Abstract
Over the past decade it has emerged that the cohesin protein complex, which functions in sister chromatid cohesion, chromosome segregation and DNA repair, also regulates gene expression and development. Even minor changes in cohesin activity alter several aspects of development. Genome-wide analysis indicates that cohesin directly regulates transcription of genes involved in cell proliferation, pluripotency, and differentiation through multiple mechanisms. These mechanisms are poorly understood, but involve both partial gene repression in concert with Polycomb group proteins, and facilitating long-range looping, both between enhancers and promoters, and between CTCF protein binding sites.
Introduction
Structural Maintenance of Chromosome (SMC) protein complexes play essential roles in chromosome mechanics, including chromosome segregation, condensation, and DNA repair, in both prokaryotes and eukaryotes [1]. Eukaryotes have three SMC complexes: condensin, Smc5/6 and cohesin. Cohesin mediates sister chromatid cohesion to ensure proper chromosome segregation in mitosis and meiosis, and condensin is required for chromosome condensation. Both Smc5/Smc6 and cohesin play roles in DNA repair. Cohesin and specialized condensin complexes also regulate gene expression, with consequences for development [2]. Here we review the roles of cohesin in gene regulation and development in higher organisms, and how recent genome-wide analyses have shed light on possible mechanisms.
Small cohesin deficits alter gene expression and development
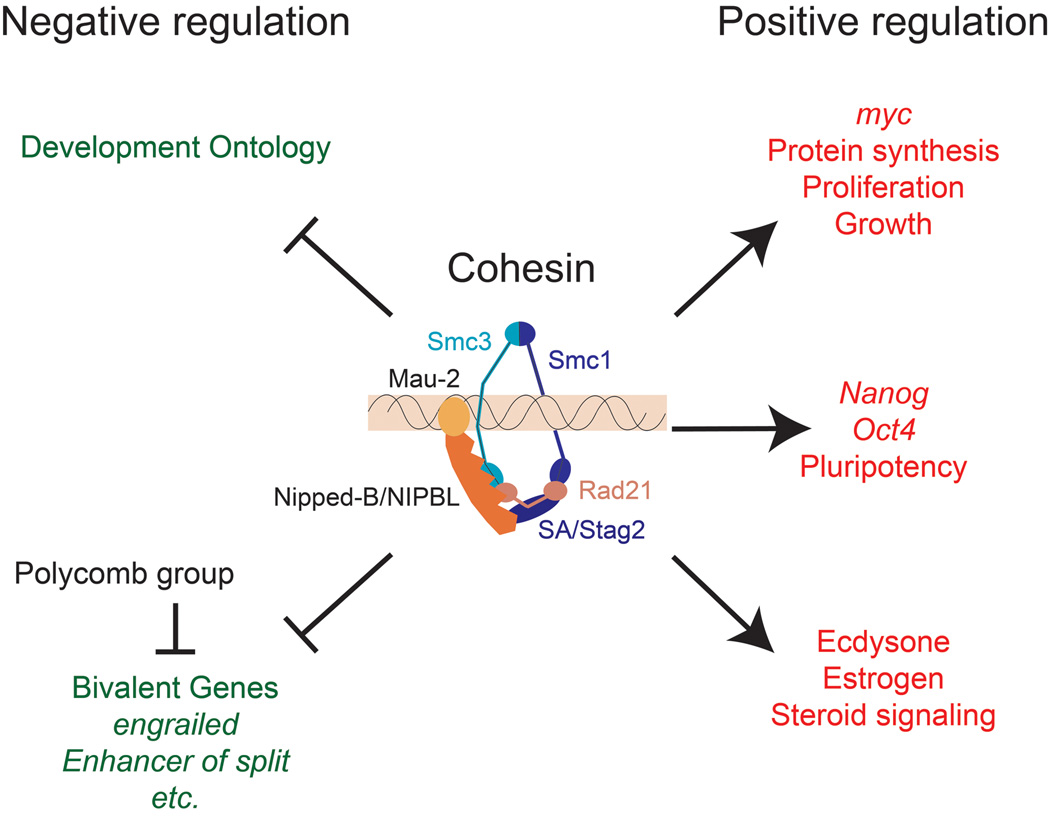
The cohesin complex consists of the Smc1, Smc3, Rad21 and Stromalin/Stag1/2 proteins, which form a ring-like structure (Figure 1) [3]. The Nipped-B – Mau-2 complex and ATPase activities of the Smc1 and Smc3 head domains are required for cohesin to bind to chromosomes [3]. There is evidence that cohesion encircles DNA topologically, but uncertainty about how cohesin actually holds sister chromatids together [3,4].
