Abstract
Estrogen-Related Receptors alpha (ERRα) and gamma (ERRγ) are orphan nuclear receptors implicated in breast cancer that function similarly in the regulation of oxidative metabolism genes. Paradoxically, in clinical studies high levels of ERRα are associated with poor outcomes whereas high levels of ERRγ are associated with a favorable course. Recent studies suggest that ERRα may indeed promote breast tumor growth. The roles of ERRγ in breast cancer progression and how ERRα and ERRγ may differentially affect cancer growth are unclear. In mammary carcinoma cells that do not express endogenous ERRγ, we found that ectopic expression of ERRγ enhanced oxidative metabolism in vitro and inhibited the growth of tumor xenografts in vivo. In contrast, ectopic expression of the ERRα coactivator PGC-1α enhanced oxidative metabolism but did not affect tumor growth. Notably, ERRγ activated expression of a genetic program characteristic of mesenchymal-to-epithelial transition (MET). This program was apparent by changes in cellular morphology, upregulation of epithelial cell markers, downregulation of mesenchymal markers, and decreased cellular invasiveness. We determined that this program was associated also with upregulation of E-cadherin, which is activated directly by ERRγ. In contrast, PGC-1α activated only a subset of genes characteristic of the MET program and, unlike ERRγ, did not upregulate E-cadherin. In conclusion, these results show that ERRγ induces E-cadherin, promotes MET and suppresses breast cancer growth. Our findings suggest that ERRγ agonists may have applications in the treatment of breast cancer.
Keywords: breast cancer, mesenchymal-to-epithelial transition, estrogen-related receptor, E-cadherin regulation
Introduction
Breast cancer is the most common cancer in American women, with an incidence of ~1 in 10 women. Estrogens play a major role in breast cancer by promoting the initiation and growth of tumors. About 60-70% of breast cancers are estrogen receptor (ER) positive, and a significant proportion of the affected patients benefits from endocrine therapies that inhibit estrogen signaling. This leaves ~30-40% of breast cancers that are ER negative and do not respond to endocrine regiments, illustrating the need to identify other breast cancer biomarkers for development of therapies. Recent studies suggest that two estrogen-related receptors (ERR), ERR alpha (ERRα) and ERR gamma (ERRγ) have prognostic marker value for clinical outcome in breast carcinoma (1, 2). The roles and mechanisms of action of these two receptors in breast cancer progression are still poorly understood.
The three ERRs, (ERRα, ERRβ and ERRγ), comprise a subfamily of orphan nuclear receptors that share sequence similarity with the ERs (3, 4). Despite this similarity, ERRs are not activated by estrogen-like molecules and remain to date orphans with no known endogenous natural ligands. Nevertheless, ERRs have a hydrophobic pocket that is accessible to synthetic ligands. There exist synthetic inverse agonists for all ERRs, such as diethylstilbestrol, or selective inverse agonists, such as 4-hydroxy-tamoxifen for ERRβ and ERRγ, and XCT790 for ERRα (3, 4). Synthetic ERR agonists have so far been identified only for ERRβ and ERRγ (3, 4). In lieu of regulation by small molecule ligands, the activity of ERRs is determined by interactions with coregulators and post-translational modifications (5-7). ERRα transcriptional activity in particular depends on interactions with members of the PGC-1 coactivator family (8). In contrast, ERRβ and ERRγ are transcriptionally active in cells lacking PGC-1 proteins, likely due to efficient interactions with other coactivators (9). The three ERRs recognize a common DNA sequence, TNAAGGTCA, termed ERR response element (ERRE), and act as transcription factors for genes bearing functional ERREs (4, 10). Though they can activate some known ER - responsive genes, such as TFF1 (9, 11, 12), ERRs are major regulators of energy metabolism pathways, a function that is not shared by the ERs (4, 8). ERRα cooperates with the coactivators PGC-1α and PGC-1β to induce the expression of genes important for mitochondrial biogenesis, lipid transport, and oxidative capacity (13-17), and ERRα-deficient mice display tissue-specific defects in oxidative metabolism (18-20). ERRγ recognizes and induces many ERRα targets with roles in energy metabolism, contributing to oxidative capacity in the heart and playing a role in ion homeostasis (17, 21, 22). ERRα and ERRγ also target and regulate the expression of genes with roles outside metabolism, suggesting that their actions are not confined to metabolic regulation (11, 12, 17).
In contrast to the overlapping metabolic functions of ERRα and ERRγ, breast cancer association studies suggest that ERRα and ERRγ play opposite roles in cancer progression (1, 2). High levels of ERRα are associated with the expression of ErbB2, increased risk of recurrence and adverse clinical outcome (1, 2). Conversely, ERRγ expression is associated with markers predictive of responsiveness to endocrine treatment and is higher in diploid (less aggressive) than in aneuploid (more aggressive) tumors (1). These associations suggest that ERRα and ERRγ might respectively promote and suppress aggressive breast cancer progression. In support for ERRα promoting tumor growth, suppression of ERRα expression leads to reduced growth of MDA-MB-231 breast cancer tumors in mice (11). Furthermore, inverse agonists of ERRα inhibit proliferation, induce cell death and reduce tumorigenicity of cancer cell lines (23-25). To date, the extent to which ERRγ can affect breast tumor progression in vivo has not been determined.
In this study, we use a xenograft model to address the role of ERRγ in breast tumor growth. We find that ectopic ERRγ expression suppresses the growth of MDA-MB-231 tumors in mice. In contrast, ectopic expression of the coactivator PGC-1α which activates endogenous ERRα does not affect xenograft growth. Both ERRγ and PGC-1α enhance oxidative metabolism, suggesting that the increased oxidative capacity cannot by itself explain the reduced growth of ERRγ expressing tumors. Instead, ERRγ activates the program of mesenchymal-to-epithelial transition (MET), up-regulating the expression of the cell adhesion molecule E-cadherin and decreasing invasiveness. Our findings show for the first time that ERRγ can play an active role in breast cancer growth. They also provide evidence for ERRα– and ERRγ– selective targets, which may explain the distinct action of the two related receptors in breast cancer.
