SUMMARY
Details are emerging on the structure and function of a remarkable class of capsid-like protein assemblies that serve as simple metabolic organelles in many bacteria. These bacterial microcompartments consist of a few thousand shell proteins, which encapsulate two or more sequentially acting enzymes in order to enhance or sequester certain metabolic pathways, particularly those involving toxic or volatile intermediates. Genomic data indicate that bacterial microcompartment shell proteins are present in a wide range of bacterial species, where they encapsulate varied reactions. Crystal structures of numerous shell proteins from distinct types of microcompartments have provided keys for understanding how the shells are assembled and how they conduct molecular transport into and out of microcompartments. The structural data emphasize a high level of mechanistic sophistication in the protein shell, and point the way for further studies on this fascinating but poorly appreciated class of subcellular structures.
Keywords: Protein assembly, capsid, molecular transport, pore, BMC, carboxysome
INTRODUCTION
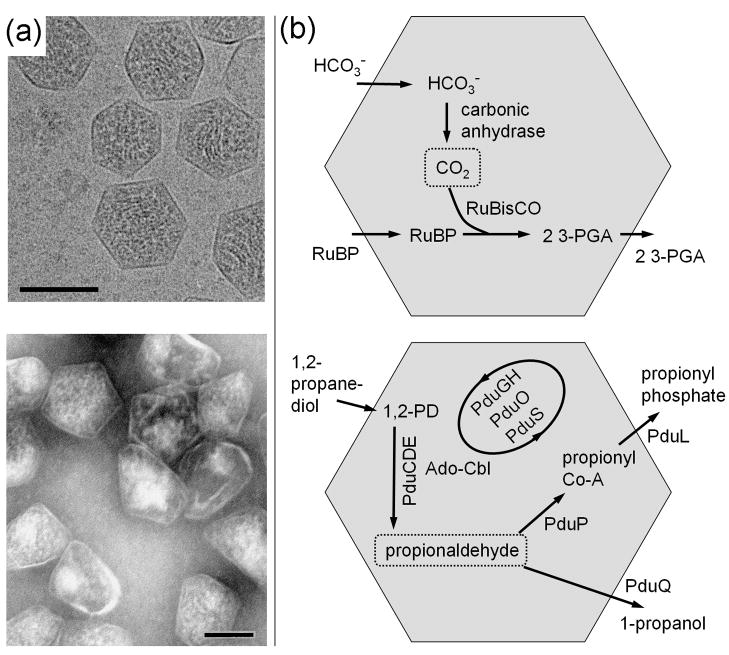
Many bacteria contain large, polyhedral, protein-based organelles referred to as bacterial microcompartments (reviewed in [1–4]). Polyhedral inclusions that had been visualized by electron microscopy in cyanobacteria and some chemoautotrophs were first isolated in 1973 and determined to contain the CO2-fixing enzyme RuBisCO[5]; they were therefore named carboxysomes, and are now recognized as the founding member of a diverse group of microcompartments (Fig. 1). Subsequent genetic and physiological studies on carboxysomes indicated that they played an active role in CO2 fixation[6–9]. As part of a mechanism for enhancing CO2 fixation, carbonic anhydrase, which dehydrates bicarbonate to CO2, is encapsulated in the carboxysome together with RuBisCO[10–12]. This arrangement is believed to provide a high concentration of CO2 for RuBisCO under growth conditions where inorganic carbon is limiting, and may confer other advantages as well[13]. Besides the carboxysome, several other types of bacterial microcompartments carrying out more complex reactions have been either studied directly or inferred from genomic data.
Figure 1.
Examples of bacterial microcompartments. (a) Electron micrographs of purified carboxysomes (top) from Halothiobacillus neapolitanus and purified propanediol utilization (Pdu) microcompartments (bottom) from Salmonella enterica. The scale bars are 100 nm. Under conditions where they are produced, several microcompartments are typically found within each bacterial cell. (b) Diagram of CO2 fixation in the carboxysome (top) and a simplified diagram of propanediol metabolism in the Pdu microcompartment (bottom). RuBP is 1,5-ribulose bisphosphate and 3-PGA is 3-phosphoglycerate. Reactions in the Pdu microcompartment involve multiple cofactors (not shown), including NAD+/NADH, ATP, coenzyme-A, and a B12 cofactor, with reactivation and replacement of the latter cofactor being carried out by the PduGH, PduS, and PduO enzymes. In each microcompartment, the key encapsulated intermediate is boxed. Carboxysomes were provided by Gordon Cannon and Sabine Heinhorst and imaged by Kelly Dryden and Mark Yeager.