Figure 1.
Cohesin regulates expression of genes that control differentiation, morphogenesis, proliferation, and pluripotency. The cohesin complex consists of the Smc1, Smc3, Rad21 and Stromalin (SA)/Stag2 subunits [3]. Cohesin is thought to bind by encircling DNA, and binding requires the Nipped-B/NIPBL – Mau-2 complex, and ATPase activity in the Smc1 and Smc3 head domains that interact with Rad21. Both the genes that increase or decrease in expression with reduced Nipped-B/NIPBL or cohesin activity are enriched for cohesin-binding genes, indicating that cohesin both represses and facilitates transcription. Developmental processes are the top gene ontology categories for genes that increase in expression when cohesin or Nipped-B/NIPBL activity is reduced in Drosophila cells derived from central nervous system [15*], and mouse embryonic stem cells [28*]. The majority of the genes that show the largest increases in expression are bivalent, with both histone H3 lysine 4 and lysine 27 trimethylation, indicating that they are also partially repressed by Polycomb group silencing proteins. Cohesin directly promotes expression of genes in the ecdysone steroid hormone signaling pathway in salivary glands and the mushroom body γ neuron [13,14,24*] and the estrogen pathway in human breast cancer cells [27*]. Nipped-B/NIPBL and cohesin also directly facilitate expression of myc genes, which promote protein synthesis and cell proliferation in all species examined [12*,15*,26*,28*,30,31,34*], and pluripotency genes in mouse embryonic stem cells [28*].
The first observations linking cohesin to gene regulation during development were the recovery of Nipped-B mutations in a genetic screen for factors that facilitate activation of the Drosophila cut and Ultrabithorax homeobox genes by distant transcriptional enhancers [5], and identification of the human ortholog, Nipped-B-Like (NIPBL), as the gene mutated in many cases of Cornelia de Lange syndrome (CdLS) [6,7]. CdLS displays diverse structural and intellectual deficits, including slow growth, upper limb and organ dysmorphologies, distinctive facial features, mental retardation and autism spectrum disorders [8].
CdLS is caused by heterozygous loss-of-function NIPBL mutations that reduce expression at most by 30%. As little as a 15% decrease in NIPBL expression is sufficient to cause CdLS [9]. Heterozygous Drosophila Nipped-B null mutations reduce Nipped-B mRNA levels by less than 30%, and decrease expression of mutant and wild-type cut alleles in the developing wing without causing cohesion or chromosome segregation defects in mitosis or meiosis [10,11]. Heterozygous Nipbl mutant mice show a 30% decrease in Nipbl mRNA and display several developmental defects overlapping those seen in CdLS, and many changes in gene expression [12*]. Some 75% of Nipbl(+/−) mice perish perinatally, but there are no effects on cohesion or chromosome segregation.
The lack of overt defects in chromatid cohesion or chromosome segregation with reduction of Nipped-B/NIPBL activity in multiple organisms argues that the developmental deficits derive primarily from altered gene expression. More evidence favoring this idea came from experiments in which cohesin function was severely disrupted in the non-dividing mushroom body γ neuron in Drosophila, which blocked axon pruning by reducing expression of the ecdysone receptor gene (EcR) [13,14].
It is unlikely that Nipped-B/NIPBL has developmental functions separate from cohesin. Genome-wide analysis reveals that knockdown of Nipped-B and cohesin in Drosophila cells alters expression of the same several hundred genes [15*]. Moreover, 5% of CdLS cases are caused by mutations affecting the SMC1A cohesin subunit, and one by an SMC3 mutation [16,17]. These cases are milder than those caused by NIPBL mutations, showing primarily intellectual deficits and the characteristic facial features. All the SMC1A mutations and the SMC3 mutation maintain the open reading frame, and cause amino acid substitutions and/or small deletions in the protein. SMC1A is X-linked, and both hemizygous male and heterozygous female individuals have been identified, indicating that the mutant proteins are functional and dominantly interfere with development. Like heterozygous NIPBL mutations [7,18], the SMC1A mutations do not affect chromatid cohesion or chromosome segregation [19].