Materials and Methods
DNA vectors
For retroviral packaging, 293T cells were cotransfected with pMSCV-puro (expressing Flag-ERRγ, Flag-PGC-1α, or EGFP), pVPack-GP and pVPack-VSV-G plasmids, and supernatants were concentrated by ultracentrifugation. For production of lentiviral particles, 293T cells were cotransfected with pLVTHM [expressing shRNAs for CDH1, ERRγ or controls (see Supplement)], psPAX2 and pMD2.G plasmids (Addgene), and supernatants were collected. For the E-cadherin ERRE luciferase constructs, oligonucleotides containing the ERREs plus 14 to 16 flanking nucleotides were cloned as trimers upstream of the minimal promoter of pΔ-Luc (26).
Cell lines
MDA-MB-231, PC-3, and BT474 cells were obtained from ATCC in 2005, 2009 and 2010, respectively, and cryopreserved. ATCC characterizes cell lines by short tandem repeat profiling, cell morphology and karyotyping. All experiments were performed with cells at less than 20 passages after receipt. Cells were cultured in DMEM (MDA-MB-231) or RPMI 1640 (PC-3, BT474) supplemented with 10% FBS and 100 units/ml penicillin/streptomycin. ERRγ, PGC-1α, and EGFP expressing cells were derived by transducing MDA-MB-231 cells with retroviruses and selecting populations in puromycin (Invivogen, 1 μg/ml). Populations were independently derived twice, to confirm reproducibility of findings. Transgene expression was confirmed by RT-qPCR, Western blot and immunofluorescence. E-cadherin and ERRγ expression were suppressed by infection with lentiviruses expressing shRNAs. Infection efficiency was monitored by microscopy, based on GFP expression by the vectors.
Western blots and immunohistochemistry
For western blots, cells or tumors were lysed in RIPA buffer. For immunocytochemistry, cells were fixed with ice-cold methanol; tumor sections were deparaffinized, rehydrated in descending ethanol series and treated with 0.01 M citrate buffer (pH 6.0). Antibodies used were anti-E-cadherin (clone 36, BD Biosciences), Flag M2 antibody (Sigma), anti-GAPDH (Chemicon International), and anti-vimentin (clone V9, Dako). Image acquisition was with an Olympus IX70 microscope and Microfire software.
Gene expression
RNA isolation, cDNA synthesis and gene expression determination were as published (18), using quantitative PCR, gene-specific primers (Supplementary Table 1) and normalization to 36B4 as a reference gene.
Oxygen Consumption
Oxygen consumption was monitored with a Clark-type oxygen electrode (Hansatech Instruments), using 2 × 106 cells and at 30°C.
Glucose Oxidation Assay
Glucose oxidation rates were determined as published (27). Briefly, cells were incubated with serum-free medium supplemented with 0.5 μCi/ml [U-14C]glucose (200.6 μCi/μmol) for 2 hours at 37°C. 14CO2 released from the medium by acidification with 60% perchloric acid was trapped on filter paper saturated with benzethonium hydroxide, and counted in aβ-counter. The results were normalized to total protein.
Cell Growth assay
Cells were plated at 4,000 cells/well in a 96-well plate and assayed using the MTT assay (Sigma), or at 5×104 cells/well in 12-well plates and counted after trypsinization using a hemocytometer.
Breast Cancer Xenografts
Protocols were approved by the TSRI Institutional Animal Care and Use Committee and performed under veterinary supervision. Cells (1 × 106 cells in 50 μl of serum-free media) were injected orthotopically into the mammary fat pad of 6 to 8-week-old female C.B17 SCID mice (Taconic Farms). Tumor growth was monitored weekly; volume was calculated as (width2 × length)/2 (mm3). At 2 months after injection, mice were sacrificed and tumors were excised, weighed and processed for RNA, protein or sections. Representative data were obtained from 8 to 9 mice per experimental group, and the entire experiment was repeated once with independently derived cell populations.
Matrigel Transwell invasion assays
Cells were shed using EDTA-free Trypsin (Sigma), supplemented with 0.5 mM CaCl2, preincubated in media containing 0.5% FBS, and transferred into Matrigel-coated Transwells (BD BioCoat Matrigel Invasion Chamber, Becton Dickinson) at 2 × 105 cells/well. The gel including non-migrating cells was removed after 22 hours, and cells invading the membrane were fixed, stained with DAPI and counted.
Luciferase assays
Cells were transfected with 40 ng luciferase reporter, 5 ng pcDNA3/ERRγ and 5 ng pcDNA3/PGC-1α, using Fugene6 (Roche). Luciferase was read 24 hours later, using BriteLite Reagent (PerkinElmer) and the Pherastar Spectrophotometer (BMG Labtech).
Chromatin immunoprecipitation (ChIP)
The CDH1 locus was scanned for putative ERREs using the MAPPER search engine (28). ChIPs were performed as published (18), using the anti-Flag antibody (Sigma) and gene-specific primers (Supplementary Table 1).
Results
Increasing ERRγ or PGC-1α levels leads to enhanced oxidative capacity in breast cancer MDA-MB-231 cells
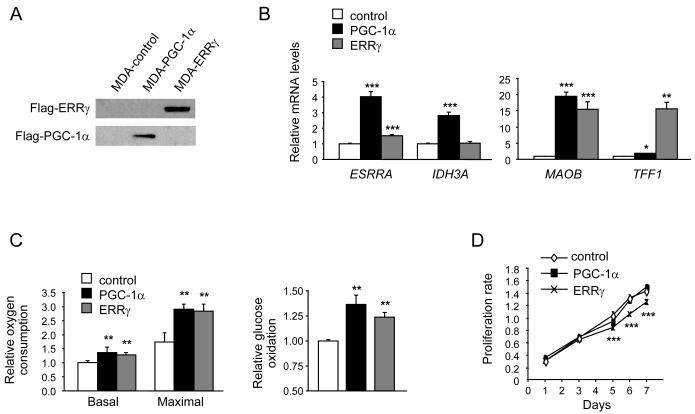
The human mammary adenocarcinoma line MDA-MB-231 is an established model for in vitro and in vivo xenograft studies of breast cancer. MDA-MB-231 cells are ER negative and express high levels of ERRα and low to undetectable levels of ERRγ, PGC-1α and PGC-1β (Supplementary Fig. S1). To elucidate the functional significance of ERRγ in breast cancer progression, we established MDA-MB-231 populations that express stably human ERRγ or EGFP as control (Fig. 1A). We also generated MDA-MB-231 cells expressing the coactivator PGC-1α, to induce and activate endogenous ERRα. ERRγ and PGC-1α were transcriptionally active in MDA-MB-231 cells, as determined by the induction of known ERR targets (Fig.1B). PGC-1α induced the expression of endogenous ERRα and of the ERR targets isocitrate dehydrogenase 3A (IDH3A), monoamine oxidase B (MAOB) and, to a lesser extent, Trefoil factor 1 (TFF1) (9, 13, 29, 30). The PGC-1α-driven induction of these genes was mediated by endogenous ERRα (Supplementary Fig. S2). ERRγ induced strongly MAOB and TFF1, had a modest effect on ERRα and no significant effect on IDH3A. Both PGC-1α and ERRγ increased significantly cellular respiration and glucose oxidation (Fig. 1C), consistent with the two proteins enhancing oxidative metabolism. This increase was due to changes in mitochondrial function and not mitochondrial biogenesis, as mitochondrial DNA copy number remained unchanged (Supplementary Fig. S3). Finally, neither PGC-1α nor ERRγ had a profound effect on cell growth in vitro, though ERRγ cell growth slowed down modestly after 5 days, upon reaching confluency (Fig. 1D; Supplementary Fig. S4).
