Abstract
NUT midline carcinoma (NMC) is a lethal pediatric tumor defined by the presence of BRD-NUT fusion proteins that arrest differentiation. Here we explore the mechanisms underlying the ability of BRD4-NUT to prevent squamous differentiation. In both gain-of and loss-of-expression assays we find that expression of BRD4-NUT is associated with globally decreased histone acetylation and transcriptional repression. Bulk chromatin acetylation can be restored by treatment of NMC cells with histone deacetylase inhibitors (HDACi), engaging a program of squamous differentiation and arrested growth in vitro that closely mimics the effects of siRNA mediated attenuation of BRD4-NUT expression. The potential therapeutic utility of HDACi differentiation therapy was established in three different NMC xenograft models, where it produced significant growth inhibition and a survival benefit. Based on these results and translational studies performed with patient-derived primary tumor cells, a child with NMC was treated with the FDA-approved HDAC inhibitor, vorinostat. An objective response was obtained after five weeks of therapy, as determined by positron emission tomography. These findings provide preclinical support for trials of HDACi in patients with NMC.
Keywords: BRD4, NUT, epigenetic, differentiation, fusion oncogene
Introduction
Mechanistic study of cancer-associated translocations has led to the development of effective therapies that target fusion oncoproteins, particularly constitutively active tyrosine kinases (1–3). In contrast, while successes such as targeting of PML-RARα fusion proteins with all-trans retinoic acid and arsenic trioxide have been described (4–6), discovery of ligands that directly target fusion oncoproteins that function as transcriptional co-factors has proven to be challenging. Due to this difficulty, there is great interest in targeting the enzymatic components of higher-order gene regulatory complexes, such as histone deacetylases (HDACs) (7), but clinical application of this approach has been limited. Examples include the use of DNA methyltransferase inhibitors (DNMTi) in myelodysplastic syndrome (MDS) (8) and HDAC inhibitors in cutaneous T-cell lymphoma (CTCL) (9).
NUT midline carcinoma (NMC) is a distinctive, aggressive human cancer defined by rearrangements of the gene NUT (10). These poorly differentiated carcinomas usually arise in midline structures of the nasopharynx or mediastinum. Although rare, NMCs occur throughout life and are often mistaken for other entities, including thymic carcinoma, squamous cell carcinoma of the head and neck, lung carcinoma, Ewing sarcoma, and acute leukemia. Advanced local disease is frequently accompanied by distant hematogenous metastases. Commonly used therapies include surgical debulking, consolidative radiotherapy and cytotoxic chemotherapy, but even with multimodality therapy the median survival from diagnosis is only 9.5 months.
In the majority of NMCs, most of the coding sequence of NUT on chromosome 15q14 is fused in-frame to the 5’ portions of BRD4 or BRD3, creating chimeric genes that encode BRD-NUT fusion proteins (11, 12). The fusion proteins retain BRD-encoded bromodomains, which bind acetylated histones and recruit chromatin remodeling complexes (13). BRD4 facilitates transcriptional elongation and is associated with mitotic chromosomes, a property that may act to preserve epigenetic marks in daughter cells (14–16). NUT encodes an unstructured polypeptide of unknown function that is highly expressed in normal spermatids (11). A major oncogenic effect of the BRD4-NUT fusion protein appears to lie in its ability to arrest the differentiation of NMC cells (12). Based on the recently reported observation (17) that NUT directly binds to the histone acetyltransferase (HAT), p300, it was hypothesized that BRD4-NUT sequesters HAT activity. Here, we present data consistent with this model in which BRD-NUT fusion proteins act by inducing global histone hypoacetylation and transcriptional repression, effects that can be reversed by HDAC inhibitors, which show promise as targeted therapeutic agents for NMC.
Materials and Methods
Mammalian cells
The NMC cell lines TC797 (18), PER-403 (19), 00–143 (20), and TY82 (21), and the non-NMC squamous cell carcinoma cell lines, HTB-43 (pharyngeal squamous cell carcinoma, (22)) and HCC-95 (lung squamous cell carcinoma, (23), have been described. Patient tumor tissue was minced, digested with collagenase, and cultured in WIT medium optimized for carcinoma cells as described (24). 293T and U2OS cells were obtained from the American Type Culture Collection (Manassas, VA). A derivative of 293Ts, 293Trex, which contains a single genomic FRT recombination site (25), was a gift from Dr. Jeffrey D. Parvin. A tetracycline-inducible isogenic derivative, 293Trex-FLAG-BRD4-NUT was created by recombination with the plasmid pcDNA5 FRT/TO-FLAG-BRD4-NUT (below) using Flp-In technology (Invitrogen, Carlsbad, CA).
TC797, 00–143, TY82, 10326, U2OS, and 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% bovine growth serum (Hyclone, Logan, UT), 2 mM L-glutamine, 100 U of penicillin G/ml, and 100 µg of streptomycin/ml (Invitrogen) at 37 °C under 5% CO2. PER-403 was maintained in the same media with 1 mM sodium pyruvate (Mediatech, Herndon, VA), 0.1 mM non-essential amino acids (Invitrogen), and 40 µM β-mercaptoethanol (American Bioanalytical, Natick, MA). U2OS and 293T cells were transfected with Lipofectamine 2000 (Invitrogen). All work with human discarded tissues and live cells was performed in accordance with IRB protocol 2000-P-001990/6; BWH. Trichostatin A (used at a concentration of 25nM (Fig. 2–3) or 100nM (Figure S1)) was was obtained from Sigma-Aldrich (St. Louis, MO), and dimethyl sulfoxide (DMSO) from American Bioanalytical (Natick, MA).
