Abstract
Somatic mutations in the EGFR tyrosine kinase (TK) domain play a critical role in the development and treatment of non-small cell lung cancer (NSCLC). Strong genetic influence on susceptibility to these mutations has been suggested. To identify the genetic factors conferring risk for the EGFR TK mutations in NSCLC, a case-control study was conducted in 141 Taiwanese NSCLC patients by focusing on three functional polymorphisms in the EGFR gene [-216G/T, intron 1(CA)n and R497K]. Allelic imbalance (AI) of the EGFR -216G/T polymorphism was also tested in the heterozygous patients as well as in the NCI-60 cancer cell lines to further verify its function. We found that the frequencies of the alleles -216T and CA-19 are significantly higher in the patients with any mutation (p=0.032 and 0.01, respectively), in particular in those with exon 19 microdeletions (p=0.006 and 0.033, respectively), but not in the patients with L858R mutation. The -216T allele is favored to be amplified in both tumor DNA of lung cancer patients and cancer cell lines. We conclude that the local haplotype structures across the EGFR gene may favor the development of cellular malignancies and thus significantly confer risk to the occurrence of EGFR mutations in NSCLC, particularly the exon 19 microdeletions.
Keywords: EGFR, mutations, polymorphisms, germline, association
Introduction
The discovery of somatic mutations in the EGFR tyrosine kinase domain constitutes one of the most important findings in lung cancer in recent years. The missense point mutation L858R in exon 21 and microdeletions in exon 19 represent approximately 85-90% of all EGFR mutations and are significantly associated with clinical response to EGFR inhibitors (1-3). Why and how somatic EGFR mutations develop during carcinogenesis, however, remains largely unknown. The observation that the mutations are found more often in individuals who have never smoked excludes the involvement of carcinogens in tobacco smoke. The mutations are more prevalent in East Asian populations (30%~50%), but are relatively rare in individuals of European and African descent (<20%) (1-3). Moreover, the prevalence of these mutations in East Asians who have migrated to other countries remains high, suggesting that the origin of these mutations is related more to ethnicity than environment (3). These observations imply a germline susceptibility to this mutagenesis event. We therefore hypothesize that lung cancer carrying EGFR mutations may be attributable to susceptibility alleles with significant ethnic differences in their frequency. Moreover, these alleles may be distinct from those associated with lung cancer in general, and the molecular epidemiology of each somatic mutation may also be different.
We and others have previously demonstrated that EGFR is highly polymorphic and its expression and activity are significantly regulated by polymorphisms (4-8). A germline (CA)n polymorphism (rs45559542) in the intron 1 of EGFR has been associated with EGFR gene expression, with the shorter CA-repeats being associated with higher transcription level of EGFR (7). We also discovered a promoter SNP (-216G>T/rs712829) that increases EGFR expression (4). A nonsynonymous common EGFR polymorphism R497K (G>A) (rs2227983) SNP was also identified, and the K (A) allele appears to decrease the activity of EGFR (8). We have demonstrated that the allele frequency of these polymorphisms differs significantly among ethnic groups (4-6). Whether these polymorphisms confer risk for development of EGFR mutations, and the ethnic distribution of these polymorphisms further leads to a higher mutation rate in Asian patients have not yet been systematically evaluated. Here we performed an association study between EGFR mutations and the three aforementioned germline polymorphisms.
Materials and Methods
EGFR mutation detection and genotyping in NSCLC patient samples
EGFR mutation data of exons 18-21 in a total of 141 NSCLC patients collected from Taiwan have been previously published (9). The three functional polymorphisms were genotyped in the germline DNA extracted from peripheral blood samples of these patients. Candidate polymorphisms were genotyped according to protocols published previously (4-6). Genotyping assays were repeated in 10% of patient samples (randomly chosen), and 100% concordance between the replicates and original data were observed. The -216G/T and Intron 1 (CA)n were also genotyped in the HapMap HCB (n=45) and CEU (n=60) samples to compare their allele frequencies with our patient population (see supplemental materials).