Figure 1.
Generation and characterization of MDA-MB-231 cells expressing ERRγ and PGC-1α. A, ERRγ and PGC-1α protein expression, determined by western blot analysis. B, Increased expression of ERR target genes ESRRA, IDH3A, MAOB and TFF1. Levels of mRNA are expressed relative to levels of each gene in MDA-control cells and are the mean ± SEM of 4 experiments performed in duplicate. C, Increased oxygen consumption [monitored in the absence (basal) or presence (maximal) of 200 nM of the uncoupler FCCP] and glucose oxidation in MDA-PGC-1α and MDA-ERRγ cells. Data are the mean change ± SD in oxygen consumption from two experiments performed in duplicate (left panel) and mean change ± SEM in [U-14C]glucose oxidation rate from 4 experiments (right panel), relative to MDA-control cells. D, In vitro growth rates of the indicated populations, measured with a colorimetric MTT assay. Data are the mean ± SD of eight replicates. *, P < 0.05; **, P<0.01; ***, P<0.001.
ERRγ but not PGC-1α inhibits tumor growth in a breast cancer xenograft mouse model
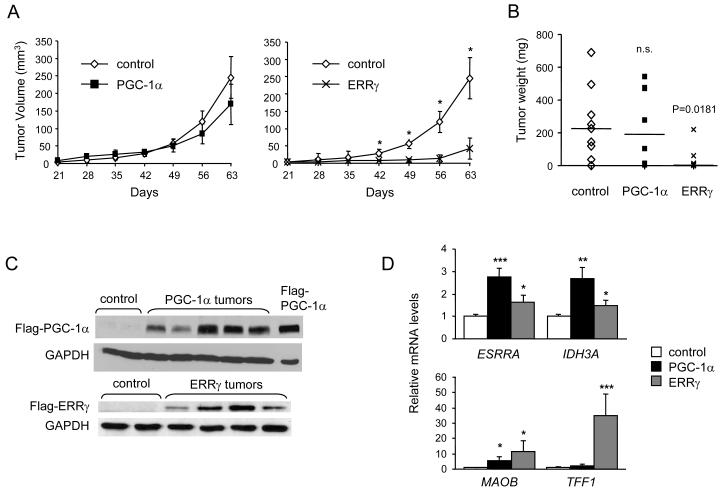
To test the effect of ERRγ and PGC-1α on the growth of MDA-MB-231 cells in vivo, MDA-EGFP (control), MDA-PGC-1α and MDA-ERRγ cells were injected orthotopically into the mammary fat pad of female SCID mice. MDA-ERRγ cells gave rise to significantly slower growing and smaller tumors than the ones seen with control cells (Fig. 2A and 2B). No significant difference was observed in PGC-1α-expressing tumors (Fig. 2A and 2B). A second cohort of mice with independently derived cell populations showed similar results (Supplementary Fig. S5), providing strong evidence that ERRγ suppresses tumor growth. To assess transgene expression and activity in vivo, tumors were excised at 63 or 70 days and analyzed for protein and mRNA levels. ERRγ and PGC-1α expression were detected in ERRγ and PGC-1α tumors respectively (Fig. 2C and Supplementary Fig. S6). We also observed sustained increased expression of ERR-regulated genes (ERRα, IDH3A, MAOB and TFF1, Fig. 2D), suggesting that ERRγ and PGC-1α remained active in vivo. Notably, the largest ERRγ tumor (outlier in Fig. 2B) had the lowest expression of ERR targets, consistent with ERRγ activity being important for suppressed tumor growth.
Figure 2.
ERRγ inhibits the growth of MDA-MB-231 xenografts. A, Tumor growth rates in SCID mice inoculated orthotopically with MDA-MB-231 populations expressing control (EGFP), PGC-1α or ERRγ. Data are tumor volumes measured at indicated days after injection and represent mean ± SEM from one of two representative experiments (n=8 to 9). B, Tumor weights at day 63, displayed as scatter plots with median values. C, Protein expression of transgenes in indicated tumors, detected by western blot analysis. D, Induced expression of ERR target genes in MDA-PGC-1α and MDA-ERRγ tumors. Data are expressed relative to mRNA levels of each gene in control tumors and are the mean ± SEM of 14 tumors for control, 12 tumors for PGC-1α and 6 tumors for ERRγ. n.s., not significant; *, P < 0.05; **, P<0.01; ***, P<0.001.