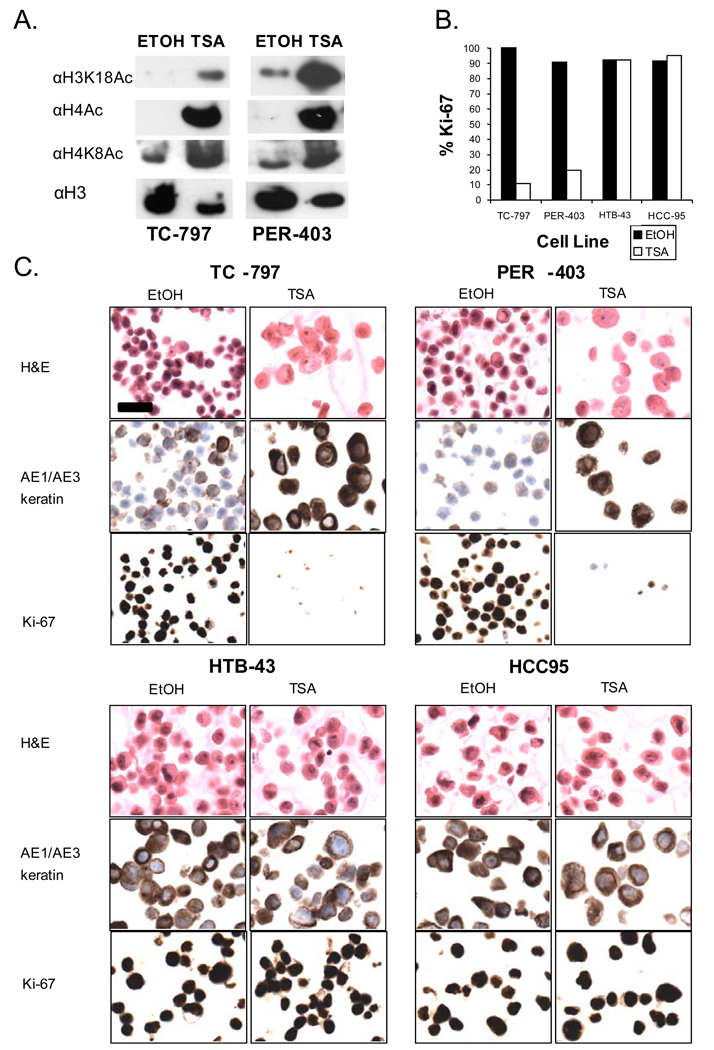
Figure 2.
Treatment of NMC cells with the HDACi, trichostatin A, restores global histone acetylation and induces squamous differentiation and arrested growth. (A) Changes in histone acetylation resulting from TSA treatment of two NMC cell lines after 24 hr. (B) Ki-67 (marker of cycling cells) fraction in two NMC cell lines (TC-797 and PER-403) and two non-NMC squamous cell carcinoma cell lines (HTB-43 and HCC-95). Value is from a denominator of 200 viable cells counted. (C) Morphologic and immunophenotypic changes following TSA (72h, 25nM) treatment of NMC cell lines, TC-797, PER-403, and non-NMC cell lines, HTB-43 and HCC-95. Magnification (scale bar, 25um) is identical for all panels.
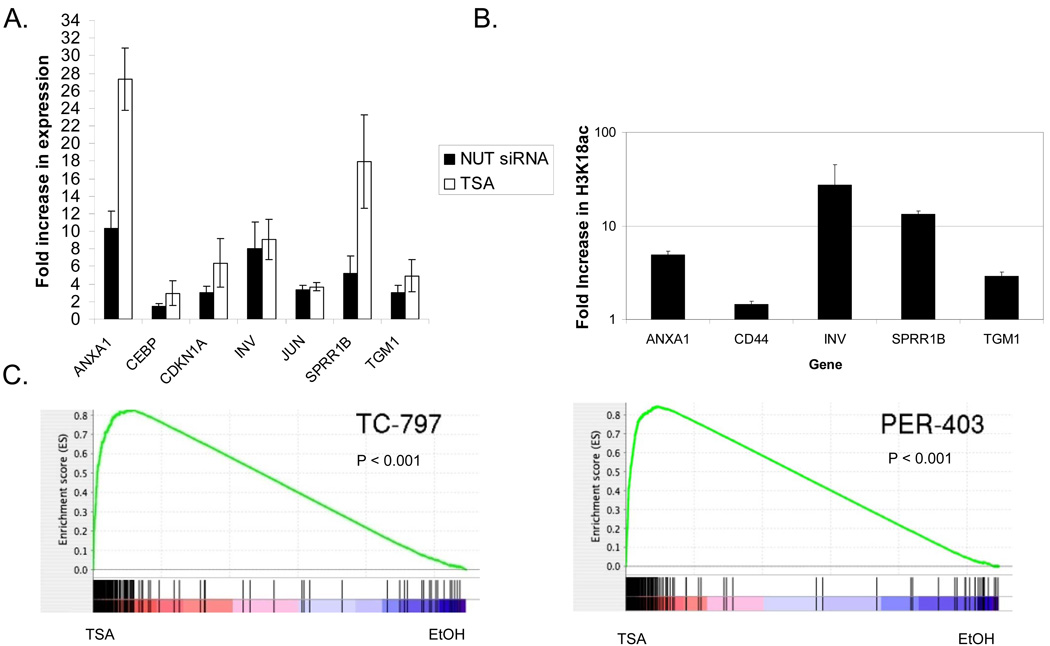
Figure 3.
Histone deacetylase inhibition induces a very similar program of differentiation with that following knockdown of BRD4-NUT in NMC cells. (A) Quantitative RT-PCR analysis of seven selected squamous differentiation-specific genes 24h following siRNA-knockdown and TSA (25nM) treatment of TC-797 NMC cells. Results are from triplicate independent samples. ct, control scrambled siRNA. (B) Quantitative PCR of ChIPd DNA from NMC TC-797 cells using primers to the promoter regions of select differentiation specific genes (from A, above), using H3K18Ac antibody. Values, obtained from independent duplicate samples, are relative to IgG ChIP control. (C) Gene set enrichment analysis (GSEA) measuring the correlation of genes up-regulated following knockdown of BRD4-NUT in both TC-797 and PER-403 NMC cell with HDACi-mediated expression changes. In these plots, vertical lines indicate the rank order of the knockdown gene set genes within the HDACi-treated cells (top: TC-797, bottom: PER-403). Red (left): genes up-regulated by TSA treatment; blue (right) down-regulated genes. The concentration of vertical lines within the TSA (red) portion of the spectrum reflected in the running enrichment score plot (green line) indicates the degree of correlation between up-regulation in response to BRD4-NUT knockdown or TSA treatment.
TRex inducible cell lines
The FLAG-BRD4-NUT-inducible derivative of 293TRex was created from commercially available 293TRex cells according the manufacturer’s instructions (Invitrogen).
Expression plasmids
cDNAs encoding proteins of interest were assembled in the vectors pcDNA5 FRT/TO (Invitrogen) and confirmed by DNA sequencing.
Histology and immunohistochemistry
Formalin-fixed, paraffin-embedded cell blocks of cultured cells were prepared as described (12, 26) using Histogel (Richard-Allan Scientific, Kalamazoo, MI). Sections were stained with hematoxylin and eosin or by immunohistochemistry (IHC), which was performed on 5 µm sections prepared from formalin-fixed, paraffin-embedded primary tumors or cell blocks. Immunohistochemical stains performed using anti-NUT rabbit polyclonal antibody (27), anti-NUT rabbit monoclonal antibody (26), Ki-67 (MIB-1 clone; DAKO USA, Carpinteria, CA) (12), and PanKeratin (12) (clone MNF116, DAKO USA) were as described.