A unifying feature of diverse bacterial microcompartments is a thin shell composed primarily of a few thousand small protein subunits belonging to a family of homologous so-called BMC (for bacterial microcompartment) shell proteins. This outer shell encapsulates the interior enzymes while allowing transport of substrates and products. BMC shell proteins, which were first identified in carboxysomes [14–16], were subsequently identified in the propanediol utilization operon (pdu) of Salmonella (a heterotroph)[17,18]. Genetic and biochemical studies demonstrated that the Pdu microcompartment encapsulates a series of enzymes that metabolize 1,2-propanediol (1,2-PD)[19,20](Fig. 1). Sequestering that metabolic pathway prevents exposure of the cytosol to propionaldehyde – an intermediate in the reaction pathway – which is toxic to the cell at high concentrations[20,21]. Likewise, genes for BMC shell proteins are found in an operon for metabolizing ethanolamine in enteric bacteria[22,23], including Salmonella and E. coli. The ethanolamine utilization (or Eut) microcompartment system shares similar chemistry and analogous enzymes with the Pdu system. Experiments on the Eut system in Salmonella indicate that the sequestered metabolism of ethanolamine prevents exposure of the cytosol to the reactive acetaldehyde intermediate[24], while also preventing the detrimental evaporative loss of that compound from the cell[25]. Compared to the carboxysome, the latter two types of microcompartments are more complex. Besides encapsulating more enzymes, the interior reactions involve numerous cofactors, including adenosyl cobalamin (B12), NAD+/NADH, acetyl-CoA, ATP, and [Fe-S] clusters (reviewed in [1,3]).
Motivated by the discovery of BMC shell proteins in diverse bacteria, searches for homologous shell proteins across the known protein sequence databases have emphasized the widespread occurrence of microcompartments across the bacterial kingdom, and their likely spread by horizontal gene transfer[2–4,26,27]. Approximately 1700 unique proteins containing BMC domains can be identified at present, covering at least 10 different bacterial phyla. Multiple paralogs of the shell proteins are essentially always found together. Experimental studies on these wide ranging systems are limited, though inferences regarding their likely functions can be made in some cases from the genomic contexts in which the BMC shell proteins are found[2,3]. Enzymes found to occur often in chromosomal proximity to BMC shell proteins have been tabulated[3]. These represent avenues for exploratory experimental work.
Until recently, the structure and mechanisms of bacterial microcompartment shells were unclear. At the architectural level, early electron microscopy studies on carboxysomes had not provided a definitive shape. Likewise, at the atomic level, three-dimensional details were lacking. How the shell proteins assembled to form a semi-permeable layer, and why multiple distinct paralogs are involved, were unknown. Crystal structures of numerous BMC shell proteins, first from the carboxysome[27–31] and then from the Pdu[32,33], Eut[34–36], and other systems[37], have clarified how BMC shell proteins assemble to form a shell, how they might facilitate molecular transport through their pores, and how distinct paralogs play specialized roles in microcompartment organelles.
ELEMENTS OF BMC SHELL PROTEIN ASSEMBLY
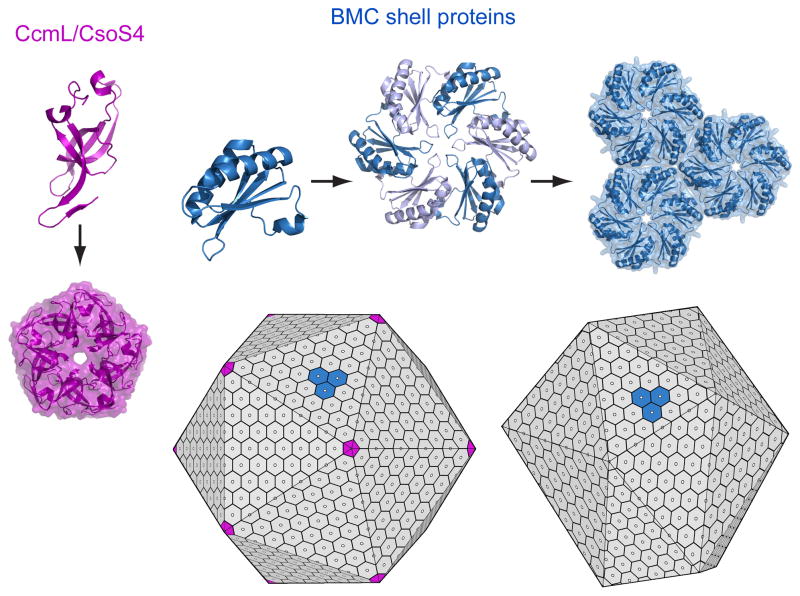
The typical BMC domain is about 90 amino acids in length and adopts an alpha/beta fold ([27], reviewed in [3]). Individual BMC proteins self-assemble to form cyclic, disc-shaped hexamers that constitute the basic building blocks of the shell (Fig. 2). Each hexamer typically presents a narrow pore through the middle, along the six-fold axis of symmetry. Although individual BMC proteins differ, as described subsequently, in most cases the two sides of the disc reveal dramatically different shape properties. One side, described as concave, generally bears a bowl-shaped depression with a relatively hydrophobic surface. The N and C-termini, which typically reside on this concave side, tend to diverge in length and structure between BMC paralogs, and often appear flexible or disordered in crystal structures. This side of the shell proteins has been argued to be the side facing towards the microcompartment interior[38].
Figure 2.