ERRγ promotes MET
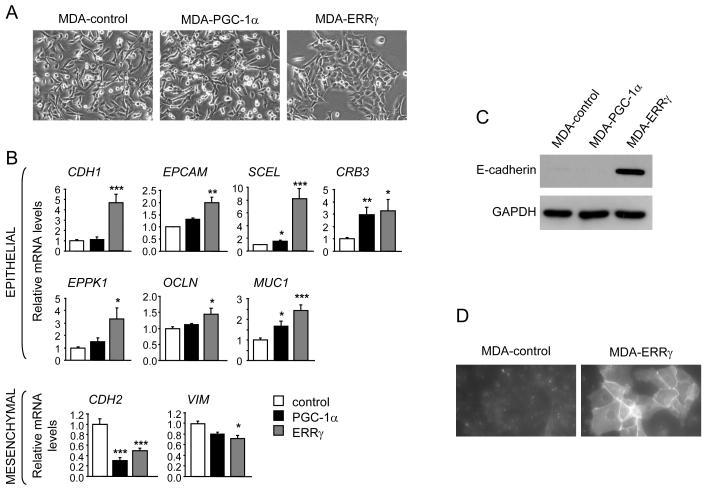
Because both ERRγ and PGC-1α enhanced oxidative metabolism but only ERRγ suppressed tumor growth, we reasoned that the increased oxidative capacity was unlikely to explain by itself the decreased in vivo growth of MDA-ERRγ tumors. We thus focused our study on features unique to the ERRγ-expressing cells. Strikingly, ERRγ expression led to changes in the morphology of MDA-MB-231 cells. Compared to control and MDA-PGC-1α cells that exhibit the typical mesenchymal phenotype of MDA-MB-231 cells, MDA-ERRγ cells formed epithelium-like clusters that were flatter and less spindle-shaped (Fig. 3A). These changes were suggestive of MET. MET and the reverse process EMT (epithelial-to-mesenchymal transition) are important factors in cancer progression, with EMT being associated with epithelial tumor dedifferentiation, and enhanced invasion of surrounding tissues and migration (31).
Figure 3.
ERRγ promotes MET. A, Cellular morphology of MDA-control, MDA-PGC-1α and MDA-ERRγ cells, examined by phase-contrast microscopy. B, Levels of mRNA for the indicated epithelial [E-cadherin (CDH1), EPCAM, Sciellin (SCEL), Crumbs3 (CRB3), Epiplakin 1 (EPPK1), Occludin (OCLN), Mucin 1 (MUC1)] and mesenchymal [N-cadherin (CDH2), vimentin (VIM)] markers in MDA-control, MDA-PGC-1α and MDA-ERRγ cells. Data are expressed as in Fig. 1 and are the mean ± SEM of 4 experiments performed in duplicate. C,D, Increased E-cadherin protein levels in MDA-ERRγ cells, determined by western blot and immunofluorescence analyses. GAPDH protein levels in (C) are shown as control for protein loading. *, P < 0.05; **, P<0.01; ***, P<0.001.
We next assessed whether changes in cell morphology were accompanied by changes in epithelial and mesenchymal marker expression. MDA-ERRγ cells showed increased expression of multiple epithelial markers, including E-cadherin (CDH1), the epithelial cell adhesion molecule EPCAM, sciellin (SCEL), epiplakin 1 (EPKK1), occludin (OCLN), mucin 1 (MUC1) and the cell polarity gene crumbs 3 (CRB3), and decreased expression of the mesenchymal markers N-cadherin (CDH2) and vimentin (VIM) (Fig. 3B). Increased expression of E-cadherin, a major characteristic of MET, was confirmed at the protein level (Figs. 3C & 3D). PGC-1α expressing cells showed a selective and weaker induction of just a subset of epithelial markers (sciellin, mucin 1 and crumbs 3), as well as repression of the mesenchymal marker N-cadherin; notably, E-cadherin expression was not induced. Importantly, the inability of PGC-1α to induce E-cadherin and other epithelial markers was not due to a weaker transcriptional activity, as PGC-1α was stronger than ERRγ or as good as ERRγ in inducing other ERR targets (e.g. ERRα, IDH3A, MAOB, Fig. 1B).
To confirm that ERRγ in MDA-ERRγ cells was driving the induction of E-cadherin, we treated cells with 4-hydroxy-tamoxifen (4OHT), which inhibits ERRγ activity (32, 33). 4OHT led to a dose-dependent inhibition of known ERRγ targets, such as MAOB and TFF1, as well as E-cadherin expression, in MDA-ERRγ cells, but not in control cells, consistent with E-cadherin being induced by ERRγ (Supplementary Fig. S7).
Next, we asked whether ERRγ could regulate the expression of epithelial and mesenchymal markers when expressed acutely, in the absence of selection of stable cell populations. ERRγ, PGC-1α, and as an additional control, ERRα, were expressed in MDA-MB-231 cells via adenoviral vectors. 24 hours after infection, ERRγ but not PGC-1α nor ERRα induced strongly E-cadherin. ERRγ and PGC-1α also induced IDH3A, MAOB and TFF1, with a similar preference as in the stable populations, and downregulated N-cadherin and vimentin expression (Supplementary Fig. S8). ERRα by itself did not activate ERR targets, consistent with the lack of ERRα activity in cells that have low endogenous PGC-1 (Supplementary Fig. S8).
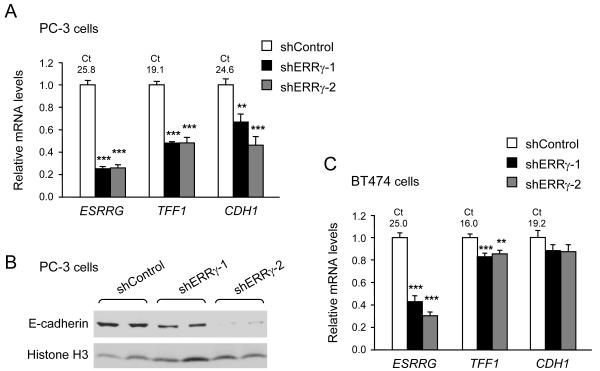
Finally, we tested whether endogenous ERRγ regulates E-cadherin, using prostate cancer PC-3 and breast cancer BT474 cells, where ERRγ is readily detectable; other breast cancer lines we have tested have low to undetectable ERRγ expression. In PC-3 cells, suppression of ERRγ via two different shRNAs led to significant decreases in the known ERR target TFF1, as well as E-cadherin (CDH1), at the mRNA and protein level (Fig. 4). In BT474 cells, suppression of ERRγ led to smaller decreases that reached significance only for TFF1, possibly due to the higher expression levels of ERRγ, TFF1 and CDH1, and the less efficient suppression of ERRγ, compared to PC-3 cells (Fig. 4).
Figure 4.
