmacological properties of betulin and its derivatives have been recently reviewed.15
Based on previous studies we were intrigued about potential activity of such compounds against BoNT/A. Therefore, a library of 40 betulin derivatives was tested for their inhibition of BoNT/A protease. The chemical structures of the betulin-derived triterpenoids are presented in Table 1; we note that we have previously reported these compounds,12a,13 with the exception of 4, 7, 12 and 26, which are now provided in supporting information. Thus, compounds were tested at a single concentration (50 μM) by LC/MS assay7b at 10 μM concentration of optimized truncated SNAP-substrate (66-mer; 141-206 aa) encompassing the key recognition elements of SNAP-25. From this lot, five positive hits 7, 8, 18, 19 and 21 (IC50 < 100 μM) were uncovered and three of these, 7, 8 and 18 were further evaluated at various concentrations of substrate and inhibitor. Obtained data were most consistent with a competitive inhibition model (Supp. Information). The inhibition constants, Ki were determined by a non-linear least squares global fit to the initial rates of product formation for matrixes of substrate and inhibitor concentrations bracketing Km and Ki, respectively (Table 2). These results revealed that 28-hemisuccinylbetulin 7 was the best inhibitor with Ki = 0.8 ± 0.2 μM. Thus, making it interesting lead structure for iterative rounds of structural modification in search of more potent small molecule antagonists of BoNT/A. The other betulin derivatives, betulinyl 28-carboxymethoxycarvacrolate 8 and betulinic acid 18 (Table 1) were about 16 and 18-fold less potent, respectively.
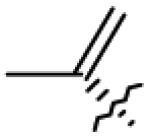
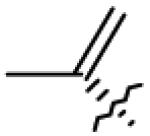
Table 1.
Structures of betulin derivatives screened against BoNT/A protease
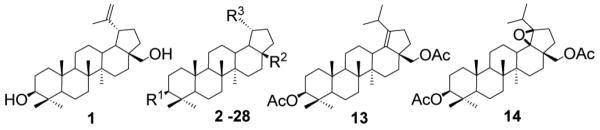 | |||
|---|---|---|---|
| Compound | R1 | R2 | R3 |
| 1 | OH | CH2OH |
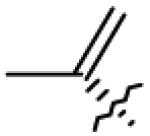
|
| 2 | OH | CH2OH |
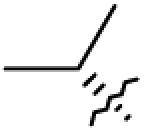
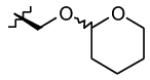
|
| 3 | OH |
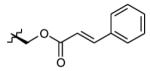
|
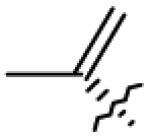
|
| 4 | OH |
|
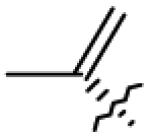
|
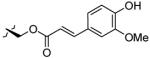
| 5 | OH |
|
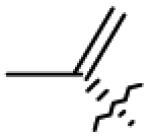
|
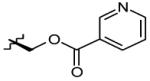
| 6 | OH |
|
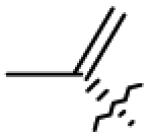
|
| 7 | OH |
|
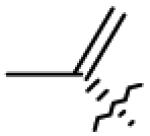
|
| 8 | OH |
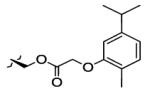
|
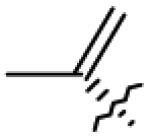
|
| 9 | OH | OAc |
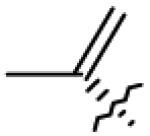
|
| 10 | OAc | OH |
|
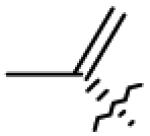
| 11 | OAc | CH2OAc |
|
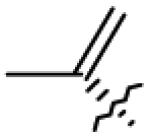
| 12 | OAc | CH2OAc |
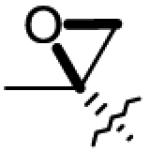
|
| 13 2 | OAc | CH2OAc |
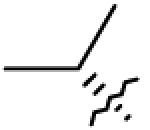
|
| 14 2 | OAc | CH2OAc |
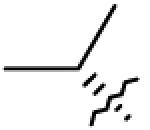
|
| 15 | OAc |
|
|
| 16 | OAc |
|
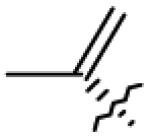
|
| 17 |
|
|
|
| 18 | OH | CO2H |
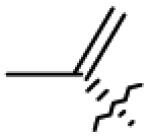
|
| 19 | OH | CO2Me |
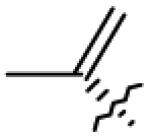
|
| 20 | O= | CHO |
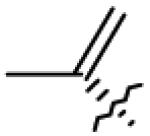
|
| 21 | O= | CO2H |
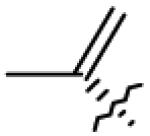
|
| 22 | O= | CO2Me |
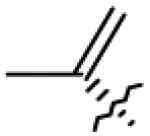
|
| 23 | O= | CO2H |
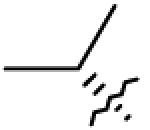
|
| 24 | O= |
|
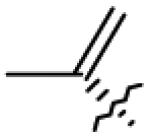
|
| 25 | O= |
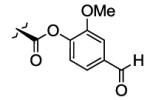
|
|
| 26 | O= |
|
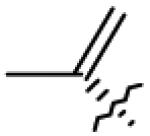
|
| 27 | OH | CH=NOH |
|
| 28 | =NOH | CH=NOH |
|
 | ||
|---|---|---|
| Compound | R3 | R4 |
| 29 | Me | Ac |
| 30 | Me | CocHex |
| 31 | Me | COPh |
| 32 | n-Bu | Ac |
| 33 | Ph | Ac |
| 34 | Bn | Ac |
| 35 | 3-MeO-Ph | Ac |
| 36 | 4-F-Ph | Ac |
| 37 | 3-NO2-Ph | Ac |
| 38 | 4-Ac-Ph | Ac |
| 39 | 4-Cl-Ph | Ac |
| 40 | indan-5-yl | Ac |
Table 2.