Fluorescence in situ hybridization (FISH)
Dual-color FISH assays for BRD4 and NUT breakpoints were performed on formalin-fixed paraffin-embedded 4µm tissue sections as described (20). Probes used for the 15q14 NUT breakpoint, flanking a 181kb region containing NUT, included the 3’ telomeric BAC clones 1H8 and 64o3, and the 5’ centromeric clones 412e10 and 3d4. Probes used for the 19p13.1 BRD4 breakpoint were the 5’ centromeric BAC clone 187l3 and the 3’ telomeric BAC clone 87m17.
High throughput screening
NMC cells were plated at a concentration of 1000 cells per well in black, clear-bottom, 384-well microtiter plates. Small-molecule HDAC inhibitors were transferred from library plates arrayed in a dose-response format using robotic pin transfer. Following compound incubation for 48 or 72 hr, cells were fixed with paraformaldehyde (3.8%) and stained with Hoechst, anti-acetyl-lysine (Cell Signaling Technologies, Danvers, MA), or anti-cytokeratin (clone AE1/AE3, DAKO USA, Carpinteria, CA). Secondary antibodies included rhodamine donkey anti-rabbit IgG (1:100, Jackson Immuno Labs) and FITC pig anti-rabbit IgG (1:100, DAKO USA). Three-color fluorescence images were acquired using automated epifluorescence microscopy (ImageXPressMICRO; Molecular Devices) and analyzed using MetaXPress (Molecular Devices), GraphPad Prizm and Spotfire DecisionSite software.
Small interfering RNA (siRNA) and transient transfection
The design, synthesis, and electroporation of siRNA duplexes specific for human NUT were as described (12). Scrambled siRNA (Silencer Negative Control #1 siRNA Template, Applied Biosystems/Ambion, Austin) was used as a negative control.
Immunoblotting
Proteins in cell extracts prepared with high-salt RIPA buffer (50 mM Tris, pH 8.0, containing 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 250mM NaCl, and 5 mM EDTA) were separated by SDS-PAGE, electrophoretically transferred to Immobilon membranes (Millipore, Billerica, MA), and stained with anti-NUT polyclonal antibody, or anti-FLAG (Sigma-Aldrich, St. Louis, MO). Acid extraction of histones was performed as per instructions (Millipore, Billerica, MA). Acid extracts were electrophoretically separated, blotted as above, and stained with anti-acetyl histone H4 (Millipore), anti-acetyl-histone H4 K8 (Abcam, Cambridge, MA), anti-acetyl-histone H3 K18 (Abcam), or anti-histone H3 polyclonal antibody (Abcam, Cambridge, MA). Staining was developed using a chemiluminescent method (SuperSignal, West Pico; Pierce, Rockford, IL).
Microarray analysis
Total RNA was isolated 24 hr post-siRNA transfection (duplicate separate samples per cell line) or treatment with trichostatin A (25nM, triplicate separate samples per cell line) from TC797 and PER-403 cells using TRIzol reagent (Invitrogen, Carlsbad, CA). RNAs were further processed, labeled, and hybridized to Human Genome U133A Plus 2.0 microarray chips (Affymetrix, Santa Clara, CA) in the Partners Healtcare Center for Genetics and Genomics Gene Chip Microarray Facility, as described in the Affymetrix GeneChip Expression Analysis Technical Manual (revised version 4). After manually inspecting the quality of arrays, GeneChip RMA (GCRMA) was used to normalize and summarize expression as log2 values. Probe sets were filtered based on minimum expression (log2 (100)) and minimum variance (interquartile range > 0.5). Probe sets satisfying those criteria were analyzed using the LIMMA (Linear Models for Microarrays Data) package (28) to identify differentially expressed probe sets using a q-value cutoff of 0.05 (Benjamini and Hochberg corrected p-value). Raw data and .cel files can be accessed online at the gene expression omnibus (GEO) website at the following link (29).
Radiologic Imaging
Standard 18(F)-FDG PET and CT were performed at the Department of Radiology at The Children’s Hospital Boston, MA.
Cell growth assay
Cells were plated in a 96-well plate at a density of 5,000 cells/well and incubated for 2, 3, or 7 days in the presence of 1 µM SAHA or DMSO. Relative cell numbers were determined in 6 replicates using Cell Titer Glo (Promega, Madison, WI) according to the manufacturer’s instructions.
Gene set enrichment analysis (GSEA)
Genes whose expression was significantly up-regulated upon knockdown of BRD4-NUT in both TC-797 and PER-403 cells were used as the comparison gene set in the GSEA (30) of expression changes in TC-797 and PER-403 cells following TSA (25nM) treatment. Gene permutation was used to estimate the significance of enrichment for the BRD4-NUT gene set within the ranked list of genes up-regulated in TSA-treated cells (1000 permutations per GSEA).
In vivo studies
Xenograft tumors were generated from 2 established NMC cell lines (TC-797 and PER-403) and from 1 low-passage primary tumor (8645). Cells were transduced with a lentivirus encoding firefly luciferase, mCherry, and puromycin phosphotransferase (31), followed by selection in 2 µg/ml of puromycin. A total of 107 cells in 100 µL of 30% Matrigel/70% PBS were injected subcutaneously into the flanks of 6 week old female NCr nude mice (Charles River Labs). Mice were serially imaged after injection of 75 mg/kg D-luciferin using an IVIS Spectrum instrument (Caliper Life Sciences). Tumor volumes were calculated from caliper measurements using the formula Vol = ½ × length × width2. Established xenograft tumors were defined as tumors with increasing bioluminescence and measureable tumor volume. Cohorts of mice with established tumors were divided into groups with statistically equivalent tumor burden, and treated daily with vehicle or 10 mg/kg of LBH589 by intraperitoneal injection. Tumor burden was determined by serial bioluminescence imaging and tumor volume measurements. For survival studies, mice were sacrificed when tumors reached 2 cm in the largest diameter. Statistical significance was determined by two-tailed Student’s t-test. Samples for histopathological analysis were fixed in 10% buffered formalin. All animal studies were performed under IACUC approved protocols.
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
Total RNA from TC-797 cells was harvested 24 hr after siRNA electroporation or Trichostatin A (25nM) treatment using TRizol (Invitrogen) and further purified with an RNeasy Mini Kit (Qiagen). One microgram of RNA was used for cDNA synthesis using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). qPCR was performed in duplicate on a Bio-Rad iCycler in 96 well plate format with IQ SYBR Green supermix (Bio-Rad) and 1uL of cDNA template per reaction. Amplification curves and Ct values were generated using MyiQ Single-Color Real-Time software (Bio-Rad). Transcript levels were normalized to the RPL3 transcript as a cDNA input control. Data from three independent experiments are shown.