Knockdown of ERRγ decreases TFF1 and E-cadherin expression. A,C, Levels of ERRγ, TFF1 and E-cadherin (CDH1) mRNA in PC3 (A) or BT474 (C) cells expressing two different shRNAs for ERRγ are expressed relative to levels of each gene in cells expressing control shRNA (set equal to 1) and are the mean ± SEM of 3 experiments performed in duplicate or triplicate. **, P<0.01; ***, P<0.001; Ct, cycle threshold value of RT-qPCR. B, E-cadherin protein levels in PC3 cells expressing control shRNA, shERRγ-1 or shERRγ-2, determined by western blot. Histone H3 protein levels are shown as control for protein loading.
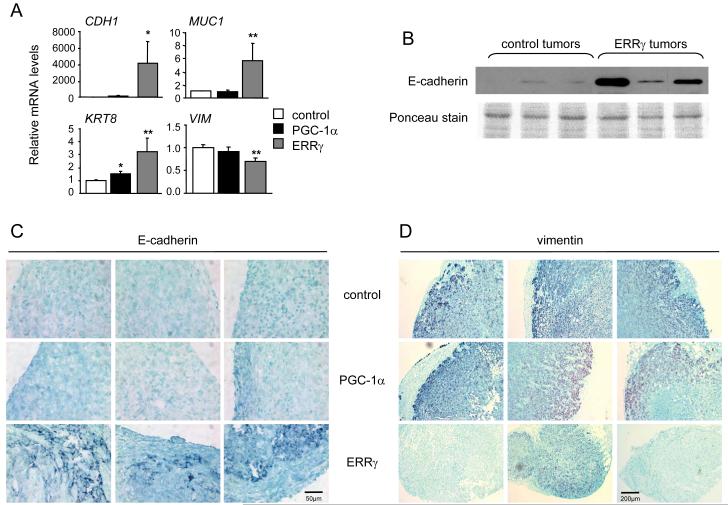
MDA-ERRγ tumors display markers of MET
We next determined the expression of epithelial and mesenchymal markers in the MDA-MB-231 tumor xenografts. E-cadherin mRNA and protein levels were significantly increased in ERRγ tumors, compared to PGC-1α and control tumors (Fig. 5A and 5B). The epithelial markers mucin 1 and keratin 8 (KRT8) were also upregulated in ERRγ tumors, while vimentin expression was decreased (Fig. 5A). Additionally, we observed positive immunostaining for E-cadherin in multiple ERRγ tumors but not in control and PGC-1α tumors (Fig. 5C). In contrast, vimentin was not detected in 2 of 3 ERRγ tumors analyzed, while all 3 control and PGC-1α tumors showed positive vimentin staining (Fig. 5D). In conclusion, the transition of mesenchymal to epithelial markers is maintained in the ERRγ tumors in vivo.
Figure 5.
MDA-ERRγ tumors display markers of MET. A, Levels of mRNA for epithelial [E-cadherin (CDH1), mucin 1 (MUC1), keratin 8 (KRT8)] and mesenchymal [vimentin (VIM)] markers in control, PGC-1α and ERRγ tumors, determined by RT-qPCR and expressed as in Fig. 2D. *, P < 0.05; **, P<0.01. B, E-cadherin protein levels in three MDA-control and three MDA-ERRγ tumors, detected by western blot analysis. Ponceau staining of the blot (lower panel) is shown as loading control. C,D, Immunohistochemical staining of E-cadherin (C) and vimentin (D) in sections of 3 tumors from each group.
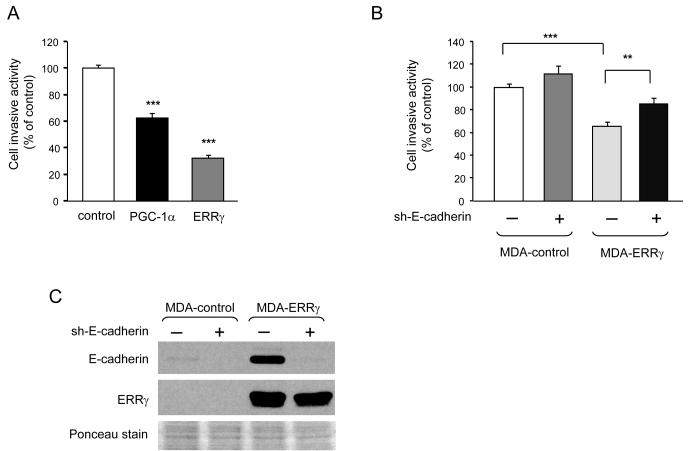
ERRγ suppresses invasiveness in vitro, in an E-cadherin-dependent manner
Mesenchymal cells display a higher invasive ability than epithelial ones. Consistent with MET, MDA-ERRγ cells showed reduced invasiveness, compared to control cells (Fig. 6A). Interestingly, PGC-1α also decreased the invasive properties of MDA-MB-231 cells, though to a lesser extent than ERRγ, suggesting that either the partial induction of the MET program seen at the molecular level (Fig. 3B) or another PGC-1α-dependent program affected cell behavior (Fig. 6A). The impaired invasiveness of MDA-ERRγ cells was at least partially dependent on E-cadherin, as silencing of E-cadherin diminished the suppressive effect of ERRγ on invasion (Fig. 6B).
Figure 6.
ERRγ decreases the invasiveness of MDA-MB-231 cells, partly via induction of E-cadherin expression. A, Decreased invasiveness of MDA-PGC-1α and MDA-ERRγ cells relative to MDA-control cells. B, E-cadherin knockdown by shRNA partially restores the decreased invasiveness of MDA-ERRγ cells. A & B, Data are expressed relative to the number of matrigel-invading MDA-control cells (set equal to 100; 1907 cells in panel A, 1085 cells in panel B) and are the mean ± SEM of 3 (A) or 2 (B) experiments performed in triplicate. C, E-cadherin and ERRγ protein expression in cells of panel B, determined as in Figures 1A and 3C. Ponceau staining of the blot (lower panel) is shown as loading control.