IC50 and Ki values of positive hits
| Compound | IC50 /(μM) | Ki / (μM) |
|---|---|---|
| 7 | 1.5 | 0.8 ± 0.2 |
| 8 | 10 | 13.3 ± 2.3 |
| 18 | 20 | 14.3 ± 2.2 |
N. D. = not determined
Potency of the best inhibitor was further investigated using an in vitro cell-based assay that monitors intracellular cleavage of SNAP-25. Thus, the cellular efficacy of compound 7 was tested using primary rat cerebellar neurons at 30 and 80 μM concentrations, respectively. However, this assay did not show inhibitory activity of this derivative against BoNT/A. Clearly, cells are complex biological systems and other factors such as permeability can influence the efficacy of small molecules within cellular models. Additionally, we highlight that betulin derivatives possess very low solubility in aqueous buffers that can also influence cellular activity (see Supp. Information for calculated physicochemical data).
In summary, a small library of 40 betulin derivatives was prepared and subsequently tested for potential inhibition of BoNT/A protease. Interestingly, five compounds within this library were discovered to exert low micromolar activities against the protease. Further detailed evaluation of the mechanism of inhibition of the three most active compounds revealed competitive inhibition, with a sub-micromolar Ki value for the best inhibitor. Disappointingly, in vitro cell-based examination did not demonstrate protection of rat cerebellar neurons against BoNT/A intoxication by 7. It is anticipated that additional modifications of the betulin structure to endow better water solubility and cell permeability will allow the discovery of betulins with cellular activity against BoNT/A.
Supplementary Material
Acknowledgments
We thank Dr. Mark S. Hixon and Adjunct Professor Salme Koskimies for very helpful discussions. This project was supported through the Skaggs Institute for Chemical Biology as well as with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract number N01-AI-30050 (C.B.S.) and award number AI082190 (K.D.J). The financial support from the Finnish Funding Agency for Technology and Innovation, Foundation for Research of Natural Resources in Finland, Marjatta ja Eino Kollin Säätiö, and the project by the European Commission (Contract No EU-KBBE-227239) are gratefully acknowledged (J.Y.-K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Experimental procedures, full characterization for compounds 4, 7, 12, 26 and biological assay conditions are available on-line.
References and notes
- 1.(a) Johnson EA, Bradshaw M. Toxicon. 2001;39:1703. doi: 10.1016/s0041-0101(01)00157-x. [DOI] [PubMed] [Google Scholar]; (b) Roxas-Duncan V, Enyedy I, Montgomery VA, Eccard VS, Carrington MA, Lai H, Gul N, Yang DCH, Smith LA. Antimicrob. Agents Chemother. 2009;53:3478. doi: 10.1128/AAC.00141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M, Lillibridge S, Osterholm MT, O’Toole T, Parker G, Perl TM, Russell PK, Swerdlow DL, Tonat K. JAMA. 2001;285:1059. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- 3.(a) Hackett R, Kam PC. Med. Chem. 2007;3:333. doi: 10.2174/157340607781024438. [DOI] [PubMed] [Google Scholar]; (b) Truong DD, Jost WH. Parkinsonism Relat. Disord. 2006;12:331. doi: 10.1016/j.parkreldis.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Willis B, Eubanks LM, Dickerson TJ, Janda KD. Angew. Chem. Int. Ed. 2008;47:8360. doi: 10.1002/anie.200705531. and references cited therein.