What form of cohesin is important for gene regulation?
The finding that mild disruption of Nipped-B/NIPBL and cohesin activity alters gene expression and development raises the question of how these disruptions affect cohesin binding to chromosomes. One possibility is that only low cohesion binding is needed for cohesion and segregation, and higher levels are needed for gene regulation. Consistent with this idea, knockdown of Nipped-B or cohesin by 80% in Drosophila cells causes no significant cohesion or segregation defects but alters expression of some genes several-fold [15*], and systematic reduction of cohesin in yeast reveals that 13% of normal levels is sufficient for cohesion and chromosome segregation, although chromosome condensation and DNA repair are affected [20].
In vivo fluorescence recovery after photobleaching (FRAP) experiments in Drosophila salivary glands reveals that cohesin binds chromosomes in two modes – a weak binding mode with a chromosomal half-life of some 20 sec, and a stable mode with a half-life of 6 min or so [21]. A heterozygous null Nipped-B mutation reduces the fraction of stable-binding cohesin by a third, suggesting that the stable binding mode is critical for gene regulation. FRAP experiments, however, do not address whether or not stable binding, which is postulated to be topological, is affected more at some genes than others.
Drosophila Nipped-B and the yeast Scc2 ortholog both show nearly identical chromosome-binding dynamics as cohesin, suggesting that a significant fraction interacts tightly with cohesin on chromosomes, although Nipped-B’s stoichiometry to cohesin varies dramatically between tissues [21,22]. More precise knowledge of how cohesin binding is affected by reduced Nipped-B/NIPBL levels and SMC1A missense mutations is needed to fully understand how cohesin regulates transcription.
Cohesin binds active genes and CTCF sites, and avoids Polycomb-silenced regions
Cohesin binding has been mapped genome-wide by ChIP-chip, DamID, or ChIPseq in Drosophila cell lines [23*], Drosophila salivary gland [24*], HeLa cells [25*], human lymphocytes [26*], MCF-7 and HepG2 tumor cell lines [27*] and mouse embryonic stem cells [28*], and in part of the genome of mouse pre-B cells and thymocytes [29*]. In Drosophila, Nipped-B and cohesin completely co-localize and bind preferentially to active genes, with peaks near the transcription start sites [23*]. Cohesin prefers active genes and peaks near transcription start sites in mammalian cells, but also co-localizes with a large fraction of the CCCTC-binding factor (CTCF) binding sites [25*–29*,30,31]. CTCF has multiple functions in gene regulation, but its insulator role in blocking enhancer-promoter interactions is the best characterized [32].
Remarkably, Nipbl co-localizes with cohesin at transcription start sites and transcriptional enhancers in mouse embryonic stem cells, but not at CTCF sites [28*]. Given that Nipbl is required to load cohesin onto chromosomes, and barring potential issues such as weak crosslinking or epitope masking of Nipbl at CTCF sites, this implies that cohesin gets to CTCF sites either by sliding from Nipbl sites, or that cohesin binds differently at CTCF sites. CTCF knockdown does not cause cohesion defects or reduce overall cohesin binding, indicating that CTCF sites are not crucial for cohesion [25*,29*]. In Drosophila, cohesion doesn’t co-localize with CTCF, but cohesin (and Nipped-B) co-localize with a fraction of sites binding the CP190 co-insulator protein [33]. Thus a relationship between cohesin and insulators may be evolutionarily conserved, even if localization with CTCF is not.
In Drosophila, cohesin and Nipped-B are excluded from regions marked by the histone H3 lysine 27 trimethylation modification made by the Enhancer of zeste [E(z)] Polycomb group (PcG) silencing protein [23*]. Most PcG-targeted genes are silenced, and genes that are silenced in one cell line can bind Nipped-B and cohesin in other cells in which they are transcribed. As described below, there are important rare exceptions in which genes bind cohesin and PcG proteins simultaneously, and are hypersensitive to Nipped-B and cohesin levels [15*].
