Abstract
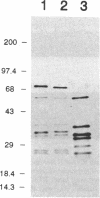
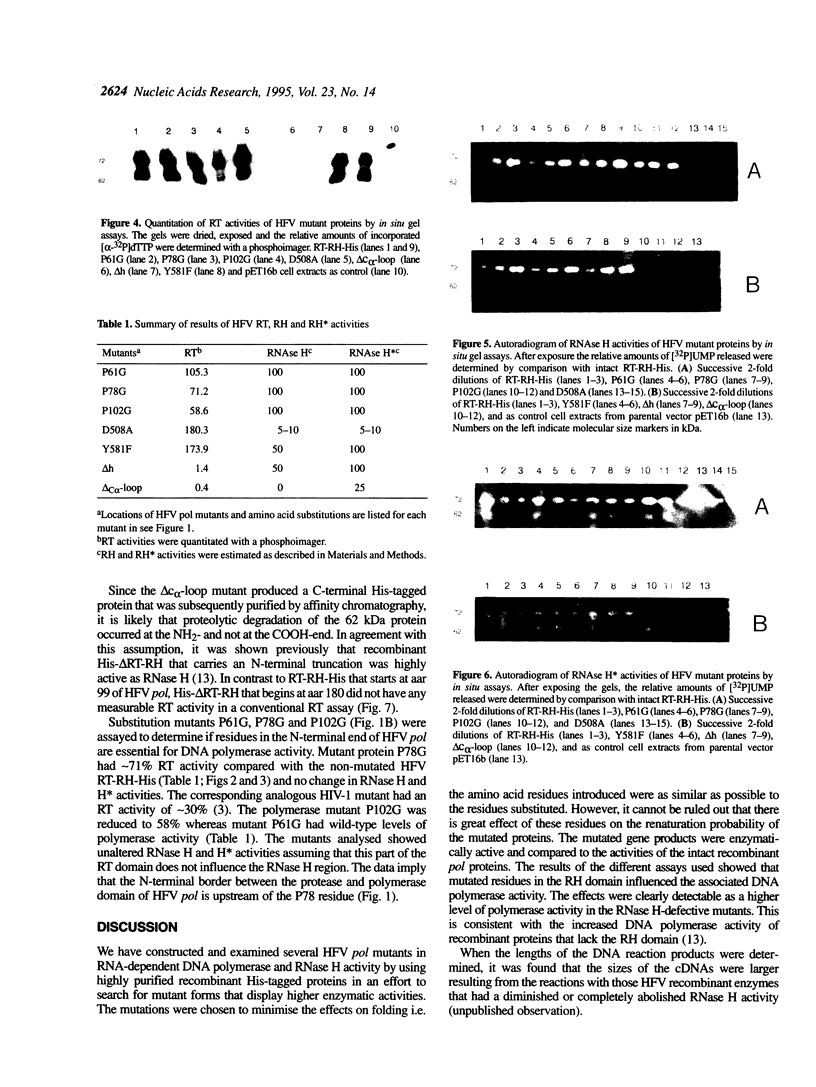
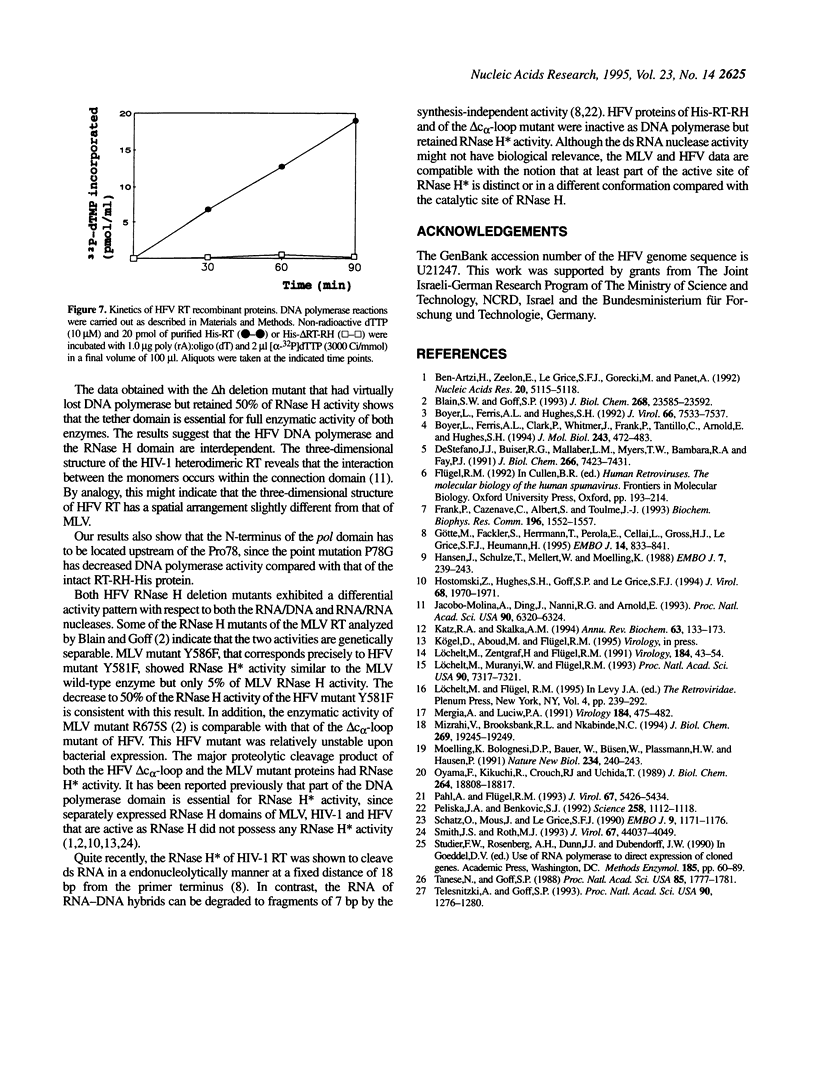
Human foamy or spuma virus (HFV) codes for a distinct set of pol gen products. To determine the minimal requirements for the HFV enzymatic activities, defined residues of the reverse transcriptase (RT) and ribo-nuclease H (RNase H) domain of the HFV pol gene were mutated by site-specific PCR mutagenesis. The mutant gene products were bacterially expressed, purified by Ni2+ chelate affinity chromatography and characterised by Western blotting. The enzymatic activities of the individual recombinant HFV pol mutant proteins were characterised by the situ RT, RNase H and RNase H assays. Two substitution mutants reached RT activity levels higher than that of the intact recombinant HFV RT-RH-His. When the catalytically essential D508 was substituted by A508, 5% of RNase H activity was retained while DNA polymerase activity increased 2-fold. A deletion of 11 amino acid residues in the hinge region completely abolished DNA polymerase while RNase H activity decreased 2-fold. A deletion mutant in the C-terminal RH domain showed no RNase H but retained RNase H activity indicating that the activities are genetically separable. The combined data reveal that the HFV DNA polymerase and RNase H activities are interdependent.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Artzi H., Zeelon E., Le-Grice S. F., Gorecki M., Panet A. Characterization of the double stranded RNA dependent RNase activity associated with recombinant reverse transcriptases. Nucleic Acids Res. 1992 Oct 11;20(19):5115–5118. doi: 10.1093/nar/20.19.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blain S. W., Goff S. P. Nuclease activities of Moloney murine leukemia virus reverse transcriptase. Mutants with altered substrate specificities. J Biol Chem. 1993 Nov 5;268(31):23585–23592. [PubMed] [Google Scholar]
- Boyer P. L., Ferris A. L., Clark P., Whitmer J., Frank P., Tantillo C., Arnold E., Hughes S. H. Mutational analysis of the fingers and palm subdomains of human immunodeficiency virus type-1 (HIV-1) reverse transcriptase. J Mol Biol. 1994 Oct 28;243(3):472–483. doi: 10.1006/jmbi.1994.1673. [DOI] [PubMed] [Google Scholar]