Analysis of allelic imbalance (AI) of the EGFR -216G/T polymorphism
The peak heights of the G and T alleles in NSCLC tumor DNAs of the heterozygous individuals were assessed using SNaPshot (Applied Biosystems). AI was defined by taking the ratio of the peak height of each allele and normalizing to that of the germline DNA. For NCI-60 cancer cell lines, we selected heterozygous samples based on our previous study (6), and used the mean value of the peak height ratio from 10 randomly selected normal germline DNA samples as controls for data normalization. A final ratio of <0.60 or >1.67 was set as the cut-off for AI.
Statistics
Chi-square test (CST) or Fisher’s exact test (FET) was used to test for Hardy-Weinberg equilibrium (HWE) and the allelic association between polymorphisms and EGFR mutations. We regarded the patients bearing EGFR mutations as ‘cases’ and those bearing EGFR wild type as ‘controls’. Because of the biological difference between the EGFR exon 19 microdeletions and the exon 21 L858R point mutation (1), we also tested for association between each polymorphism and each of these two mutations. In addition, we conducted an exploratory analysis of the combined effect of the “risk” alleles on the presence of EGFR mutations. Odds ratios (OR), 95% confidence intervals (CI) and p values (two-sided) were calculated for each allelic association. P=0.05 was set as the cutoff for statistical significance, without adjustment for multiple testing.
Results
With the exception of the EGFR intron 1 (CA)n polymorphism that appeared ambiguous in three samples, all polymorphisms were successfully genotyped in all samples. No significant deviation from HWE was observed (CST, p>0.05 for all tests, data not shown).
Both the -216T and 19 CA-repeat (CA-19) alleles of the intron 1 (CA)n polymorphism were significantly associated with the exon 19 mutations, but not with the L858R mutations. No statistically significant differences were found between the R497K polymorphism and EGFR mutations (Table 1).
Table 1.
Allelic association between EGFR polymorphisms and mutations.
| Polymorphism | Allele | All MT | All WT | OR (95%CI) | p | Ex19 del+ | Ex19 del- | OR (95%CI) | p | 858 L>R+ | 858 L>R - | OR (95%CI) | p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -216G/T | T | 8 | 6 | 3.13 | 0.03 | 6 | 8 | 4.28 | 0.006 | 2 | 12 | 1.07 | 1.00 |
| G | 80 | 188 | (1.05-9.33) | 40 | 228 | (1.41-12.98) | 36 | 232 | 0.23-5.00 | ||||
| R497K | G | 42 | 89 | 1.08 | 0.77 | 26 | 105 | 1.62 | 0.14 | 14 | 117 | 0.63 | 0.20 |
| A | 46 | 105 | 0.65-1.79 | 20 | 131 | 0.86-3.07 | 24 | 127 | 0.31-1.28 | ||||
| Intron 1 (CA)n | CA-19 | 12 | 9 | 3.43 | 0.01 | 7 | 14 | 2.77 | 0.03 | 4 | 17 | 1.53 | 0.51 |
| others | 76 | 179 | (1.27-7.76) | 39 | 216 | (1.05-7.30) | 34 | 221 | 0.49-4.82 | ||||
| CA-15 | 3 | 8 | - | 0.18 | 2 | 9 | - | 0.60 | 1 | 10 | - | 0.38 | |
| CA-16 | 14 | 35 | 7 | 42 | 6 | 43 | |||||||
| CA-17 | 3 | 5 | 2 | 6 | 0 | 8 | |||||||
| CA-18 | 4 | 6 | 2 | 8 | 2 | 8 | |||||||
| CA-19 | 12 | 9 | 7 | 14 | 4 | 17 | |||||||
| CA-20 | 44 | 109 | 23 | 130 | 20 | 133 | |||||||
| CA-21 | 4 | 13 | 2 | 15 | 2 | 15 | |||||||
| CA-22 | 4 | 3 | 1 | 6 | 3 | 4 | |||||||
Note: MT=mutant; WT=wild type; OR=odds ratio; CI=confidence interval; Ex19 del=exon 19 deletion.