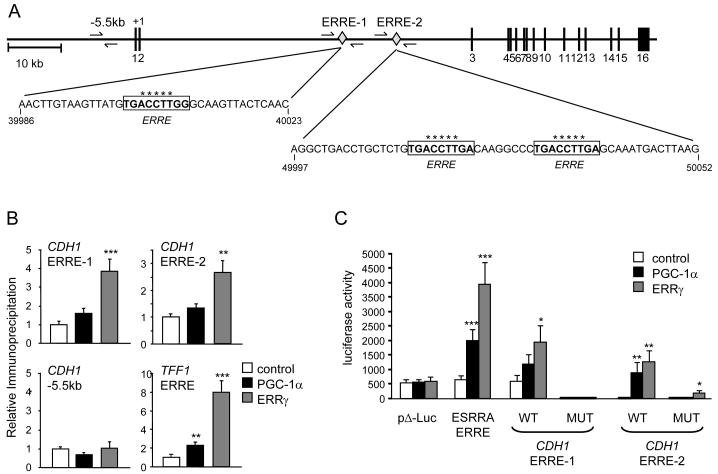
ERRγ is a direct regulator of E-cadherin expression
Given the central role of E-cadherin in MET and tumor growth, we next asked whether ERRγ regulated E-cadherin expression directly. Putative ERREs in the E-cadherin gene were screened for their association with ERRγ using ChIP assays. Binding of ERRγ was detected at two sites in the second intron (Fig. 7A and 7B), which is important for expression in vivo (34). As controls, ERRγ was also detected at the ERRE of TFF1 (9) but not at a region upstream of the E-cadherin promoter that does not harbor ERREs (Fig. 7B).
Figure 7.
ERRγ is a direct regulator of E-cadherin. A, Diagram of the human CDH1 (E-cadherin) locus showing ERRE-1 and ERRE-2 (boxed sequences) in intron 2. Numbered bars in diagram represent exons. Small arrows indicate the regions amplified in the ChIP experiments of panel B. B, ERRγ and PGC-1α recruitment to ERREs, determined by ChIP. Immunoprecipitated products were amplified by qPCR, using primers indicated in (A) or flanking the ERRE of the TFF1 gene. Data are expressed relative to the amount of DNA immunoprecipitated in control cells and are the mean ± SEM of three experiments performed in quadruplicate. The amounts of CDH1-ERRE-1, CDH1-ERRE-2 and TFF1-ERRE – containing DNA in the ERRγ IP were 0.13%, 0.10% and 0.16% of input DNA, respectively. C, ERRE-1 and ERRE-2 of CDH1 mediate ERRγ-dependent activation of transcription. Luciferase reporter constructs bearing just the basal promoter (pΔ-Luc) or having trimers of ERRE-1 or ERRE-2 of CDH1 [wild-type or bearing mutations in the nucleotides marked with a star (*) in panel A], were cotransfected in MDA-MB-231 with control plasmid (pcDNA3), PGC-1α or ERRγ expression constructs. The ERRE-containing regulatory sequence of the ESRRA gene was used as a positive control. Data are the mean ± SEM luciferase activity of three independent experiments performed in quadruplicate. *, P < 0.05; **, P<0.01; ***, P<0.001.
To determine the ability of the E-cadherin (CDH1) ERREs to respond to ERRγ, we subcloned these ERREs upstream of a basal luciferase reporter construct. ERRγ activated CDH1-ERRE-1 and CDH1-ERRE-2 containing reporters, as well as the positive control ESRRA-ERRE reporter, but had no effect on the ERRE-less basal promoter (Fig. 7C). Point mutations in the CDH1 ERREs abolished or diminished responsiveness to ERRγ, consistent with these sequences mediating the effect of ERRγ. PGC-1α also activated reporter activity in a CDH1-ERRE- and ERRα-dependent manner (Fig. 7C and Suppl. Fig. 9). In conclusion, ERRE-1 and ERRE-2 of the E-cadherin gene are bound by ERRγ in MDA-ERRγ cells and mediate ERRγ-dependent transcription in reporter assays, suggesting that ERRγ is a direct positive regulator of E-cadherin expression.
Discussion
ERRγ has been suggested as a marker of favorable clinical course in human breast cancer (1). In support of such a role, a survey of the Oncomine cancer database shows significant correlations between decreased ERRγ levels and higher breast cancer grade, metastasis, recurrence and unfavorable outcome (Supplementary Table 2). The extent to which these correlations reflect an active role of ERRγ in breast cancer progression had not been tested so far. Using MDA-MB-231 breast cancer cells expressing human ERRγ, we show here that ERRγ suppresses cell invasiveness in vitro and breast cancer xenograft growth in mice. Moreover, we provide evidence that ERRγ acts by inducing the expression of epithelial genes, thereby promoting MET.
In contrast to ERRγ, high levels of ERRα in breast cancer have been associated with increased risk of recurrence and adverse clinical outcome (1, 2). Recent studies support a role of ERRα in suppressing tumor growth (11, 23, 24, 25). In our study, enhancing ERRα levels and activity, via PGC-1α expression, did not enhance tumor growth. It is possible that the tumor promoting role of ERRα involves PGC-1α-independent activities, and is thus not affected by PGC-1α. Alternatively, a tumor promoting role of ERRα may be antagonized by ERRα-independent tumor suppressive effects of PGC-1α. Notably, low levels of PGC-1α have been associated with poor clinical outcome in breast cancer (35, 36). Additional studies will be needed to test a potential role of PGC-1α in breast cancer, as well as to understand the cellular context in which ERRα promotes tumor growth.
EMT plays an important role in tumorigenesis (31). The ability of epithelial cancer cells to invade surrounding tissues and escape from the primary tumor is associated with loss of intercellular adhesion, loss of contact inhibition, loss of apical-basal polarity and enhanced migratory potential. During this process, cells transit from a polarized epithelial phenotype to a highly motile mesenchymal one. Loss of E-cadherin expression is often considered a common indicator of EMT, while the reverse process, MET, is usually associated with E-cadherin re-expression. ERRγ promoted multiple aspects of MET, including increased levels of several epithelial markers, decreased levels of mesenchymal markers, morphological changes, and decreased invasiveness. Interestingly, ectopic expression of ERRα (Suppl. Fig. 8) or activation of endogenous ERRα (via expression of PGC-1α) led to either no effect or weaker and gene-selective induction of epithelial markers, suggesting that ERRγ is a more efficient activator than ERRα with respect to the epithelial program. Notably, PGC-1α/ERRα were as efficient as ERRγ or more efficient than ERRγ in inducing other ERR targets, such as MAOB and IDH3A. These findings suggest that ERRα and ERRγ, which bind common target genes (17), can act differentially at select targets. The underlying mechanisms for such differential action are not yet clear but could reflect differential recruitment of coregulators.