Chromatin immunoprecipitation quantitative polymerase change reaction (ChIP-qPCR)
ChIP was performed using the SimpleChIP Enzymatic Chromatin IP Kit from Cell Signaling Technologies (Danvers, MA). For each IP, approximately 1.5× 107 cells were crosslinked with 1% formaldehyde for ten minutes at 37°C and processed as per manufacturer instructions. Total H3 was immunoprecipitated with 10uL of H3 antibody provided in the SimpleChIP Kit, and acetylated histone H3K18 was immunoprecipitated with 10uL of ab1191 from Abcam (Cambridge, MA). Negative control ChIP was performed using rabbit IgG (Jackson Immunoresearch, West Grove, PA). qPCR was performed as described above using primers designed to amplify promoter-proximal regions of the target genes. For each PCR reaction, 2uL of ChIP DNA was used. The ratio of H3K18Acet to total H3 was calculated for each promoter and used to determine relative changes in H3 acetylation upon Trichostain A (25nM) treatment.
Results
Expression of BRD4-NUT is associated with a global decrease in histone acetylation and overall repression of gene expression
Recent findings indicating that NUT binds to, and activates the histone acetyltransferase (HAT) activity of p300 (17), prompted us to determine the effects of BRD4-NUT on bulk chromatin acetylation. We found that expression of BRD4-NUT in 293T cells markedly diminished H3K18, H4 and H4K8 acetylation, histone marks associated with gene expression. Conversely, siRNA-mediated knockdown of BRD4-NUT in NMC cell lines increased, to a more variable degree, acetylation of these same residues within 24 hr (Figure 1A). Changes in chromatin marks induced by BRD4-NUT knockdown preceded morphologic changes indicative of squamous differentiation (Figure 1B), which is marked by increases in nuclear size, open chromatin, and cytoplasmic volume. In line with the observed increases in activating histone marks, transcriptional profiling 24 hr after siRNA knockdown of BRD-NUT, prior to morphologic changes indicative of squamous differentiation, revealed that among 369 genes that changed expression significantly in the NMC line Per-403, a disproportionate fraction (87%) was upregulated, consistent with a repressive effect on gene transcription. Similarly, 68% of genes that changed expression following the knockdown of BRD4-NUT in the NMC line TC-797 were upregulated.
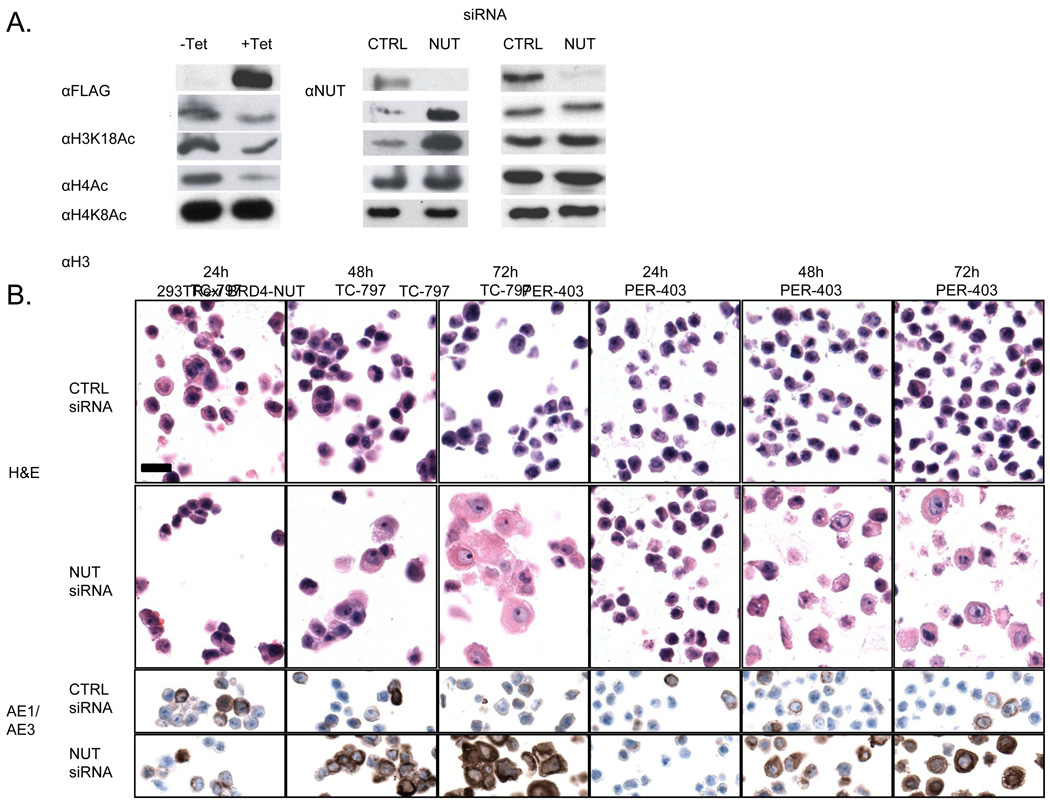
Figure 1.
BRD4-NUT expression is associated with blocked differentiation, and repression of histone acetylation. (A) Effects on histone acetylation resulting from induced expression of flag-BRD4-NUT in 293TRex cells, and siRNA-induced knockdown of BRD4-NUT in two NMC cell lines after 24 hr.. (B) siRNA-induced knockdown of BRD4-NUT using an siRNA targeting NUT in two NMC cell lines. Hematoxylin and eosin staining (H & E) as well as the immunohistochemical (IHC) detection of keratin with antibody AE1/AE3 are shown for each cell line 24h, 48h, and 72h following knockdown. Magnification (scale bar, 25um) is identical for all panels.