Cohesin directly controls expression of many genes
Hundreds of genes are altered in expression in lymphocytes derived from CdLS individuals with NIPBL or SMC1A mutations [26*], in Nipbl(+/−) mouse tissues [12*], in Drosophila cells [15*] and mouse embryonic stem cells [28*] when Nipped-B/NIPBL or cohesin are knocked down by RNAi, and in Drosophila salivary glands when cohesin is removed from chromosomes [24*]. Roughly equal numbers of genes increase and decrease in expression, and most effects are less than 2-fold. In Drosophila cells and salivary glands, however, large effects on the order of 100-fold also occur.
Comparison of genome-wide binding of cohesin to the genome-wide effects of reducing Nipped-B/NIPBL or cohesin activity on transcript levels reveals that the genes that change in expression with altered cohesin activity are highly enriched for cohesin-binding genes [15*,24*,26*,28*]. In Drosophila cells and mouse embryonic stem cells, there is a strong correlation between the effects of Nipped-B/Nipbl and cohesin knockdown. This is compelling evidence that Nipped-B/NIPBL regulates genes through control of cohesin chromosome binding, and that cohesin directly regulates gene transcription. Potential indirect effects, however, make it difficult to be sure how much of the effect on any individual gene is direct.
Recent experiments provide strong evidence for direct regulation of specific genes by cohesin in Drosophila salivary glands [24*]. By replacing the native Rad21 subunit with a form that can be cleaved by TEV protease to remove cohesin from chromosomes, and using a temperature-sensitive system to induce TEV protease, decreases in expression of some genes in the ecdysone steroid signaling pathway were detected within 4 hours, and in ecdysone-induced chromosome puffing within 2 hrs. By immunostaining, the EcR activator remains bound to the Eip74EF gene while it decreases 10-fold in expression, although EcR expression is also reduced.
Cohesin regulates genes controlling development, proliferation and pluripotency
Cohesin preferentially regulates genes important for development and cell proliferation (Figure 1). The top ontology categories for genes that increase in expression with cohesin or Nipped-B/Nipbl knockdown involve development in Drosophila and mouse embryonic stem cells [15*,28*]. The top categories for genes that increase in expression with proteolysis of Rad21 in Drosophila salivary glands are metabolic processes, followed by development, but in these experiments, cohesin removal was nearly complete, triggering acute changes in cellular physiology which likely affect metabolism [24*].
As described above, cohesin facilitates expression of genes in the ecdysone steroid hormone signaling pathway in the salivary gland and the mushroom body γ neuron (Figure 1) [13,14,24*]. Ecdysone is a key regulator of morphogenesis and molting in Drosophila, raising the possibility that cohesin might also play a vital role in these developmental processes. Cohesin also facilitates steroid hormone signaling in human MCF-7 breast cancer cells, where it co-localizes with the estrogen receptor on target genes (Figure 1) [27*]. Cohesin knockdown decreases re-entry of these cells into the cell cycle in response to estrogen treatment, indicating that cohesin regulates estrogen function. The findings that cohesin influences both ecdysone and estrogen signaling raises the question if it also regulates other steroid hormone pathways.
A remarkably consistent finding is that cohesin binds and facilitates expression of the myc gene in all species examined, including Drosophila, zebrafish, mouse, and human (Figure 1) [12*,15*,26*,28*,30,31,34*]. Myc is a key regulator of protein synthesis and cell proliferation, and this may explain why protein translation is the top ontology category for genes that decrease in expression in Drosophila cells when Nipped-B or cohesin is knocked down [15*]. Many genes directly activated by Myc that don’t bind cohesin also decrease in expression with Nipped-B or cohesin knockdown in Drosophila cells, and the list of affected genes is nearly identical to that determined for myc mutant larvae [34*,35].
In addition to myc, Nipbl and cohesin also promote expression of genes required for pluripotency in mouse embryonic stem cells (Figure 1) [28*]. Nipbl, cohesin, and the Mediator transcriptional coactivator complex were identified in an RNAi screen for factors required for stem cell maintenance, and found to directly facilitate expression of the Oct4 and Nanog pluripotency genes. Upregulation of myc and pluripotency genes by cohesin, combined with downregulation of several differentiation genes makes it tempting to speculate that cohesion provides input into the decision between proliferation versus differentiation.