Structure and assembly of microcompartment shells. BMC shell proteins (top) form hexamers that constitute the main building blocks of the shell. These assemble further to form tightly packed layers with pores in the middle of the hexamers, some of which have gated openings. BMC shell proteins are found widely across the bacteria. Where they occur, they provide a signature for the presence of a microcompartment organelle. In the carboxysome, pentameric proteins (CcmL and CsoS4) likely form the vertices of the shell, which is nearly icosahedral in shape (lower left). Pdu microcompartments and some other types form less geometrically regular shells (lower right) from hexameric BMC proteins; the presence of pentameric shell proteins in those cases has not been established.
Crystal structures show that individual BMC hexamers are tailored to further assemble side-by-side, thereby forming a tightly packed molecular layer[27,30,35,36,38](Fig. 2). Multiple lines of evidence indicate that the molecular layers visualized within the context of three-dimensional crystals represent the arrangement of proteins in the facets of natural biological shells. The specific mode of packing is conserved in at least six of the crystal structures reported to date, and key intermolecular hydrogen bonds are conserved at the interfaces in cases where hexamers pack tightly together.
Furthermore, true two-dimensional layers of BMC proteins can be formed at the air-water interface of a liquid droplet, where they recapitulate the packing seen in three-dimensional crystals[39]. From these extended arrays, a picture of the shell emerges as a tightly packed molecular layer perforated by narrow protein pores spaced just less than 70 Å apart (Fig. 2).
The question of how an otherwise flat layer of BMC shell protein hexamers bends or folds up to make a closed shell was answered in part by structural studies on another family of conserved proteins that, along with the BMC protein family, also appears to be present in all microcompartments. Homologous proteins CcmL and CsoS4A from two different types of carboxysome were both shown to be pentamers whose size and shape are compatible with their placement at the vertices of an icosahedral shell built from many hexamers and 12 pentamers[29](Fig. 2). An icosahedral structure for the carboxysome was anticipated by the hexameric structure of the BMC shell proteins[27], and confirmed by EM cryotomography studies[40,41]. A scenario in which the CcmL or CsoS4 pentamers occupy icosahedral vertices is consistent with the low abundance of these proteins in the shell, and with mutational experiments in which deletion of the genes (or their homologs in other systems) led to extended assemblies that generally failed to close up[42,43]. However, recent mutagenesis experiments on the carboxysome suggest that the situation may be more complicated[44]. A further complication is presented by homologs of this protein family from other types of microcompartments. The homologous protein in the Eut microcompartment is EutN, which forms hexamers instead of pentamers when expressed and crystallized by itself[29]; whether EutN and other members of this protein family from different microcompartment types are pentamers in the context of an intact shell is unknown. It is notable however that the Pdu and Eut microcompartments do not resemble a regular icosahedron as closely as the carboxysome does; this may correlate with the different behavior of the CcmL/CsoS4/EutN/PduN family of proteins in different systems.
THE ROLES OF BMC PROTEIN FOLD VARIATIONS
Crystal structures of numerous BMC proteins have revealed a surprising array of conformational and topological variations within a small protein domain (reviewed in [3])(Table 1). Some BMC proteins are related to the canonical type by a circular permutation, so that a similar tertiary structure arises from secondary structures occurring in a different order[28,32–34,36,37]. This produces N and C termini at different spatial locations, which may be an important feature. In permuted BMC proteins PduU and EutS, the occurrence of the N-terminus on the other side of the disk makes it possible for an extended tail to create a 6-stranded beta barrel that blocks the pore[33,36].
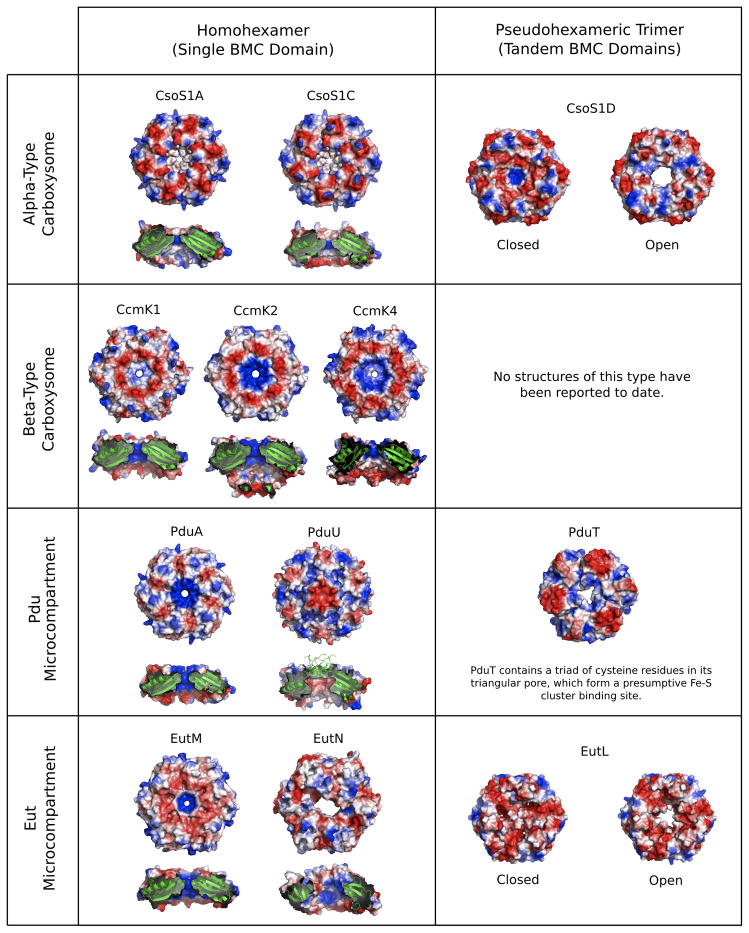
Table 1.