Restoration of acetylation using histone deacetylase inhibitors induces a program of squamous differentiation of NMC cells, phenocopying siRNA-induced silencing of BRD4-NUT
Based on these data, we postulated that global repression of acetylation and transcription by BRD4-NUT may prevent the expression of genes required for differentiation. If this idea is correct, it follows that restoration of histone acetylation with HDACi should induce NMC cell differentiation. To test this idea, we treated NMC cells with the tool HDACi compound, trichostatin A (TSA). Indeed, we found that TSA caused global increases in histone acetylation (Figure 2A), growth arrest (Figure 2B), and squamous differentiation marked by flattening of cells, accumulation of abundant, keratin-positive, eosinophilic cytoplasm, nuclear enlargement, and decreased nuclear staining consistent with an increase in euchromatin (Figure 2C), all features that are also induced by BRD4-NUT knockdown. The induction of differentiation and arrested growth by TSA was unique to NMC cells, as non-NMC squamous cell carcinoma cell lines were unresponsive to treatment (Figure 2B–C). Even a higher concentration of TSA (100nM) that was lethal to the NMCs caused no changes in differentiation or growth of the non-NMCs (Figure S1). Comparative analysis of seven selected genes associated with squamous differentiation revealed that all seven changed in expression similarly following BRD4-NUT knockdown and TSA treatment in TC-797 NMC cells as measured by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) (Figure 3A). The mechanism by which HDACi increased expression of these genes appears to be due to promoter acetylation, as quantitative chromatin immunoprecipitation-PCR (ChIP-qPCR) with α acetyl H3K18 antibody revealed enrichment of five of the squamous differentiation associated gene promoters in TSA-treated NMC chromatin extracts (Fig. 3B).
To gain a broader view of the effects of siRNA and TSA on gene expression, we performed gene set enrichment analysis (GSEA) using microarray data from NMC cell lines treated with BRD4-NUT siRNA or TSA. GSEA revealed that genes that increased in expression following BRD4-NUT knockdown were highly enriched among those that increased following TSA treatment (Figure 3C, p-value < 0.001 for both cell lines). Together, these findings suggest that the NMC response to TSA is mediated through interference with BRD4-NUT function.
HDACi abrogate the growth of NMC cells in vitro and in vivo
The correlates above suggested that HDACi should only promote NMC differentiation at concentrations that cause histone hyperacetylation, irrespective of their potency against individual HDACs. To test this idea, we scored a library of structurally dissimilar HDACi for effects on NMC cells using a miniaturized high throughput assay that quantifies changes in bulk chromatin acetylation and expression of keratin. Figure S2A shows an example of this analysis using NMC cells treated with the FDA-approved HDAC inhibitor vorinostat (SAHA), a clinically-approved substance with prior pediatric experience. NMC cells treated with SAHA exhibit increased histone hyperacetylation and keratin expression by immunofluorescence microscopy (Figure S2A). Automated measurement of these phenotypes revealed a dose-dependent induction of hyperacetylation and differentiation that was inversely associated with cellular proliferation (Figure S2B). The effects of nine structurally unrelated HDAC inhibitors on histone acetylation and cell growth in two different NMC cell lines (Table 1) were highly correlated (R2 = 0.96; Figure S2C, Table 1), strongly suggesting that the growth inhibitory effects of these drugs is mediated through increased histone acetylation.
Table 1.
Dose-responses of NMC cell lines to treatment with 9 structurally different HDAC inhibitors.
| AcLys EC50 (µM) | Viability IC50 (µM) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Rank | Compound | TC-797 | 143 | Ty-82 | TC-797 | 143 | Ty-82 | PER-403 | 10326 |
| 1 | LBH-589 | 0.010 | 0.026 | 0.009 | 0.006 | 0.036 | 0.009 | 0.038 | 0.001 |
| 2 | TSA | 0.002 | 0.031 | 0.015 | 0.003 | 0.043 | 0.015 | 0.025 | 0.003 |
| 3 | CRA-024781 | 0.07 | 0.42 | 0.17 | 0.04 | 0.44 | 0.20 | 0.35 | 0.07 |
| 4 | ITF-2357 | 1.71 | 1.28 | 0.57 | 0.03 | 1.17 | 0.52 | 1.14 | 0.13 |
| 5 | PXD101 | 0.28 | 1.38 | 0.88 | 0.04 | 2.09 | 0.68 | 0.48 | 0.21 |
| 6 | SAHA | 0.21 | 2.45 | 1.48 | 0.26 | 3.02 | 1.78 | 1.94 | 0.65 |
| 7 | MGCD0103 | 1.29 | 3.47 | 0.87 | 0.32 | 3.54 | 1.44 | 2.38 | 0.18 |
| 8 | MS-275 | 0.57 | 4.09 | 1.96 | 0.28 | 4.74 | 2.96 | 5.56 | 0.39 |
| 9 | CI-994 | 3.2 | 28.2 | 10.1 | 3.3 | 19.9 | 15.3 | 19.3 | 2.1 |
AcLys, nuclear acetyl-lysine.
EC50, concentration of drug that increases nuclear acetylation by 50% (1.5 fold induction) relative to vehicle control.
IC50, concentration of drug that results in a 50% reduction in cell number relative to vehicle control.
TC-797, 143, Ty-82, PER-403, NMC cell lines which express BRD4-NUT.
10326, NMC cell line which expresses BRD3-NUT.
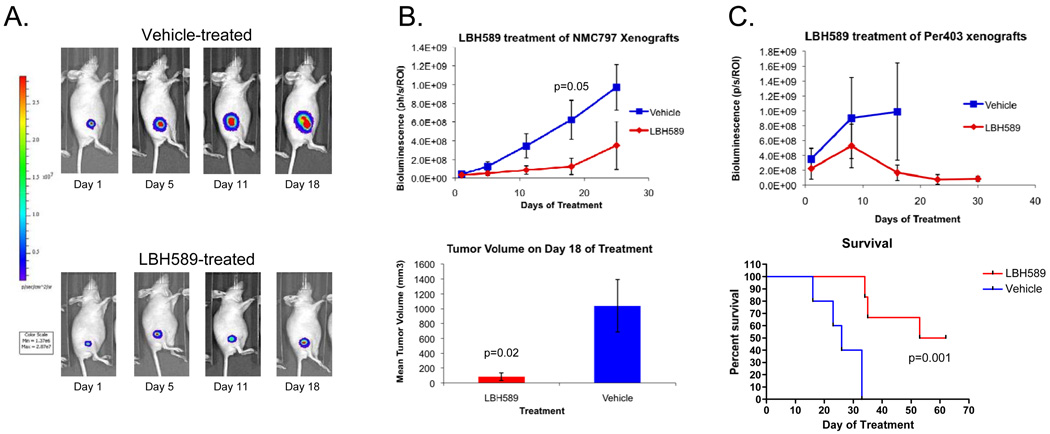
To determine the in vivo efficacy of HDACi in NMC, we created 2 luciferized NMC cell line xenograft models (TC-797, PER-403). Tumor burden was assessed by bioluminescence imaging (Figure 4) and tumor volume. Mice with established xenografts, defined as increasing bioluminescence and measurable tumor volumes, were divided into groups that were treated with vehicle or HDACi. For this study, we used the the HDACi, LBH-589 (Novartis, Cambridge, MA), an advanced second-generation investigational HDACi used extensively in mice (32) that is formulated for intra-venous injection, and thus easier to administer compared with SAHA (an oral medication). Significant suppression of tumor growth, as assessed by tumor volume or bioluminescence, was apparent in both models (Figures 4A–C). In the PER-403 xenografts where survival analysis was performed, significant survival benefit was seen in HDACi-treated mice (p = 0.001, Figure 4C (bottom graph)). Tumors in LBH-589 treated mice showed increased levels of histone acetylation and morphologic and immunohistochemical changes (increased keratin expression) consistent with squamous differentiation (Figure 5). Taken together, the findings indicate that HDACi induce differentiation and growth arrest of NMC cells in vitro and in vivo, providing a strong rationale for their therapeutic use in this disease.