Cohesin facilitates enhancer-promoter and CTCF-mediated DNA looping
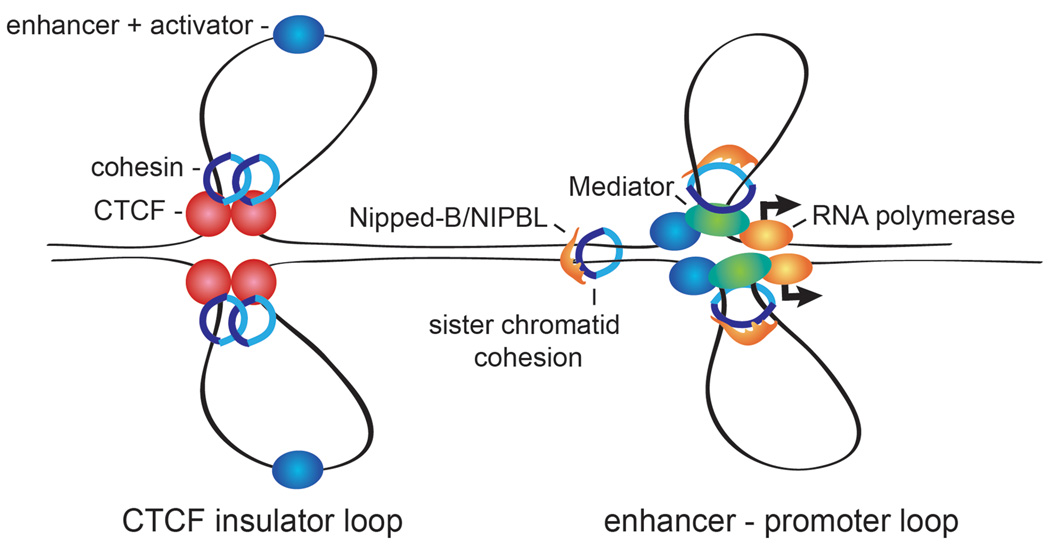
The mechanisms by which Nipped-B/NIPBL and cohesin regulate gene expression are not well understood, but there is growing evidence that cohesin facilitates long distance DNA looping over several kilobases (Figure 2). This idea was put forth to explain genetic effects of Nipped-B mutations on Drosophila cut expression in the developing wing margin [5], but ironically, new evidence described below suggests that partial repression by cohesin might also be involved. Direct evidence for a role for cohesin in looping comes from the finding that cohesin knockdown reduces long-range interactions between CTCF sites in the IFNG, apolipoprotein, Igf2-H19 and β globin genes measured by chromosome conformation capture (3C) (Figure 2) [36*–39*]. In most cases, the reduced long-range interactions are accompanied by modest changes in gene expression. The effects on looping range from modest reduction to complete loss. Given the experimental variables, which include potential effects of reduced protein binding on formaldehyde crosslinking in 3C experiments, and incomplete cohesin knockdown, these results must be interpreted cautiously, but they suggest that cohesin performs a secondary stabilization role in some cases, but is essential for looping in others.
Figure 2.
Cohesin facilitates DNA looping. The diagram shows two sister chromatids. On the left is a hypothetical model for how cohesin could support intrachromosomal looping between two CTCF binding sites, which occurs at several loci in mammalian cells [36*–39*]. In this example, the loop functions as an insulator, and sequesters a transcriptional enhancer, preventing it from activating flanking genes. The Nipped-B/NIPBL cohesin-loading factor is not present at CTCF binding sites [28*], suggesting that cohesin binds differently than at sites of sister chromatid cohesion. The right shows a model for how cohesin could stabilize a loop between an enhancer and promoter, facilitating transcriptional activation, such as occurs at the Nanog gene in mouse embryonic stem cells. The Mediator coactivator complex and Nipped-B/NIPBL are present [28*], and cohesin might function intrachromosomally in a manner similar to the way it mediates cohesion between sister chromatids.
Cohesin binding predict enhancer-promoter loops in several active genes in mouse embryonic stems cells (Figure 2) [28*]. In these cases there are overlapping peaks of the Mediator coactivator, Nipbl and cohesin on both the enhancer and promoter, and an interaction between them is detected by 3C. The cohesin/Mediator peaks and loops are absent in embryonic fibroblasts in which these genes are inactive. Cohesin knockdown reduces the enhancer-promoter interaction in the Nanog gene in stem cells, correlating with a substantial decrease in Nanog expression.