- Boyer P. L., Ferris A. L., Hughes S. H. Mutational analysis of the fingers domain of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1992 Dec;66(12):7533–7537. doi: 10.1128/jvi.66.12.7533-7537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano J. J., Buiser R. G., Mallaber L. M., Myers T. W., Bambara R. A., Fay P. J. Polymerization and RNase H activities of the reverse transcriptases from avian myeloblastosis, human immunodeficiency, and Moloney murine leukemia viruses are functionally uncoupled. J Biol Chem. 1991 Apr 25;266(12):7423–7431. [PubMed] [Google Scholar]
- Frank P., Cazenave C., Albert S., Toulmé J. J. Sensitive detection of low levels of ribonuclease H activity by an improved renaturation gel assay. Biochem Biophys Res Commun. 1993 Nov 15;196(3):1552–1557. doi: 10.1006/bbrc.1993.2428. [DOI] [PubMed] [Google Scholar]
- Götte M., Fackler S., Hermann T., Perola E., Cellai L., Gross H. J., Le Grice S. F., Heumann H. HIV-1 reverse transcriptase-associated RNase H cleaves RNA/RNA in arrested complexes: implications for the mechanism by which RNase H discriminates between RNA/RNA and RNA/DNA. EMBO J. 1995 Feb 15;14(4):833–841. doi: 10.1002/j.1460-2075.1995.tb07061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Schulze T., Mellert W., Moelling K. Identification and characterization of HIV-specific RNase H by monoclonal antibody. EMBO J. 1988 Jan;7(1):239–243. doi: 10.1002/j.1460-2075.1988.tb02805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostomsky Z., Hughes S. H., Goff S. P., Le Grice S. F. Redesignation of the RNase D activity associated with retroviral reverse transcriptase as RNase H. J Virol. 1994 Mar;68(3):1970–1971. doi: 10.1128/jvi.68.3.1970-1971.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobo-Molina A., Ding J., Nanni R. G., Clark A. D., Jr, Lu X., Tantillo C., Williams R. L., Kamer G., Ferris A. L., Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- Löchelt M., Muranyi W., Flügel R. M. Human foamy virus genome possesses an internal, Bel-1-dependent and functional promoter. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7317–7321. doi: 10.1073/pnas.90.15.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löchelt M., Zentgraf H., Flügel R. M. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel 1 gene. Virology. 1991 Sep;184(1):43–54. doi: 10.1016/0042-6822(91)90820-2. [DOI] [PubMed] [Google Scholar]