Figure 4.
Growth inhibition of xenograft models of two NMCs by the HDACi LBH-589. (A) Representative bioluminescence images of TC-797 xenografts treated with vehicle or LBH589 at 10 mg/kg IP. (B) Effects of LBH589 treatment on TC-797 (n=6 per group) xenograft growth measured by bioluminescence and tumor volume. (C) Bioluminescence imaging and survival analysis of PER-403 xenografts (n=5 per group) treated with LBH589. Mice were sacrificed when tumors reached 2 cm. Due to rapid tumor growth, the bioluminescence data for vehicle-treated animals (upper panel) is confounded by the need to sacrifice animals bearing large tumors beginning on treatment day 18 (lower panel).
Figure 5.
Morphologic and immunophenotypic squamous differentiation correlates with increased acetylation of TC-797 xenografts treated with HDACi (LBH-589). AcL, immunohistochemistry using anti-acetyl-lysine antibody (Cell Signaling Technologies); Veh1, vehicle control (DMSO) treated mouse 1; Veh2, vehicle treated mouse 2; LBH1, LBH-589 treated test mouse 1; LBH2, LBH-589 treated test mouse 2. Magnification (scale bar, 25um) is identical for all panels.
An FDA-approved HDAC inhibitor, vorinostat, exhibits anti-tumor activity in a pediatric patient with NMC
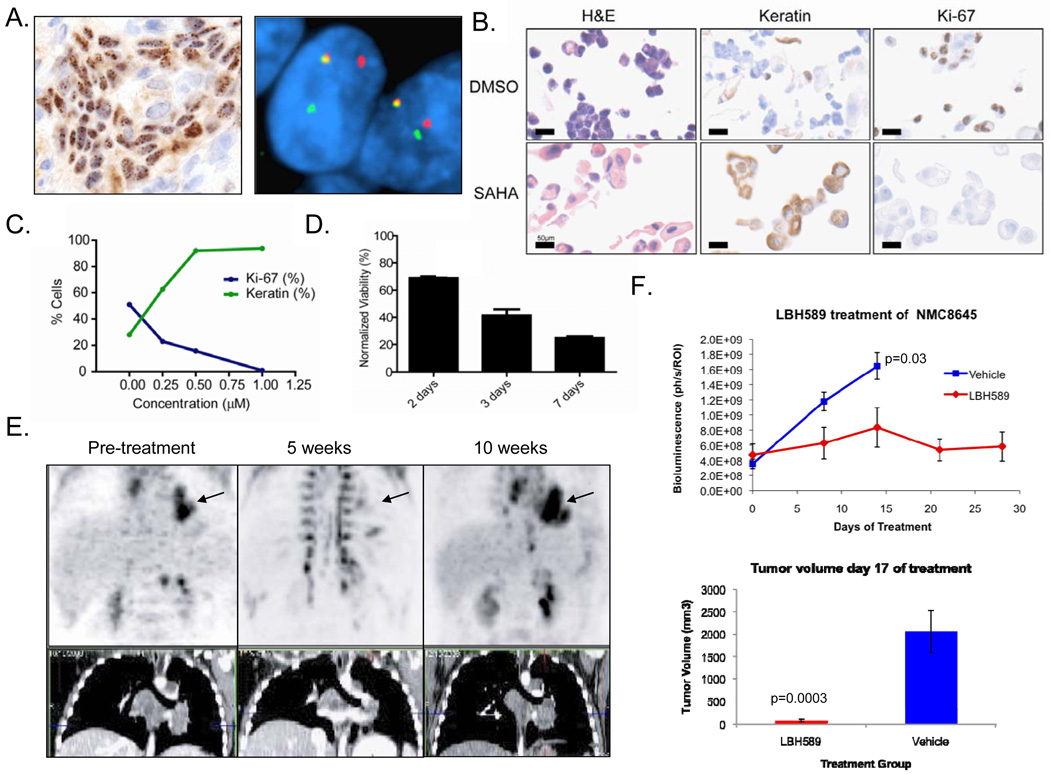
NMC commonly presents in pediatric patients and invariably progresses through cytotoxic chemotherapy. Metastases frequently threaten vital structures and therapeutic options are extremely limited. As the research above was being completed, a 10 year-old male was transferred to the Children’s Hospital of Boston for management of an aggressive mediastinal mass invading the left atrium and pulmonary vein. Partial resection of this mass led to the diagnosis of NMC, based on positive immunohistochemical staining for NUT and the presence of a BRD4-NUT fusion gene by fluorescence in situ hybridization (Figure 6A). Based on the above findings, treatment of this patient with HDACi was considered, and the only clinically-approved HDACi with experience in children was vorinostat (SAHA). Culture of patient-derived NMC cells (designated 8645) established in a medium optimized for the growth of carcinoma cells (24) confirmed that vorinostat induced 8645 cells to undergo squamous differentiation (Figure 6B and C) and growth arrest (Figure 6B–D) as assessed by Ki-67 staining and cell counts. The IC50 (based on Ki-67 fraction) and EC50 (based on keratin expression) were calculated to be 250nM, well below the known Cmax of vorinostat of 1.5–2µM. Following institutional approval and informed consent, the open-label administration of single-agent, oral vorinostat was initiated (400 mg daily). At the conclusion of five weeks of drug therapy, a marked decrease in tumor avidity for 18F-fluorodeoxyglucose was observed by positron emission tomography (PET; Figure 6E). The responsiveness of the patient’s tumor to HDACi was confirmed by treating xenografted tumor cells from the patient with LBH-589, which produced strong growth inhibition (p = 0.0003, Figure 6F). Tumor response in the patient was accompanied by marked thrombocytopenia (17,000/µl; normal range 150,000–400,000/µl), an established dose-limiting toxicity of vorinostat (33). Due to severe (NCI Grade 3) nausea and emesis, the patient was unable to tolerate further vorinostat therapy, and tumor recurrence was noted on PET scans performed five weeks later (Figure 6E). He was subsequently treated with a combination chemotherapy protocol, and died due to recurrent and metastatic disease 11 months after initial diagnosis.