The mechanisms by which cohesin facilitates the long-range interactions between CTCF sites and between enhancers and promoters is unknown, but it has been speculated that it might mediate “intrachromosomal cohesion” to stabilize loops (Figure 2) [40]. The finding that Nipbl is not present at the cohesin/CTCF peaks in mouse embryonic stem cells [28*], however, suggests that the mechanism may be different for CTCF and enhancer-promoter loops.
Cohesin participates in partial repression of some Polycomb targeted genes
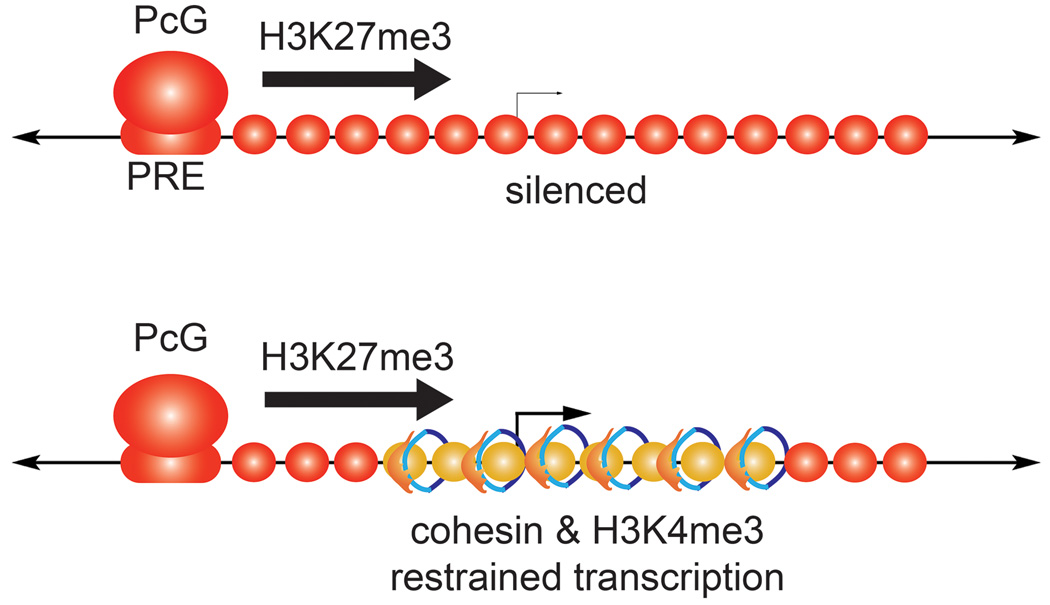
In a Drosophila cell line derived from central nervous system, genes that increase in expression with Nipped-B or cohesin knockdown are more enriched for cohesin-binding than the genes that decrease [15*]. Thus, cohesin has more direct repressive than activating effects. While most effects are modest, some genes increase dramatically in expression with cohesin knockdown, up to a hundred-fold. Most strongly affected genes, which include the Enhancer of split [E(spl)-C] and invected-engrailed complexes, are rare exceptions where a cohesin-binding domain overlaps a region targeted by Polycomb group (PcG) silencing proteins (Figure 3). In contrast to genes that are targeted only by PcG proteins, the co-targeted genes are expressed at modest to moderate levels. Knockdown of Polycomb also increases E(spl)-C expression, indicating that PcG proteins and cohesin are both needed to restrain transcription. It is unlikely that cohesin and PcG proteins target the genes in different cells in the population because genes that are targeted only by PcG proteins are unaffected by cohesin knockdown, and the E(spl)-C is targeted only by cohesin in another cell line and does not respond to cohesin knockdown. It is unknown if this partial repression affects activator binding, or another step in transcription.
Figure 3.
Cohesin acts in concert with Polycomb group (PcG) silencing proteins to restrain gene expression. In Drosophila cells, most PcG targeted genes are fully silenced and marked by histone H3 lysine 27 trimethylation (H3K27me3). They do not bind Nipped-B/NIPBL or cohesin [23*]. In rare cases, PcG targeted genes also bind cohesin, and all are expressed at low to moderate levels [15*]. All such genes encode transcription factors that control development. Nearly all genes targeted by both cohesin and PcG proteins are bivalent, having both the H3K27me3 mark, and the H3K4me3 modification associated with active genes. Reduction of either cohesin or Polycomb proteins strongly increases transcription of genes targeted simultaneously by cohesin and PcG proteins. The majority of genes that increase the most in expression with cohesin knockdown in mouse embryonic cells are also bivalent [28*,44], indicating that cohesin also contributes to repression of many PcG-targeted genes in mammalian cells.