Known structures of shell proteins from bacterial microcompartments.
| Protein | Structure | Properties |
|---|---|---|
| α-Carboxysome (e.g. Halothiobacillus neapolitanus) | ||
| CsoS1A | Single-BMC domain hexamer | Major shell component. |
| CsoS1C | Single-BMC domain hexamer | Major shell component. |
| CsoS1D | Tandem-BMC domain (permuted) trimer | Trimers stack to form a hexamer with D3 symmetry; open and closed conformations suggest gated transport. |
| CsoS4A/B | Pentamer (non-BMC domain) | Minor shell component; believed to form icosahedral vertices. |
| β-Carboxysome (e.g. Synechocystis PCC6803) | ||
| CcmK1 | Single-BMC domain hexamer | Major shell component. |
| CcmK2 | Single-BMC domain hexamer | Major shell component. |
| CcmK4 | Single-BMC domain hexamer | Minor shell component. |
| CcmL | Pentamer (non-BMC domain) | Minor shell component; believed to form icosahedral vertices. |
| Pdu Microcompartment (e.g. Salmonella enterica) | ||
| PduA | Single-BMC domain hexamer | Major shell component. |
| PduT | Tandem-BMC domain trimer | Pore likely involved in electron transport or Fe-S cluster transport. |
| PduU | Single-BMC domain hexamer (permuted) | Minor shell component; pore is blocked by 6-stranded β-barrel in crystal structure. |
| Eut Microcompartment (e.g. S. enterica & E. coli) | ||
| EutK | Single-BMC domain fused to helix-turn-helix (HTH) domain | HTH motif with positively charged surface suggests role in nucleic acid binding. |
| EutL | Tandem-BMC domain trimer | Pore observed in open and closed conformations; probable role in gated transport. |
| EutM | Single-BMC domain hexamer | Probable major shell component. |
| EutN | Non-BMC domain hexamer | Crystallizes as a hexamer despite homology to pentameric CcmL/CsoS4 family; role unclear. |
| EutS | Single-BMC domain (permuted) hexamer | Hexamer forms bent structure with C2 symmetry; possible role in edge formation within shell. |
| Other Microcompartments | ||
| EtuB | Tandem-BMC domain (permuted) trimer | Part of presumptive ethanol utilization microcompartment in Clostridium kluyveri. |
| PduT homolog | Tandem-BMC domain trimer | Part of microcompartment of uncharacterized function in Desulfitobacterium halfniense. |
Numerous BMC proteins, including at least one from all the microcompartments that have been analyzed to date, contain tandem BMC domains. In such cases, three copies of the protein assemble to make a symmetric trimer with pseudo six-fold symmetry[28,32,34,36,37]. Examples of such structures have been obtained from four types of microcompartments. In two of these cases (CsoS1D and EutL), crystal structures have been visualized of two alternate forms related by dramatic conformational changes that open and nearly close the central pore[28,35,36]. The symmetry breaking that follows from domain duplication appears to facilitate conformational variation and flexibility[3,37]. Also notable is the finding that different tandem BMC domain proteins can have their consecutive domains arranged in opposite orders in the pseudohexamer[36]. Furthermore, some tandem domain proteins are built from two circularly permuted domains (CsoS1D, EutL and EtuB), while some are built from two canonical BMC domains (PduT)[32], further highlighting the evolution of structural variety in this protein family.
Another form of symmetry breaking was observed in the structure of the permuted, single BMC domain protein, EutS. Here, six chemically identical subunits form a hexagonal disc that is bent or creased by about 40°[36]. Mutagenesis and sequence comparisons identify a glycine substitution as critical for formation of the bent structure. Such deviations from symmetry are rare in nature, and often have functional significance[45,46, reviewed in 47]. This particular BMC paralog could form edges in the context of an intact shell, or generally promote curvature of the BMC protein layer.
Further structural variation appears in the form of double disk structures, which have been seen in one carboxysome protein (CsoS1D)[28] and one protein (homologous to PduT) from Desulfitobacterium hafniense (PDBID 3NWG). CsoS1D displays open and nearly closed conformations, which could allow alternating access to the chamber formed by two opposing discs[28]. Alternatively, it was suggested that the observed two-tier structure could either indicate that shells are actually comprised of double protein layers, or could represent the way adjacent microcompartment shells might be bound together. However, the presence of a relative twist between the two discs appears incompatible with formation of two layers in an extended register, which weighs against the latter two scenarios.
Finally, numerous proteins contain BMC domains as fusions with other domains. The full range of fused domains has not been characterized, and only one structure has been elucidated to date. In the Eut microcompartment, the EutK protein contains a C-terminal domain of approximately 60 amino acids. A crystal structure of the fusion domain revealed a helix-turn-helix motif, which is common in nucleic acid binding proteins, and a positively charged surface consistent with that putative function[36]. The unexpected prediction of a connection between BMC proteins and nucleic acids presents an open line for investigation.