From the genes we have tested, E-cadherin is one of the most differentially affected by ERRγ and PGC-1α. In breast cancer, dysfunction of E-cadherin correlates with loss of epithelial characteristics, acquisition of invasiveness, increased tumor grade, metastatic behavior and poor prognosis (37, 38). Overexpression of E-cadherin in MDA-MB-231 cells decreases invasiveness (39) and growth in soft agar (40). Additional cellular and animal models support E-cadherin as a suppressor of tumor growth and invasion (41, 42). The mechanism by which ERRγ induces E-cadherin is likely to be direct, via ERREs in the second intron, which is important for expression (34). Interestingly, when taken out of their genomic context, these ERREs also mediate PGC-1α/ERRα-activated transcription, suggesting that epigenetic regulation at the CDH1 locus may keep the endogenous gene refractory to PGC-1α. Consistent with this notion, the CDH1 locus in MDA-MB-231 cells is methylated (38). Although we cannot exclude yet additional mechanisms by which ERRγ induces E-cadherin, we have not seen significant differences in the levels of other known E-cadherin regulators (43) between ERRγ and PGC-1α expressing MDA-MB-231 cells (Supplementary Table 3).
A role of ERRγ for epithelial function may not be restricted to breast cancer. ERRγ is highly expressed in epithelia of several tissues, including intestine, stomach, kidney and salivary gland, where it may contribute to developmental processes, normal epithelial function, and/or pathophysiological processes where EMT/MET transitions have been implicated [e.g. organ fibrosis, (44)]. Moreover, ERRγ expression has been correlated with a better prognosis in prostate, endometrial and ovarian carcinomas (45-47), and overexpression of ERRγ suppresses prostate cancer cell proliferation and tumor growth (48). In conclusion, ERRγ may be involved in the pathology of different tumors and development of ERRγ agonists may have general applications in cancer.
Supplementary Material
Acknowledgments
We thank Melissa Ritland, Sherry Niessen and Yoshitake Cho for technical advice and assistance, Dr. Cravatt for expression vectors and Drs. Saez and Felding-Habermann for discussions. The study was supported by funding from the California Breast Cancer Research Program (12IB-0010) and the Department of Defense Breast Cancer Research Program (W81XWH-09-1-0327).
Funded by the California Breast Cancer Research Program (12IB-0010) and the Department of Defense Breast Cancer Research Program (W81XWH-09-1-0327).
References
- 1.Ariazi EA, Clark GM, Mertz JE. Estrogen-related receptor alpha and estrogen-related receptor gamma associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res. 2002;62:6510–6518. [PubMed] [Google Scholar]
- 2.Suzuki T, Miki Y, Moriya T, et al. Estrogen-related receptor alpha in human breast carcinoma as a potent prognostic factor. Cancer Res. 2004;64:4670–4676. doi: 10.1158/0008-5472.CAN-04-0250. [DOI] [PubMed] [Google Scholar]
- 3.Ariazi EA, Jordan VC. Estrogen-related receptors as emerging targets in cancer and metabolic disorders. Curr Top Med Chem. 2006;6:203–215. doi: 10.2174/1568026610606030203. [DOI] [PubMed] [Google Scholar]
- 4.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 5.Ariazi EA, Kraus RJ, Farrell ML, Jordan VC, Mertz JE. Estrogen-related receptor alpha1 transcriptional activities are regulated in part via the ErbB2/HER2 signaling pathway. Mol Cancer Res. 2007;5:71–85. doi: 10.1158/1541-7786.MCR-06-0227. [DOI] [PubMed] [Google Scholar]
- 6.Vu EH, Kraus RJ, Mertz JE. Phosphorylation-dependent sumoylation of estrogen-related receptor alpha1. Biochemistry. 2007;46:9795–9804. doi: 10.1021/bi700316g. [DOI] [PubMed] [Google Scholar]
- 7.Tremblay AM, Wilson BJ, Yang XJ, Giguere V. Phosphorylation-dependent sumoylation regulates estrogen-related receptor-alpha and -gamma transcriptional activity through a synergy control motif. Mol Endocrinol. 2008;22:570–584. doi: 10.1210/me.2007-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villena JA, Kralli A. ERRalpha: a metabolic function for the oldest orphan. Trends Endocrinol Metab. 2008;19:269–276. doi: 10.1016/j.tem.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu D, Kiriyama Y, Lee KY, Giguere V. Transcriptional regulation of the estrogen-inducible pS2 breast cancer marker gene by the ERR family of orphan nuclear receptors. Cancer Res. 2001;61:6755–6761. [PubMed] [Google Scholar]
- 10.Tremblay AM, Giguere V. The NR3B subgroup: an ovERRview. Nucl Recept Signal. 2007;5:e009. doi: 10.1621/nrs.05009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein RA, Chang CY, Kazmin DA, et al. Estrogen-related receptor alpha is critical for the growth of estrogen receptor-negative breast cancer. Cancer Res. 2008;68:8805–8812. doi: 10.1158/0008-5472.CAN-08-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deblois G, Hall JA, Perry MC, et al. Genome-wide identification of direct target genes implicates estrogen-related receptor alpha as a determinant of breast cancer heterogeneity. Cancer Res. 2009;69:6149–6157. doi: 10.1158/0008-5472.CAN-09-1251. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber SN, Emter R, Hock MB, et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mootha VK, Handschin C, Arlow D, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huss JM, Torra IP, Staels B, Giguere V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrier JC, Deblois G, Champigny C, Levy E, Giguere V. Estrogen-related receptor alpha (ERRalpha) is a transcriptional regulator of apolipoprotein A-IV and controls lipid handling in the intestine. J Biol Chem. 2004;279:52052–52058. doi: 10.1074/jbc.M410337200. [DOI] [PubMed] [Google Scholar]