Some cohesin-PcG genes co-targeted genes show a biphasic response to cohesin levels - when cohesin is reduced by only 30%, E(spl)-C expression decreases, but when cohesin is reduced by 50 to 80% expression increases [15*]. This led to the hypothesis that cohesin-PcG co-targeted domains have a unique structure that depends on a balance between cohesin and PcG proteins. With a slight cohesin reduction, PcG silencing activity may become stronger, decreasing transcription, but when cohesin is strongly reduced, the structure is lost, leading to unrestrained transcription.
Remarkably, genetic effects reminiscent of this biphasic effect are seen in vivo, with partial Nipped-B and cohesin reduction having opposite effects on expression of cut in the wing margin, and on an E(spl)-C dependent mutant eye phenotype [10,15*,41]. Even more remarkably, cut is active and bound by cohesin over a domain extending from the wing margin enhancer to the end of the transcribed region in one Drosophila cell line, but in contrast to the developing wing margin, Nipped-B or cohesin knockdown has no effect on cut transcription in these cells [15*]. Thus cohesin binding alone is insufficient for cut to be sensitive to cohesin dosage.
Genetic experiments suggest that PcG proteins regulate but do not silence cut in the developing wing margin [42], and PcG proteins fully silence cut in some Drosophila cell lines [23*]. Combined with the opposite in vivo effects of Nipped-B and cohesin mutations these findings raise the possibility that cohesion act in concert with PcG proteins to restrain, but not silence cut expression in the wing margin, which does not exclude the possibility that cohesin also facilitates long-range activation by the wing margin enhancer.
Nearly all genes co-targeted by cohesin and PcG proteins in Drosophila cells are bivalent, having both histone H3 lysine 4 trimethylation (H3K4me3) characteristic of active genes, and the histone H3 lysine 27 trimethylation (H3K27me3) modification made by the E(z) PcG protein [15*]. Bivalent genes are common in embryonic stem cells, and like the Drosophila E(spl)-C and engrailed genes, many encode transcription factors, and are expressed at low levels [43]. Current thought is that they represent an uncommitted multipotent state, but it is interesting to speculate that the bivalent state can also ensure that a gene is expressed at an appropriate level that is not too low or too high. Notably, more than half of the 200 genes that increase the most in expression with cohesion knockdown in mouse embryonic stem cells, many of which bind cohesin [28*], are bivalent [44]. Thus partial repression of select PcG-targeted genes may also be an important mechanism for gene regulation by cohesin in embryonic stem cells, although it remains possible some of these effects reflect reduced pluripotency.
Key Questions
The evidence summarized above raises many intriguing questions: What are the molecular mechanisms by which cohesin contributes to repression of selected PcG-targeted genes, and how is it determined that a gene is targeted only by PcG proteins in one cell type, and by both cohesin and PcG proteins in another? What are the molecular mechanisms by which cohesin facilitates long-range looping, and do they differ for enhancer-promoter loops and loops between CTCF sites? Is stable topologically bound cohesin important for looping, and does it support looping by mechanisms similar to those that mediate sister chromatid cohesion? How much input does cohesin have in the decision between differentiation and proliferation, and does it represent a critical control point for pluripotency? How do effects on gene expression result in the diverse developmental deficits in Cornelia de Lange syndrome? Are large changes in gene expression at critical junctures responsible, or do most developmental changes reflect synergism between multiple small changes? Answers to these questions will vastly improve our understanding of how modest changes in cohesin function alter organism development.