PROTEIN PORES AND MOLECULAR TRANSPORT
The small pores visualized in the centers of typical BMC shell proteins are presumed to be the routes of transport for the substrates, products, and cofactors for varied microcompartments[27,30,32]. Size, electrostatic and hydrogen bonding features of the pores have been identified as potentially important in facilitating the transport of substrates and products more readily than the sequestered metabolic intermediates (Fig. 3). BMC proteins that represent major components of their respective shells all appear to possess these pores, whereas some other minor BMC shell components (e.g. PduU) have occluded pores. This trend suggests that the porosity conferred by the system of small pores is an important property of the shells[30]. Considerably larger pores in BMC shell proteins have been visualized in two cases so far[28,34,36]. In both cases, the observation of alternate, nearly closed conformations, suggests the likelihood of gated transport, possibly as a mechanism to allow the transport of larger substrates or cofactors while somehow limiting the escape of sequestered intermediates.
Figure 3.
BMC pore variations and transitions. Diagrams illustrate the structural diversity of several different pores in BMC shell proteins from experimentally characterized microcompartments (carboxysomes, Pdu, and Eut). Varied shell proteins exhibit differences in size, shape and electrostatic properties (blue=positive, red=negative). Some shell proteins that have tandem BMC domains are able to alter their porosity through conformational changes that open and close the pore. In addition to the presence of gated pores, other interesting pore features include: a beta-barrel that plugs an otherwise wide and negatively charged pore in PduU; a 4Fe-4S cluster binding site at the center of the PduT pore, and a distorted pore at the center of EutN, which is not made up of BMC proteins, but is instead a hexamer of CcmL/CsoS4-type protein subunits. The diverse properties illustrated are responsible for the ability of BMC shells to transport a wide range of substrates, products, and cofactors, while simultaneously limiting the efflux of metabolic intermediates. In each panel, the shell proteins are oriented so that the top surface represents the side predicted to represent the outside of the microcompartment (i.e. the side facing the cytosol). Known shell protein structures not illustrated here include CcmL, CsoS4A, EutS and EtuB.
A recent structure of the PduT shell protein from Salmonella provides the first indication of how the redox state inside the Pdu microcompartment might be balanced[32]. Spectroscopic data indicated that PduT binds a [4Fe-4S] cluster[48], which was determined by crystallography to be bound in the center of the pore by cysteine residues contributed by three protein subunits[32]. It has been proposed that the metal cluster in the pore either (1) facilitates electron flow, presumably out of the microcompartment in order to regenerate NAD+ required to oxidize propanediol, or (2) provides a route for transporting intact iron-sulfur clusters into the microcompartment to replace damaged metal clusters in interior enzymes, including PduS. The latter hypothesis is supported by the recent structure of a PduT homolog (PDBID 3NWG) with an alternate pore conformation lacking a metal cluster.
Despite the tantalizing clues regarding molecular transport offered by the structural findings, experimental data are lacking. In some crystal structures, electron density features in the central pores have suggested the presence of substrates or products, but the evidence has been inconclusive [30,32,38]. Those observations could reflect weak affinity for molecules that need to move through the pores. The structural data do however provide a framework for investigating the proposed transport phenomena by mutagenesis and physiological studies. Those experiments will be vital for advancing our understanding of molecular transport.
TARGETING ENZYMES TO THE SHELL INTERIOR
Some of the details concerning how enzymes are targeted to the interior surfaces of their respective microcompartments are beginning to emerge. Interestingly, at least two different kinds of mechanism appear to operate in different microcompartments. In carboxysomes belonging to the so-called beta type, a highly unusual protein, CcmM, appears to serve as a scaffold for establishing interactions to both shell proteins and enzymes[49,50]. CcmM bears an N-terminal domain that carries a redox-sensitive carbonic anhydrase activity[51] and a C-terminal region that carries multiple tandem domains with homology to the small subunit of RuBisCO[43,52]; these presumably mimic the native interactions of the small subunit with large subunits of RuBisCO inside the carboxysome.
In the Pdu microcompartment, and likely the Eut and other closely related types, recent experiments have demonstrated that some of the internal enzymes are directed to the interior by special N-terminal targeting sequences[42,53]. The simplicity of this system makes it attractive for engineering studies aimed at encapsulating distinct enzymes in order to create novel reaction chambers. The feasibility of such a goal is supported by the demonstration that the Pdu shell can be assembled heterologously in the absence of native interior enzymes[42]; similarly, carboxysomes have been shown to assemble in vivo when RuBisCO has been deleted[54]. The timing of events in the formation of a shell and the encapsulation of enzymes has been partially clarified by examination of electron microscopy images of carboxysomes in various stages of apparent assembly, which suggest that enzymes (i.e. RuBisCO) form layers together with the shell proteins early in the process of forming facets of the shell, so that the shell and its interior materialize simultaneously[55].