- 17.Dufour CR, Wilson BJ, Huss JM, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Villena JA, Hock MB, Chang WY, Barcas JE, Giguere V, Kralli A. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc Natl Acad Sci U S A. 2007;104:1418–1423. doi: 10.1073/pnas.0607696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huss JM, Imahashi K, Dufour CR, et al. The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Sonoda J, Laganiere J, Mehl IR, et al. Nuclear receptor ERR alpha and coactivator PGC-1 beta are effectors of IFN-gamma-induced host defense. Genes Dev. 2007;21:1909–1920. doi: 10.1101/gad.1553007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alaynick WA, Kondo RP, Xie W, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Alaynick WA, Way JM, Wilson SA, et al. ERR{gamma} Regulates Cardiac, Gastric, and Renal Potassium Homeostasis. Mol Endocrinol. 2009 doi: 10.1210/me.2009-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bianco S, Lanvin O, Tribollet V, Macari C, North S, Vanacker JM. Modulating estrogen receptor-related receptor-alpha activity inhibits cell proliferation. J Biol Chem. 2009;284:23286–23292. doi: 10.1074/jbc.M109.028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chisamore MJ, Wilkinson HA, Flores O, Chen JD. Estrogen-related receptor-alpha antagonist inhibits both estrogen receptor-positive and estrogen receptor-negative breast tumor growth in mouse xenografts. Mol Cancer Ther. 2009;8:672–681. doi: 10.1158/1535-7163.MCT-08-1028. [DOI] [PubMed] [Google Scholar]
- 25.Wu F, Wang J, Wang Y, Kwok TT, Kong SK, Wong C. Estrogen-related receptor alpha (ERRalpha) inverse agonist XCT-790 induces cell death in chemotherapeutic resistant cancer cells. Chem Biol Interact. 2009;181:236–242. doi: 10.1016/j.cbi.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Kressler D, Schreiber SN, Knutti D, Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J Biol Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 27.Wende AR, Huss JM, Schaeffer PJ, Giguere V, Kelly DP. PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism. Mol Cell Biol. 2005;25:10684–10694. doi: 10.1128/MCB.25.24.10684-10694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marinescu VD, Kohane IS, Riva A. MAPPER: a search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinformatics. 2005;6:79. doi: 10.1186/1471-2105-6-79. Available from: http://bio.chip.org/mapper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) J Biol Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 30.Willy PJ, Murray IR, Qian J, et al. Regulation of PPARgamma coactivator 1alpha (PGC-1alpha) signaling by an estrogen-related receptor alpha (ERRalpha) ligand. Proc Natl Acad Sci U S A. 2004;101:8912–8917. doi: 10.1073/pnas.0401420101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 32.Coward P, Lee D, Hull MV, Lehmann JM. 4-Hydroxytamoxifen binds to and deactivates the estrogen-related receptor gamma. Proc Natl Acad Sci U S A. 2001;98:8880–8884. doi: 10.1073/pnas.151244398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tremblay GB, Bergeron D, Giguere V. 4-Hydroxytamoxifen is an isoform-specific inhibitor of orphan estrogen-receptor-related (ERR) nuclear receptors beta and gamma. Endocrinology. 2001;142:4572–4575. doi: 10.1210/endo.142.10.8528. [DOI] [PubMed] [Google Scholar]
- 34.Stemmler MP, Hecht A, Kemler R. E-cadherin intron 2 contains cis-regulatory elements essential for gene expression. Development. 2005;132:965–976. doi: 10.1242/dev.01662. [DOI] [PubMed] [Google Scholar]
- 35.Jiang WG, Douglas-Jones A, Mansel RE. Expression of peroxisome-proliferator activated receptor-gamma (PPARgamma) and the PPARgamma co-activator, PGC-1, in human breast cancer correlates with clinical outcomes. Int J Cancer. 2003;106:752–757. doi: 10.1002/ijc.11302. [DOI] [PubMed] [Google Scholar]
- 36.Watkins G, Douglas-Jones A, Mansel RE, Jiang WG. The localisation and reduction of nuclear staining of PPARgamma and PGC-1 in human breast cancer. Oncol Rep. 2004;12:483–488. [PubMed] [Google Scholar]
- 37.Berx G, Van Roy F. The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res. 2001;3:289–293. doi: 10.1186/bcr309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lombaerts M, van Wezel T, Philippo K, et al. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Cancer. 2006;94:661–671. doi: 10.1038/sj.bjc.6602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong AS, Gumbiner BM. Adhesion-independent mechanism for suppression of tumor cell invasion by E-cadherin. J Cell Biol. 2003;161:1191–1203. doi: 10.1083/jcb.200212033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soto E, Yanagisawa M, Marlow LA, Copland JA, Perez EA, Anastasiadis PZ. p120 catenin induces opposing effects on tumor cell growth depending on E-cadherin expression. J Cell Biol. 2008;183:737–749. doi: 10.1083/jcb.200805113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Derksen PW, Liu X, Saridin F, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006;10:437–449. doi: 10.1016/j.ccr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 42.Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 2008;27:6920–6929. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baranwal S, Alahari SK. Molecular mechanisms controlling E-cadherin expression in breast cancer. Biochem Biophys Res Commun. 2009;384:6–11. doi: 10.1016/j.bbrc.2009.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 45.Fujimura T, Takahashi S, Urano T, et al. Differential expression of estrogen-related receptors beta and gamma (ERRbeta and ERRgamma) and their clinical significance in human prostate cancer. Cancer Sci. 2009 doi: 10.1111/j.1349-7006.2009.01451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao M, Sun P, Wang J, Zhao D, Wei L. Expression of estrogen receptor-related receptor isoforms and clinical significance in endometrial adenocarcinoma. Int J Gynecol Cancer. 2006;16:827–833. doi: 10.1111/j.1525-1438.2006.00527.x. [DOI] [PubMed] [Google Scholar]
- 47.Sun P, Sehouli J, Denkert C, et al. Expression of estrogen receptor-related receptors, a subfamily of orphan nuclear receptors, as new tumor biomarkers in ovarian cancer cells. J Mol Med. 2005;83:457–467. doi: 10.1007/s00109-005-0639-3. [DOI] [PubMed] [Google Scholar]
- 48.Yu S, Wang X, Ng CF, Chen S, Chan FL. ERRgamma suppresses cell proliferation and tumor growth of androgen-sensitive and androgen-insensitive prostate cancer cells and its implication as a therapeutic target for prostate cancer. Cancer Res. 2007;67:4904–4914. doi: 10.1158/0008-5472.CAN-06-3855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.