Figure 6.
Diagnosis and response of an NMC patient to single-agent treatment with the HDAC inhibitor vorinostat (SAHA). (A) Diagnosis of NMC in a 10 year old boy by immunohistochemical (IHC) staining with a monoclonal antibody to NUT (27) and by dual color, split-apart fluorescence in situ hybridization (FISH) (35) using probes flanking the NUT locus. (B) Morphologic and immunophenotypic effects of 72h treatment with SAHA (1µM) versus DMSO control. IHC was performed for keratins (AE1/AE3 antibodies) and the proliferation marker, Ki-67. (C) Dose-response curve of the patient’s tumor cells to SAHA (IC50=250 nM), as measured by Ki-67 fraction and keratin-positivity. (D) Cell growth in SAHA (1 µM), as measured by ATP content relative to DMSO control. (E) P18PF-fluorodeoxyglucose–positron emission tomography (FDG-PET) and CT scan of the patient’s mediastinal tumor (arrow). (F) Growth response of NMC 8645 xenografts (n=6 vehicle, and n=7 LBH589 groups) derived from this patient as measured by bioluminescence and tumor volume.
Discussion
NMC is an incurable cancer with an average survival of nine months or less that is defined by the presence of NUT fusion genes (11). We show here that BRD4-NUT expression is associated with global histone hypoacetylation and transcriptional repression. . The data are consistent with a recently postulated model (17) in which BRD4-NUT binds to and activates p300, thereby sequestering histone aceyltransferase (HAT) activity to localized regions of BRD4-NUT-acetyl-chromatin binding. This in turn results in a relative overabundance of HDAC activity outside of these regions, leading to global hypoacetylation and inadequate expression of genes required for differentiation. We hypothesize that HDACi correct this imbalance by favoring HAT activity, restoring chromatin acetylation and increasing the transcription of pro-differentiative genes.
This sequestration model is novel within the context of epithelial cancer, but precedents are found in certain leukemias (34) and neuronal degenerative diseases. Expanded polyglutamine repeats in the Huntingtin and androgen receptor proteins bind and sequester CBP/p300, resulting in global histone hypoacetylation and transcriptional repression (35–38). Moreover, the sequestration and inactivation of transcriptional co-factors in these complexes is reversible by HDAC inhibitors (36, 37).
Regardless of the precise mechanism, the reversal of BRD4-NUT-induced global hypoacetylation with HDACi is a novel means of targeted “differentiation therapy” in cancer. The striking in vitro and in vivo induction of differentiation and inhibition of NMC growth by HDACi is of particular importance in this disease because there are currently two FDA-approved HDACi reagents, vorinostat and romidepsin, which are available for immediate clinical investigation. There is an urgent need for novel therapy in this disease because there is currently no effective therapy for NMC, which has been refractory to a number of different chemotherapeutic regimens (39). Increased recognition of NMC, enabled in part by new diagnostic tests that rely on routine immunohistochemistry (26), should enhance the clinical evaluation of HDAC inhibitor drugs in this malignancy, alone and in combination with other cytotoxic agents.
Another recently reported potential means to target BRD4-NUT are small molecule bromodomain inhibitors (Brdi,(40)). Brdi specifically abrogate binding of acetylated histones to the bromodomains of BRD4 and BRD3, and cause differentiation of NMC cells in vitro and in mice. Nevertheless, it is not clear what toxicity these molecules will have, as they also inhibit acetyl-histone-binding of native BRD4 and BRD3, which are ubiquitously expressed proteins that may confer epigenetic “memory” on cells (41, 42). This new class of molecules is in the earliest stages of investigation and, unlike HDACi, has not been used in humans.
The most specific targeting of BRD4-NUT would be directed at NUT, which is not normally expressed outside testis or ovaries (11). This will require mapping of protein:protein interactions involving NUT at high resolution, and development of NUT-directed inhibitors. Although the development of deliverable small molecule inhibitors of this type has proven difficult, the advent of stapled peptides shows some promise to facilitate progress in this field (43).
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. Stuart Schreiber and the Chemical Biology Program, Broad Institute of Harvard and MIT, Cambridge, MA, for use of equipment. We would like to extend special thanks to Sara Guterman, of the clinical trials office of the Dana-Farber Cancer Institute, for helping expedite internal review board review on a moment's notice. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Service, nor does the mention of trade names, commercial products or organizations imply endorsement by the US government.
Financial Support: This works was supported by a Dana Farber/ Harvard Cancer Center Nodal Award 5P30CA06516-44 (C.A.F. and J.E.B.), US National Institutes of Health grant 1R01CA124633 (C.A.F.), the Stanley L. Robbins Memorial Award to M.D.H., the National Institutes of Health grant 1K08CA128972 (J.E.B.), and the Burroughs-Wellcome Foundation (J.E.B.), and funds from the National Cancer Institute’s Initiative for Chemical Genetics (Contract No. N01-CO-12400).