OPEN QUESTIONS AND FUTURE DIRECTIONS
Myriad questions remain concerning the function and evolution of bacterial microcompartments. Recent studies indicate that active cellular mechanisms may control the arrangement and movement of microcompartments in the cell[42,56], and that microcompartments may interact with other cellular components and inclusions[55]. Further studies are required to understand these cellular interactions. Likewise, little is known about how various enzymes are organized within microcompartments. Those details will be critical to understand how metabolic flux is enhanced.
Despite the obvious architectural similarity, no evolutionary linkage can be drawn at the present time between bacterial microcompartments and viruses; the BMC shell proteins do not resemble known viral capsid proteins. This is notable in contrast to the recent discovery in archaea of encapsulins, 60-subunit protein shells that encapsulate enzymes using a shell protein whose structure reveals a common ancestry with the capsid protein from the HK97 family of viruses[57]. With a single known exception, BMC shell proteins have been reported only in bacteria. They are not detected in plants or even primitive algae [58], which is notable given the origin of the chloroplast via endosymbiosis of an ancestral cyanobacterium that may have contained carboxysomes. Among eukaryotes, carboxysome shell proteins have only been reported in the unusual amoeboid, Paulinella chromatophora [59], which acquired a photosynthetic inclusion via a relatively recent endosymbiosis of a cyanobacterium.
The varied proteinaceous organelles described here all possess shells that derived from a common ancestral shell gene. Their appearance therefore represents a singular evolutionary development in the bacterial lineage. On the other hand, a genomic context study hints that other analogous systems might await discovery[60].
Acknowledgments
The authors thank Dr. Christopher Crowley for a critical reading of the manuscript. This work was supported by NIH grant AI081146 (TOY and TAB) and NSF grant MCB-0843065 (TOY). MCT is supported by a Ruth L. Kirschstein National Research Service Award GM007185.
Footnotes
CONFLICTS
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cheng S, Liu Y, Crowley CS, Yeates TO, Bobik TA. Bacterial microcompartments: their properties and paradoxes. Bioessays. 2008;30:1084–1095. doi: 10.1002/bies.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerfeld CA, Heinhorst S, Cannon GC. Bacterial microcompartments. Annu Rev Microbiol. 2010;64:391–408. doi: 10.1146/annurev.micro.112408.134211. [DOI] [PubMed] [Google Scholar]
- 3.Yeates TO, Crowley CS, Tanaka S. Bacterial microcompartment organelles: protein shell structure and evolution. Annu Rev Biophys. 2010;39:185–205. doi: 10.1146/annurev.biophys.093008.131418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeates TO, Kerfeld CA, Cannon GC, Heinhorst S, Shively JM. Protein-based organelles in bacteria: Carboxysomes and related microcompartments. Nature Rev Micro. 2008;6:681–691. doi: 10.1038/nrmicro1913. [DOI] [PubMed] [Google Scholar]
- 5**.Shively JM, Ball F, Brown DH, Saunders RE. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science. 1973;182:584–586. doi: 10.1126/science.182.4112.584. The polyhedral bodies were purified from a chemoautotroph and shown to contain RuBiSCO, surrounded by an outer layer or barrier. The name carboxysome was proposed. [DOI] [PubMed] [Google Scholar]
- 6.Buedeker RF, Cannon GC, Kuenen JG, Shively JM. Relations between D-ribulose-1, 5 bisphosphate carboxylase, carboxysomes, and CO2 fixing capacity in the obligate chemolithotroph Thiobacillus neapolitanus grown under different limitations in the chemostat. Arch Microbiol. 1980;124:185–189. [Google Scholar]
- 7.Cannon GC, English RS, Shively JM. In situ assay of ribulose-1,5-bisphosphate carboxylase/oxygenase in Thiobacillus neapolitanus. J Bacteriol. 1991;173:1565–1568. doi: 10.1128/jb.173.4.1565-1568.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price GD, Badger MR. Expression of human carbonic anhydrase in the cyanobacterium Synechococcus PCC7942 creates a high CO2-requiring phenotype: evidence for a central role for carboxysomes in the CO2 concentrating mechanism. Plant Physiol. 1989;91:505–513. doi: 10.1104/pp.91.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turpin DH, Miller AG, Canvin DT. Carboxysome content of Synechococcus leopoliensis (cyanophyta) in response to inorganic carbon. J Phycol. 1984;20:249–253. [Google Scholar]
- 10*.Cannon GC, Shively JM. Characterization of homogenous preparation of carboxysomes from Thiobacillus neapolitanus. Arch Microbiol. 1983;134:52–59. The protein composition of purified carboxysomes was determined. [Google Scholar]
- 11.Price GD, Coleman JR, Badger MR. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 1992;100:784–793. doi: 10.1104/pp.100.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.So AK, Espie GS, Williams EB, Shively JM, Heinhorst S, Cannon GC. A novel evolutionary lineage of carbonic anhydrase (epsilon class) is a component of the carboxysome shell. J Bacteriol. 2004;186:623–630. doi: 10.1128/JB.186.3.623-630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot. 2003;54:609–622. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- 14.Cannon GC, Heinhorst S, Bradburne CE, Shively JM. Carboxysome genomics: A status report. Funct Plant Biol. 2002;29:175–182. doi: 10.1071/PP01200. [DOI] [PubMed] [Google Scholar]