The association between the number of “risk” alleles and EGFR mutations was further tested by combining -216T and intron 1 CA-19 alleles with and without the 497G allele, using the patients with no risk alleles as a reference. As a result, an enhanced association between the combined alleles and EGFR mutations, in particular the exon 19 microdeletions was demonstrated. Although 497G/A alone is not associated with EGFR mutations, combination of the 497G allele with -216T and CA-19 alleles showed a significant increase of the OR compared to the patients with no risk alleles (Table 2). To further evaluate the function of the -216T allele, AI analysis of -216G/T alleles was successfully performed in 9 of 12 heterozygous NSCLC patients. Significant AI was observed in 4 tumors (44%) and showed a relative gain of the T allele or loss of the G allele. This preference for retention of the T-allele was also observed in the NCI-60 cancer cell lines (n=58) where Al was observed in 12 (55%) out of 22 heterozygous cell lines, 10 of which (83%) contained a relative gain or retention of the T allele (Fig. 1).
Table 2.
Association between the EGFR mutations and the number of risk alleles combining the genotypes.
| Combination of polymorphisms | Number of risk alleles | All WT | All MT | OR | p | Ex19 del- | Ex19 del+ | OR | p | 858 L>R- | 858 L>R+ | OR | p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -216G/T+ Intron 1 (CA)n | 0 | 83 | 29 | 1.00 (referent) | - | 98 | 14 | 1.00 (referent) | - | 98 | 14 | 1.00 (referent) | - |
| 1 | 13 | 10 | 2.20 (0.87-5.56) | 0.13 | 18 | 5 | 1.94 (0.62-6.07) | 0.32 | 19 | 4 | 1.47 (0.44-4.97) | 0.51 | |
| 2 | 1 | 5 | 14.30 (1.60-127.10) | 0.008 | 2 | 4 | 14.00 (2.34-83.67) | 0.005 | 5 | 1 | 1.40 (0.15-12.88) | 0.57 | |
| -216G/T+ Intron 1 (CA)n+ R497K | 0 | 25 | 9 | 1.00 (referent) | 31 | 3 | 1.00 (referent) | - | 29 | 5 | 1.00 (referent) | - | |
| 1 | 46 | 16 | 0.97 (0.37-2.50) | 1 | 54 | 8 | 1.53 (0.38-6.20) | 0.74 | 54 | 8 | 0.86 (0.26-2.87) | 1.00 | |
| 2 | 20 | 13 | 1.81 (0.64-5.08) | 0.31 | 26 | 7 | 2.78 (0.65-11.86) | 0.19 | 27 | 6 | 1.29 (0.35-4.72) | 0.75 | |
| 3 | 6 | 4 | 1.85 (0.42-8.11) | 0.45 | 7 | 3 | 4.43 (0.73-26.76) | 0.12 | 10 | 0 | 2.26 (0.01-5.03) | 0.57 | |
| 4 | 0 | 2 | 13.42 (0.59-306.10) | 0.09 | 0 | 2 | 45.00 (1.78-1140.00) | 0.02 | 2 | 0 | 1.07 (0.05-25.56) | 1.00 | |
Note: The risk alleles refer to -216T, Intron 1 CA-19 and 497R (G). MT=mutant; WT=wild type; OR=odds ratio; CI=confidence interval; Ex19 del=exon 19 deletion.
Fig. 1.