References
- 1.Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nature medicine. 1996;2(5):561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 2.Cools J, DeAngelo DJ, Gotlib J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. The New England journal of medicine. 2003;348(13):1201–1214. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- 3.Di Croce L, Raker VA, Corsaro M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science (New York, NY. 2002;295(5557):1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 4.Redner RL, Corey SJ, Rush EA. Differentiation of t(5;17) variant acute promyelocytic leukemic blasts by all-trans retinoic acid. Leukemia. 1997;11(7):1014–1016. doi: 10.1038/sj.leu.2400661. [DOI] [PubMed] [Google Scholar]
- 5.Soignet SL, Maslak P, Wang ZG, et al. Complete remission after treatment of acute promyelocytic leukemia with arsenic trioxide. The New England journal of medicine. 1998;339(19):1341–1348. doi: 10.1056/NEJM199811053391901. [DOI] [PubMed] [Google Scholar]
- 6.Hu J, Liu YF, Wu CF, et al. Long-term efficacy and safety of all-trans retinoic acid/arsenic trioxide-based therapy in newly diagnosed acute promyelocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(9):3342–3347. doi: 10.1073/pnas.0813280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Redner RL, Wang J, Liu JM. Chromatin remodeling and leukemia: new therapeutic paradigms. Blood. 1999;94(2):417–428. [PubMed] [Google Scholar]
- 8.Cheson BD. Standard and low-dose chemotherapy for the treatment of myelodysplastic syndromes. Leuk Res. 1998;22 Suppl 1:S17–S21. doi: 10.1016/s0145-2126(98)00039-3. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Manero G, Yang H, Bueso-Ramos C, et al. Phase 1 study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid [SAHA]) in patients with advanced leukemias and myelodysplastic syndromes. Blood. 2008;111(3):1060–1066. doi: 10.1182/blood-2007-06-098061. [DOI] [PubMed] [Google Scholar]
- 10.French CA. Demystified molecular pathology of NUT midline carcinomas. Journal of clinical pathology. 63(6):492–496. doi: 10.1136/jcp.2007.052902. [DOI] [PubMed] [Google Scholar]
- 11.French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003;63(2):304–307. [PubMed] [Google Scholar]
- 12.French CA, Ramirez CL, Kolmakova J, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27(15):2237–2242. doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- 13.Dyson MH, Rose S, Mahadevan LC. Acetyllysine-binding and function of bromodomain-containing proteins in chromatin. Front Biosci. 2001;6:D853–D865. doi: 10.2741/dyson. [DOI] [PubMed] [Google Scholar]
- 14.Dey A, Ellenberg J, Farina A, et al. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Molecular and cellular biology. 2000;20(17):6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Yik JH, Chen R, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19(4):535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 16.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 17.Reynoird N, Schwartz BE, Delvecchio M, et al. Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. The EMBO journal. doi: 10.1038/emboj.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toretsky JA, Jenson J, Sun CC, et al. Translocation (11;15;19): a highly specific chromosome rearrangement associated with poorly differentiated thymic carcinoma in young patients. Am J Clin Oncol. 2003;26(3):300–306. doi: 10.1097/01.COC.0000020960.98562.84. [DOI] [PubMed] [Google Scholar]
- 19.Kees UR, Mulcahy MT, Willoughby ML. Intrathoracic carcinoma in an 11-year-old girl showing a translocation t(15;19) Am J Pediatr Hematol Oncol. 1991;13(4):459–464. doi: 10.1097/00043426-199124000-00011. [DOI] [PubMed] [Google Scholar]
- 20.French CA, Miyoshi I, Aster JC, et al. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t (15;19) Am J Pathol. 2001;159(6):1987–1992. doi: 10.1016/S0002-9440(10)63049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuzume T, Kubonishi I, Takeuchi S, et al. Establishment and characterization of a thymic carcinoma cell line (Ty-82) carrying t(15;19)(q15;p13) chromosome abnormality. Int J Cancer. 1992;50(2):259–264. doi: 10.1002/ijc.2910500216. [DOI] [PubMed] [Google Scholar]
- 22.Kandaswami C, Perkins E, Drzewiecki G, Soloniuk DS, Middleton E., Jr Differential inhibition of proliferation of human squamous cell carcinoma, gliosarcoma and embryonic fibroblast-like lung cells in culture by plant flavonoids. Anti-cancer drugs. 1992;3(5):525–530. doi: 10.1097/00001813-199210000-00013. [DOI] [PubMed] [Google Scholar]
- 23.Park KS, Kim HK, Lee JH, et al. Transglutaminase 2 as a cisplatin resistance marker in non-small cell lung cancer. Journal of cancer research and clinical oncology. 136(4):493–502. doi: 10.1007/s00432-009-0681-6. [DOI] [PubMed] [Google Scholar]
- 24.Ince TA, Richardson AL, Bell GW, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12(2):160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Broach JR, Guarascio VR, Jayaram M. Recombination within the yeast plasmid 2mu circle is site-specific. Cell. 1982;29(1):227–234. doi: 10.1016/0092-8674(82)90107-6. [DOI] [PubMed] [Google Scholar]
- 26.Herbert Haack LAJ, Fry ChristopherJ, Crosby Katherine, Polakiewicz RobertoD, Stelow EdwardB, Hong Seung-Mo, Schwartz BrianE, Cameron MichaelJ, Rubin MarkA, Chang MartinC, Aster JonC, French ChristopherA. Diagnosis of NUT Midline Carcinoma Using a NUT-Specific Monoclonal Antibody. American Journal of Surgical Pathology. 2009 doi: 10.1097/PAS.0b013e318198d666. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stelow EB, Bellizzi AM, Taneja K, et al. NUT rearrangement in undifferentiated carcinomas of the upper aerodigestive tract. The American journal of surgical pathology. 2008;32(6):828–834. doi: 10.1097/PAS.0b013e31815a3900. [DOI] [PubMed] [Google Scholar]
- 28.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 29.French CA. Online expression profiling of NMCs 797 and PER-403 cell lines in response to siRNA knockdown of BRD4-NUT, and trichostatin A (24h) Boston (MA): Gene Expression Omnibus (GEO); 2010 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE18668.
- 30.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimbrel EA, Davis TN, Bradner JE, Kung AL. In vivo pharmacodynamic imaging of proteasome inhibition. Mol Imaging. 2009;8(3):140–147. [PubMed] [Google Scholar]
- 32.Crisanti MC, Wallace AF, Kapoor V, et al. The HDAC inhibitor panobinostat (LBH589) inhibits mesothelioma and lung cancer cells in vitro and in vivo with particular efficacy for small cell lung cancer. Molecular cancer therapeutics. 2009;8(8):2221–2231. doi: 10.1158/1535-7163.MCT-09-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duvic M, Talpur R, Ni X, et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109(1):31–39. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adya N, Stacy T, Speck NA, Liu PP. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Molecular and cellular biology. 1998;18(12):7432–7443. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helmlinger D, Hardy S, Abou-Sleymane G, et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS biology. 2006;4(3):e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCampbell A, Taye AA, Whitty L, Penney E, Steffan JS, Fischbeck KH. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(26):15179–15184. doi: 10.1073/pnas.261400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steffan JS, Bodai L, Pallos J, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413(6857):739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 38.McCampbell A, Taylor JP, Taye AA, et al. CREB-binding protein sequestration by expanded polyglutamine. Human molecular genetics. 2000;9(14):2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- 39.French CA, Kutok JL, Faquin WC, et al. Midline carcinoma of children and young adults with NUT rearrangement. J Clin Oncol. 2004;22(20):4135–4139. doi: 10.1200/JCO.2004.02.107. [DOI] [PubMed] [Google Scholar]
- 40.Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010 doi: 10.1038/nature09504. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mochizuki K, Nishiyama A, Jang MK, et al. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. The Journal of biological chemistry. 2008;283(14):9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishiyama A, Dey A, Miyazaki J, Ozato K. Brd4 is required for recovery from antimicrotubule drug-induced mitotic arrest: preservation of acetylated chromatin. Mol Biol Cell. 2006;17(2):814–823. doi: 10.1091/mbc.E05-08-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moellering RE, Cornejo M, Davis TN, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.