- Mergia A., Luciw P. A. Replication and regulation of primate foamy viruses. Virology. 1991 Oct;184(2):475–482. doi: 10.1016/0042-6822(91)90417-a. [DOI] [PubMed] [Google Scholar]
- Mizrahi V., Brooksbank R. L., Nkabinde N. C. Mutagenesis of the conserved aspartic acid 443, glutamic acid 478, asparagine 494, and aspartic acid 498 residues in the ribonuclease H domain of p66/p51 human immunodeficiency virus type I reverse transcriptase. Expression and biochemical analysis. J Biol Chem. 1994 Jul 29;269(30):19245–19249. [PubMed] [Google Scholar]
- Mölling K., Bolognesi D. P., Bauer H., Büsen W., Plassmann H. W., Hausen P. Association of viral reverse transcriptase with an enzyme degrading the RNA moiety of RNA-DNA hybrids. Nat New Biol. 1971 Dec 22;234(51):240–243. doi: 10.1038/newbio234240a0. [DOI] [PubMed] [Google Scholar]
- Oyama F., Kikuchi R., Crouch R. J., Uchida T. Intrinsic properties of reverse transcriptase in reverse transcription. Associated RNase H is essentially regarded as an endonuclease. J Biol Chem. 1989 Nov 5;264(31):18808–18817. [PubMed] [Google Scholar]
- Pahl A., Flügel R. M. Endonucleolytic cleavages and DNA-joining activities of the integration protein of human foamy virus. J Virol. 1993 Sep;67(9):5426–5434. doi: 10.1128/jvi.67.9.5426-5434.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peliska J. A., Benkovic S. J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992 Nov 13;258(5085):1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- Schatz O., Mous J., Le Grice S. F. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3'----5' exonuclease activity. EMBO J. 1990 Apr;9(4):1171–1176. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. S., Roth M. J. Purification and characterization of an active human immunodeficiency virus type 1 RNase H domain. J Virol. 1993 Jul;67(7):4037–4049. doi: 10.1128/jvi.67.7.4037-4049.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tanese N., Goff S. P. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky A., Goff S. P. RNase H domain mutations affect the interaction between Moloney murine leukemia virus reverse transcriptase and its primer-template. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1276–1280. doi: 10.1073/pnas.90.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]