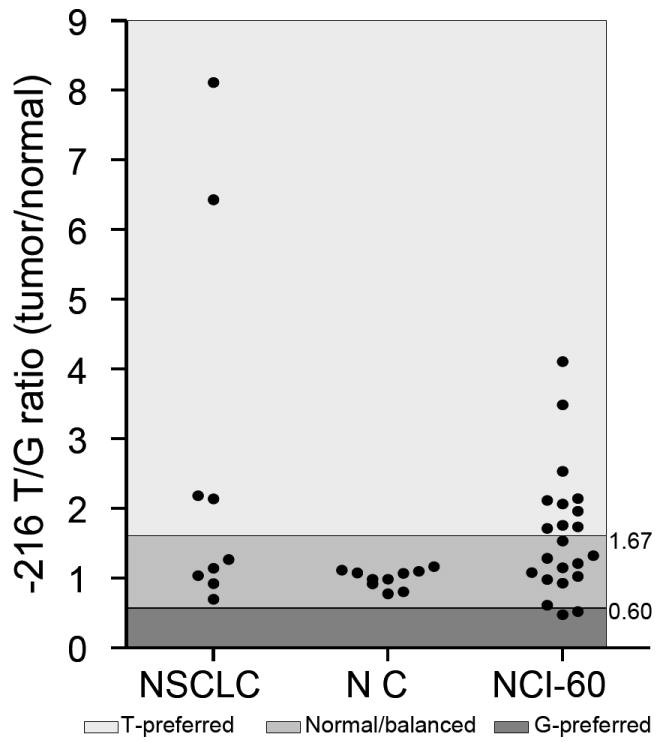
AI of the EGFR -216G/T polymorphism in NSCLC cancer patients and the NCI-60 cancer cell lines. The relative T/G ratio in each sample and the threshold of AI are shown. The imbalance of each allele was indicated with different color shades. NSCLC=non-small cell lung cancer; NC= normal control; NCI-60=NCI 60 cancer cell line panel.
Discussion
The ethnic differences in the incidence of EGFR mutations in NSCLC remain incompletely understood. In this study, we chose a population of Taiwanese patients to perform a “case-control” based association study aiming at elucidating the relationship between functional EGFR polymorphisms and somatic mutations. The Taiwanese population consisted of >98% Han Chinese (10), and is genetically close to other major ethnicities in East Asia (11). We confirmed this by showing that the allele frequencies of all three tested polymorphisms are similar to previous reports in Asian populations (see Table S1 in supplemental materials). We also found a high incidence rate of EGFR mutations (31.2%) in our NSCLC patients, consistent with data reported for other East Asian populations (2).
Our results suggest that local functional polymorphisms at the EGFR locus together play a major role in the development of EGFR mutations. Given the significant difference in the allele frequencies of these polymorphisms between Asian and Caucasian populations (6, 7), our data may further explain the ethnic differences in the EGFR mutation rate. Previous studies demonstrated that exon 19 microdeletions and the L858R point mutation have different biological properties, e.g. a differential response to EGFR inhibitors (1). A recent study further suggested that EGFR amplification is specifically associated with exon 19 deletions (12). Our study, however, suggests that these two types of mutations may also have a different genetic basis. This is consistent with the previous observation of an association between exon 19 deletions and the shorter alleles of intron 1 (CA)n polymorphism (13). These lines of evidence collectively demonstrate that EGFR exon 19 deletions have a pathogenic process distinct from other mutations. Our findings have significant clinical implications and may shed light on the pathogenesis of lung cancer as well.