- 15*.English RS, Lorbach SC, Qin X, Shively JM. Isolation and characterization of a carboxysome shell gene from Thiobacillus neapolitanus. Mol Microbiol. 1994;12:647–654. doi: 10.1111/j.1365-2958.1994.tb01052.x. The N-terminal sequence of a shell protein was determined and used to identify genes coding for the protein shell and their sequences, setting the stage for later genetic, structural, and comparative genomic studies on microcompartments. [DOI] [PubMed] [Google Scholar]
- 16.Price GD, Sültemeyer D, Klughammer B, Ludwig M, Badger MR. The functioning of the CO2 concentrating mechanism in several cyanobacterial strains: a review of general physiological characteristics, genes, proteins and recent advances. Can J Bot. 1998;76:973–1002. [Google Scholar]
- 17.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1, 2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Chen P, Andersson DI, Roth JR. The control region of the pdu/cob regulon in Salmonella typhimurium. J Bacteriol. 1994;176:5474–5482. doi: 10.1128/jb.176.17.5474-5482.1994. The identification of a gene homologous to carboxysome shell proteins in an operon for degrading propanediol in Salmonella suggested that microcompartments exist for metabolic functions beyond CO2 fixation, thereby expanding the roles of bacterial microcompartments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Havemann GD, Bobik TA. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol. 2003;185:5086–5095. doi: 10.1128/JB.185.17.5086-5095.2003. The protein composition of the Pdu microcompartment was analyzed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havemann GD, Sampson EM, Bobik TA. PduA is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol. 2002;184:1253–1261. doi: 10.1128/JB.184.5.1253-1261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampson EM, Bobik TA. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol. 2008;190:2966–2971. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stojiljkovic I, Baumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rondon MR, Kazmierczak R, Escalante-Semerena JC. Glutathione is required for maximal transcription of the cobalamin biosynthetic and 1,2-propanediol utilization (cob/pdu) regulon and for the catabolism of ethanolamine, 1,2-propanediol, and propionate in Salmonella typhimurium LT2. J Bacteriol. 1995;177:5434–5439. doi: 10.1128/jb.177.19.5434-5439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penrod JT, Roth JR. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol. 2006;70:517–525. doi: 10.1007/s00253-005-0295-0. [DOI] [PubMed] [Google Scholar]
- 27**.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. The first crystal structures of bacterial microcompartment shell proteins were determined, from a beta-type carboxysome. Disk-like hexameric protein assemblies, bearing central pores for molecular transport, and forming tightly packed molecular layers, were established as the fundamental building blocks of microcompartment shells. [DOI] [PubMed] [Google Scholar]
- 28**.Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol. 2009;392:319–333. doi: 10.1016/j.jmb.2009.03.056. The structure was reported of an overlooked carboxysome shell protein comprised of tandem BMC domains, and revealing an open vs. closed conformational change, as well as a double-disk assembly. [DOI] [PubMed] [Google Scholar]
- 29**.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. Atomic-level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. doi: 10.1126/science.1151458. The elucidation of pentameric structures for previously uncharacterized proteins encoded in carboxysome operons provided an explanation for curvature of the shell. Rough models of a complete carboxysome shell were proposed. [DOI] [PubMed] [Google Scholar]
- 30.Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 2007;5:e144. doi: 10.1371/journal.pbio.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai Y, Sawaya MR, Yeates TO. Analysis of lattice-translocation disorder in the layered hexagonal structure of carboxysome shell protein CsoS1C. Acta Crystallogr D Biol Crystallogr. 2009;65:980–988. doi: 10.1107/S0907444909025153. [DOI] [PubMed] [Google Scholar]
- 32*.Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO. Structural insights into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell. J Biol Chem. 2010;285:37838–46. doi: 10.1074/jbc.M110.160580. Structural data were used to locate an iron-sulfur cluster in the pore of a shell protein from the Pdu microcompartment, providing evidence for electron or metal cluster transport across the shell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Crowley CS, Sawaya MR, Bobik TA, Yeates TO. Structure of the PduU shell protein from the Pdu microcompartment of Salmonella. Structure. 2008;16:1324–1332. doi: 10.1016/j.str.2008.05.013. The first structure is reported for a shell protein from a microcompartment other than a carboxysome. The protein fold was seen to be a circular permutation of the typical BMC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Sagermann M, Ohtaki A, Nikolakakis K. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc Natl Acad Sci U S A. 2009;106:8883–8887. doi: 10.1073/pnas.0902324106. The first structure of a shell protein from the Eut microcompartment is reported. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takenoya M, Nikolakakis K, Sagermann M. Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the Eut-BMC. J Bacteriol. 2010 doi: 10.1128/JB.00652-00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Tanaka S, Sawaya MR, Yeates TO. Structure and mechan8isms of a protein-based organelle in Escherichia coli. Science. 2010;327:81–84. doi: 10.1126/science.1179513. Crystal structures of the varied shell proteins of the Eut microcompartment, present in Salmonella and some strains of E. coli, highlight their diverse roles. An open vs. closed conformational change was visualized in one shell protein, EutL. [DOI] [PubMed] [Google Scholar]