- 5.Oguma K, Fujinaga Y, Inoue K. Microbiol. Immunol. 1995;39:161. doi: 10.1111/j.1348-0421.1995.tb02184.x. [DOI] [PubMed] [Google Scholar]
- 6.(a) Burnett JC, Ruthel G, Stegmann CM, Panchal RG, Nguyen TL, Hermone AR, Stafford RG, Lane DJ, Kenny TA, McGrath CF, Wipf P, Stahl AM, Schmidt JJ, Gussio R, Brunger AT, Bavari S. J. Biol. Chem. 2007;282:5004. doi: 10.1074/jbc.M608166200. [DOI] [PubMed] [Google Scholar]; (b) Burnett JC, Wang C, Nuss JE, Nguyen TL, Hermone AR, Schmidt JJ, Gussio R, Wipf P, Bavari S. Bioorg. Med. Chem. Lett. 2009;19:5811. doi: 10.1016/j.bmcl.2009.01.111. [DOI] [PubMed] [Google Scholar]; (c) Pang YP, Vummenthala A, Mishra RK, Park JG, Wang S, Davis J, Millard CB, Schmidt JJ. PLoS One. 2009;4:e7730. doi: 10.1371/journal.pone.0007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Boldt GE, Kennedy JP, Janda KD. Org. Lett. 2006;8:1729. doi: 10.1021/ol0603211. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Capkova K, Hixon MS, McAllister LA, Janda KD. Chem. Commun. 2008:3525. doi: 10.1039/b808305c. [DOI] [PubMed] [Google Scholar]; (c) Park JG, Sill PC, Makiyi EF, Garcia-Sosa AT, Millard CB, Schmidt JJ, Pang YP. Bioorg. Med. Chem. 2006;14:395. doi: 10.1016/j.bmc.2005.08.018. [DOI] [PubMed] [Google Scholar]; (d) Tang J, Park JG, Millard CB, Schmidt JJ, Pang YP. PLoS One. 2007;2:e761. doi: 10.1371/journal.pone.0000761. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Stowe GN, Silhar P, Hixon MS, Silvaggi NR, Allen KN, Moe ST, Jacobson AR, Barbieri JT, Janda KD. Org. Lett. 2010;12:756. doi: 10.1021/ol902820z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silhar P, Capkova K, Salzameda NT, Barbieri JT, Hixon MS, Janda KD. J. Am. Chem. Soc. 2010;132:2868. doi: 10.1021/ja910761y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim DSHL, Chen Z, Nguyen T, Pezzuto JM, Qiu S, Lu Z-Z. Synth. Commun. 1997;27:1607. [Google Scholar]
- 10.Pisha E, Chai H, Lee IS, Chagwedera TE, Farnsworth NR, Cordell GA, Beecher CWW, Fong HHS, Kinghorn AD, Brown DM, Wani MC, Wall ME, Hieken TJ, Das Gupta TK, Pezzuto JM. Nat. Med. 1995;1:1046. doi: 10.1038/nm1095-1046. [DOI] [PubMed] [Google Scholar]
- 11.Soler F, Poujade C, Evers M, Carry JC, Henin Y, Bousseau A, Huet T, Pauwels R, De Clercq E, Mayaux JF, Le Pecq JB, Dereu N. J. Med. Chem. 1996;39:1069. doi: 10.1021/jm950669u. [DOI] [PubMed] [Google Scholar]
- 12.(a) Alakurtti S, Heiska T, Kiriazis A, Sacerdoti-Sierra N, Jaffe CL, Yli-Kauhaluoma J. Bioorg. Med. Chem. 2010;18:1573. doi: 10.1016/j.bmc.2010.01.003. [DOI] [PubMed] [Google Scholar]; (b) Alakurtti S, Bergström P, Sacerdoti-Sierra N, Jaffe CL, Yli-Kauhaluoma J. J. Antibiot. 2010;63:123. doi: 10.1038/ja.2010.2. [DOI] [PubMed] [Google Scholar]
- 13.Pohjala L, Alakurtti S, Ahola T, Yli-Kauhaluoma J, Tammela P. J. Nat. Prod. 2009;72:1917. doi: 10.1021/np9003245. [DOI] [PubMed] [Google Scholar]
- 14.Salin O, Alakurtti S, Pohjala L, Siiskonen A, Maass V, Maass M, Yli-Kauhaluoma J, Vuorela P. Biochem. Pharmacol. 2010;80:1141. doi: 10.1016/j.bcp.2010.06.051. [DOI] [PubMed] [Google Scholar]
- 15.(a) Yogeeswari P, Sriram D. Curr. Med. Chem. 2005;12:657. doi: 10.2174/0929867053202214. [DOI] [PubMed] [Google Scholar]; (b) Alakurtti S, Mäkelä T, Koskimies S, Yli-Kauhaluoma J. Eur. J. Pharm. Sci. 2006;29:1. doi: 10.1016/j.ejps.2006.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