Increasing evidence has strongly suggested that germline polymorphisms can confer susceptibility to the retention of somatic mutations during cancer development. Known examples include the R72P polymorphism in the TP53 gene (14); melanocortin-1 receptor gene polymorphisms and BRAF gene mutations in melanoma (15); JAK2 SNP (rs10974944) and JAK2V617F mutation in myeloproliferative neoplasms (16) as well as a 5’-distal SNP and FGFR3 mutations in urinary bladder cancer (17). Our findings consistently highlight the crucial role of the combined effects of functional germline polymorphisms in the development of EGFR mutations. Given the proto-oncogenic nature of EGFR, it is possible that certain haplotypes of the EGFR gene might have a selective advantage due to increased EGFR activity, and thus are more likely to contribute to neoplasia. Previous studies have demonstrated that the chromosome/haplotype bearing the EGFR mutations, in particular the exon 19 mutations, tends to be selectively amplified in NSCLC tumors (12, 18), suggesting that once certain haplotypes are mutated, they may create an “EGFR addicted” environment and hence tend to be positively selected in the tumorigenesis process. The polymorphisms tested in this study are all functional, and have been associated with higher EGFR activity (8) and increased EGFR expression (4, 6). We further observed a preferential gain of the T allele or loss of the G allele. Since amplification of the EGFR gene is commonly observed in multiple human cancers as we previously reported in NCI60 (6), we infer that our observed AI is most likely due to selective amplification of the -216T-containing allele. Since the NCI60 consists of various cancer types, the AI of -216G/T in these cells may suggest that specific EGFR haplotypes (e.g. the ones carrying these functional alleles) may benefit the cell transformation in general. While specifically in lung cancer, this intrinsic capacity confers higher capability for retaining EGFR mutations (especially exon 19 deletions).
The EGFR intron 1 (CA)n polymorphism has also been associated with EGFR gene expression (7). It was demonstrated that selective amplification of the shorter alleles occurred frequently in tumors harboring EGFR mutations, especially in patients of East Asian ethnicity (19). Another study also found a genetic association between exon 19 mutations and the shorter alleles (<17 repeats in the shorter allele) (13). However, this was not confirmed in our study (data not shown). The intron 1 (CA)n polymorphism has more than 10 alleles (5), and it is challenging to set an appropriate ‘cutoff’ for shorter and longer alleles due to the lack of clear biological rationale, or even evidence of a monotonic relationship with any phenotype. For instance, the study mentioned above used the median repeat length (17 repeats) in shorter alleles as a cutoff (13). In our population, however, the median is 19 repeats. Moreover, our previous studies suggested that -216G/T and the (CA)n polymorphism are in linkage disequilibrium, and the shorter CA alleles tend to co-segregate with the -216T allele (4, 6). This may confound the association between the (CA)n polymorphism and EGFR mutations, and the previous observation (13) between ‘shorter’ CA alleles and EGFR mutations may be due to their linkage with the -216T allele. Following this reasoning, instead of testing for an association between ‘shorter’ alleles and mutations, we tested for an association between each of the CA-repeat alleles (vs all other alleles) and EGFR mutations. As a result, we observed significant association between the allele CA-19 and EGFR mutations. However, no clear biological function has been specifically associated with CA-19 allele thus far. There is also no LD between the -216T and CA-19 alleles (data not shown). Thus, it appears that CA-19 (or an unknown functional polymorphism in LD with CA-19), alone or in combination with the -216T allele, may predispose to the EGFR mutagenesis.
We recognize certain limitations of our study. Since the EGFR gene is large (>188kb) and a large number of SNPs (>600) have been identified in the region, a comprehensive study in a much larger sample set will be necessary to completely elucidate the somatic and germline genetics at the EGFR locus. In addition, the absence of a replication data set does not exclude the possibility that the associations we report are false discovery, or are limited to a subpopulation due to the potential heterogeneity among East Asian populations. Independent validation is warranted, ideally in the context of a prospective study, since germline DNA has been infrequently collected in conjunction with tumor DNA samples, and further emphasizing the potential value of large-scale sample collection of matching blood and tumor samples. Nevertheless, the data collected in this study consistently support the hypothesis that certain haplotypes consisting of cis-acting functional polymorphisms may play a critical role in the accumulation of the EGFR exon 19 deletions during lung cancer development, which provides a strong rationale for further investigation.
Supplementary Material
Acknowledgments
This work was supported by the NIH/NIGMS grant U01GM61393 (M.J.R) and NIH R01CA125541-03 (R.S).