- 37*.Heldt D, Frank S, Seyedarabi A, Ladikis D, Parsons JB, Warren MJ, Pickersgill RW. Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. Biochem J. 2009;423:199–207. doi: 10.1042/BJ20090780. A crystal structure of a tandem BMC shell protein is reported from a yet uncharacterized microcompartment believed to metabolize ethanol. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka S, Sawaya MR, Phillips M, Yeates TO. Insights from multiple structures of the shell proteins from the beta-carboxysome. Protein Sci. 2009;18:108–120. doi: 10.1002/pro.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dryden KA, Crowley CS, Tanaka S, Yeates TO, Yeager M. Two-dimensional crystals of carboxysome shell proteins recapitulate the hexagonal packing of three-dimensional crystals. Protein Sci. 2009;18:2629–2635. doi: 10.1002/pro.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40*.Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J Mol Biol. 2007;372:764–773. doi: 10.1016/j.jmb.2007.06.059. Electron cryotomography showed that alpha-type carboxysomes from a cyanobacterium are essentially icosahedral. Interior enzymes, presumably RuBisCO, were visualized. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol. 2006;364:526–535. doi: 10.1016/j.jmb.2006.09.024. Electron cryotomography showed that alpha-type carboxysomes from a chemoautotroph are essentially icosahedral. Interior enzymes, presumably RuBisCO, were visualized. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Parsons JB, Frank S, Bhella D, Liang M, Prentice MB, Mulvihill DP, Warren MJ. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol Cell. 2010;38:305–315. doi: 10.1016/j.molcel.2010.04.008. Empty Pdu-type microcompartments were expressed heterologously and assembled in E. coli. Together with protein targeting studies, the result supports a strategy for designing novel microcompartments. [DOI] [PubMed] [Google Scholar]
- 43.Price GD, Howitt SM, Harrison K, Badger MR. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J Bacteriol. 1993;175:2871–2879. doi: 10.1128/jb.175.10.2871-2879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One. 2009;4:e7521. doi: 10.1371/journal.pone.0007521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 48.Parsons JB, Dinesh SD, Deery E, Leech HK, Brindley AA, Heldt D, Frank S, Smales CM, Lunsdorf H, Rambach A, et al. Biochemical and structural insights into bacterial organelle form and biogenesis. J Biol Chem. 2008;283:14366–14375. doi: 10.1074/jbc.M709214200. [DOI] [PubMed] [Google Scholar]
- 49.Cot SS, So AK, Espie GS. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J Bacteriol. 2008;190:936–945. doi: 10.1128/JB.01283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Long BM, Badger MR, Whitney SM, Price GD. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem. 2007;282:29323–29335. doi: 10.1074/jbc.M703896200. [DOI] [PubMed] [Google Scholar]
- 51.Pena KL, Castel SE, de Araujo C, Espie GS, Kimber MS. Structural basis of the oxidative activation of the carboxysomal gamma-carbonic anhydrase, CcmM. Proc Natl Acad Sci U S A. 2010;107:2455–2460. doi: 10.1073/pnas.0910866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ludwig M, Sultemeyer D, Price GD. Isolation of ccmKLMN genes from the marine cyanobacterium, Synechococcus sp. PCC7002 (Cyanophyceae), and evidence that CcmM is essential for carboxysome assembly. J Phyco. 2000;36:1109–1119. [Google Scholar]
- 53*.Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci U S A. 2010;107:7509–7514. doi: 10.1073/pnas.0913199107. It was demonstrated that an N-terminal sequence extension on a Pdu enzyme (PduP) is necessary for encapsulating PduP, and can be used to encapsulate other proteins by fusing them to the tail. This provides a mechanism for encapsulating novel enzyme components. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Menon BB, Dou Z, Heinhorst S, Shively JM, Cannon GC. Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric RubisCO species. PLoS One. 2008;3:e3570. doi: 10.1371/journal.pone.0003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ. Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells. J Mol Biol. 2010;396:105–117. doi: 10.1016/j.jmb.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327:1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- 57.Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, Ban N. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat Struct Mol Biol. 2008;15:939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 58.Burey SC, Poroyko V, Ergen ZN, Fathi-Nejad S, Schuller C, Ohnishi N, Fukuzawa H, Bohnert HJ, Loffelhardt W. Acclimation to low [CO2] by an inorganic carbon-concentrating mechanism in Cyanophora paradoxa. Plant Cell Environ. 2007;30:1422–1435. doi: 10.1111/j.1365-3040.2007.01715.x. [DOI] [PubMed] [Google Scholar]
- 59.Marin B, Nowack EC, Glockner G, Melkonian M. The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like gamma-proteobacterium. BMC Evol Biol. 2007;7:85. doi: 10.1186/1471-2148-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beeby M, Bobik TA, Yeates TO. Exploiting genomic patterns to discover new supramolecular protein assemblies. Protein Sci. 2009;18:69–79. doi: 10.1002/pro.1. [DOI] [PMC free article] [PubMed] [Google Scholar]