References
- 1.Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S24–31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–46. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 3.Tsao AS, Tang XM, Sabloff B, et al. Clinicopathologic characteristics of the EGFR gene mutation in non-small cell lung cancer. J Thorac Oncol. 2006;1:231–9. doi: 10.1016/s1556-0864(15)31573-2. [DOI] [PubMed] [Google Scholar]
- 4.Liu W, Innocenti F, Wu MH, et al. A functional common polymorphism in a Sp1 recognition site of the epidermal growth factor receptor gene promoter. Cancer Res. 2005;65:46–53. [PubMed] [Google Scholar]
- 5.Liu W, Innocenti F, Chen P, et al. Interethnic difference in the allelic distribution of human epidermal growth factor receptor intron 1 polymorphism. Clin Cancer Res. 2003;9:1009–12. [PubMed] [Google Scholar]
- 6.Liu W, Wu X, Zhang W, et al. EGFR mutations, expression, amplification, polymorphisms and their interrelationship with sensitivity/resistance to EGFR inhibitors in the NCI60 cell lines. Clin Cancer Res. 2007;13:6788–95. doi: 10.1158/1078-0432.CCR-07-0547. [DOI] [PubMed] [Google Scholar]
- 7.Brandt B, Meyer-Staeckling S, Schmidt H, et al. Mechanisms of egfr gene transcription modulation: relationship to cancer risk and therapy response. Clin Cancer Res. 2006;12:7252–60. doi: 10.1158/1078-0432.CCR-06-0626. [DOI] [PubMed] [Google Scholar]
- 8.Moriai T, Kobrin MS, Hope C, et al. A variant epidermal growth factor receptor exhibits altered type α transforming growth factor binding and transmembrane signaling. Proc Natl Acad Sci U S A. 1994;91:10217–10221. doi: 10.1073/pnas.91.21.10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnaswamy S, Kanteti R, Duke-Cohan JS, et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res. 2009;15:5714–23. doi: 10.1158/1078-0432.CCR-09-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spain D. Taiwan: country profile. Int Demogr. 1984;3(3):4–8. [PubMed] [Google Scholar]
- 11.Tian C, Kosoy R, Lee A, et al. Analysis of East Asia genetic substructure using genome-wide SNP arrays. PLoS One. 2008;3:e3862. doi: 10.1371/journal.pone.0003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sholl LM, Yeap BY, Iafrate AJ, et al. Lung adenocarcinoma with EGFR amplification has distinct clinicopathologic and molecular features in never-smokers. Cancer Res. 2009;69:8341–8. doi: 10.1158/0008-5472.CAN-09-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sueoka-Aragane N, Imai K, Komiya K, et al. Exon 19 of EGFR mutation in relation to the CA-repeat polymorphism in intron 1. Cancer Sci. 2008;99:1180–7. doi: 10.1111/j.1349-7006.2008.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marin MC, Jost CA, Brooks LA, et al. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet. 2000;25:47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- 15.Landi MT, Bauer J, Pfeiffer RM, et al. MC1R germline variants confer risk for BRAF-mutant melanoma. Science. 2006;313:521–2. doi: 10.1126/science.1127515. [DOI] [PubMed] [Google Scholar]
- 16.Campbell PJ. Somatic and germline genetics at the JAK2 locus. Nat Genet. 2009;41:385–6. doi: 10.1038/ng0409-385. [DOI] [PubMed] [Google Scholar]
- 17.Kiemeney LA, Sulem P, Besenbacher S, et al. A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat Genet. 2010;42:415–9. doi: 10.1038/ng.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takano T, Ohe Y, Sakamoto H, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:6829–37. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 19.Nomura M, Shigematsu H, Li L, et al. Polymorphisms, mutations, and amplification of the EGFR gene in non-small cell lung cancers. PLoS Med. 2007;4:e125. doi: 10.1371/journal.pmed.0040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