Acknowledgements
Work on the role of cohesin in gene expression and development in the author’s laboratory is supported by grants from the NIH (R01 GM055683, P01 HD052860). The author thanks former and current members of his laboratory, including Robert Rollins, Patrick Morcillo, Ziva Misulovin, Maria Gause, Cheri Schaaf, and Avery Fay, and many colleagues, including Ian Krantz, Arthur Lander, Anne Calof, Antonio Musio, Julia Horsfield, Tom Strachan, Matthias Merkenschlager, Jennifer Gerton, Andrea Pauli, Kim Nasmyth, Tatsuya Hirano, Vinnie Guacci, Kirsten Wendt and Jan-Michael Peters for enjoyable discussions on these topics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 2.Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 4.Skibbens RV. Buck the establishment: reinventing sister chromatid cohesion. Trends Cell Biol. 2010;20:507–513. doi: 10.1016/j.tcb.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 8.Dorsett D, Krantz ID. On the molecular etiology of Cornelia de Lange syndrome. Ann N Y Acad Sci. 2009;1151:22–37. doi: 10.1111/j.1749-6632.2008.03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borck G, Zarhrate M, Cluzeau C, Bal E, Bonnefont JP, Munnich A, Cormier-Daire V, Colleaux L. Father-to-daughter transmission of Cornelia de Lange syndrome caused by a mutation in the 5' untranslated region of the NIPBL gene. Hum Mutat. 2006;27:731–735. doi: 10.1002/humu.20380. [DOI] [PubMed] [Google Scholar]
- 10.Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila Nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gause M, Webber HA, Misulovin Z, Haller G, Rollins RA, Eissenberg JC, Bickel SE, Dorsett D. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma. 2008;117:51–66. doi: 10.1007/s00412-007-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *12. Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/−) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650.. This study shows that mice with a heterozygous Nipbl mutation have multiple developmental deficits overlapping those seen in CdLS, and 75% perinatal lethality. It also shows that hundreds of genes are dysregulated, and that low body fat can be correlated with decreased expression of key differentiation genes.
- 13.Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *15. Schaaf CA, Misulovin Z, Sahota G, Siddiqui AM, Schwartz YB, Kahn TG, Pirrotta V, Gause M, Dorsett D. Regulation of the Drosophila Enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS One. 2009;4:e6202. doi: 10.1371/journal.pone.0006202.. This study finds that Nipped-B and cohesin regulate hundreds of genes important for development and protein translation, and that genes regulated by cohesin are highly enriched for cohesin binding genes. It also shows that the rare cases where genes are targeted simultaneously by both cohesin and PcG silencing proteins are hypersensitive to cohesin dosage, and that cohesin helps to restrain their expression.
- 16.Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- 17.Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodríguez C, Arnedo M, Loeys B, Kline AD, et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vrouwe MG, Elghalbzouri-Maghrani E, Meijers M, Schouten P, Godthelp BC, Bhuiyan ZA, Redeker EJ, Mannens MM, Mullenders LH, Pastink A, Darroudi F. Increased DNA damage sensitivity of Cornelia de Lange syndrome cells: evidence for impaired recombinational repair. Hum Mol Genet. 2007;16:1478–1487. doi: 10.1093/hmg/ddm098. [DOI] [PubMed] [Google Scholar]
- 19.Revenkova E, Focarelli ML, Susani L, Paulis M, Bassi MT, Mannini L, Frattini A, Delia D, Krantz I, Vezzoni P, et al. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum Mol Genet. 2009;18:418–427. doi: 10.1093/hmg/ddn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol. 2010;20:957–963. doi: 10.1016/j.cub.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gause M, Misulovin Z, Bilyeu A, Dorsett D. Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5 and Wapl. Mol Cell Biol. 2010 doi: 10.1128/MCB.00642-10. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNairn AJ, Gerton JL. Intersection of ChIP and FLIP, genomic methods to study the dynamics of the cohesin proteins. Chromosome Res. 2009;17:155–163. doi: 10.1007/s10577-008-9007-9. [DOI] [PubMed] [Google Scholar]
- *23. Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1.. This study mapped Nipped-B and cohesin genome-wide in multiple Drosophila cell lines, showing that they co-localize, bind preferentially to active genes with peaks near the transcription start sites, and are usually excluded from genes silenced by Polycomb group proteins.
- *24. Pauli A, van Bemmel JG, Oliveira RA, Itoh T, Shirahige K, van Steensel B, Nasmyth K. A direct role for cohesin in gene regulation and Ecdysone-response in Drosophila salivary glands. Curr Biol. 2010 doi: 10.1016/j.cub.2010.09.006. in press.. This paper shows that cohesin directly regulates expression of key genes in the ecdysone steroid hormone signaling pathway in Drosophila, including the ecdysone receptor gene. Together with Schmidt et al. [27*] it shows that cohesin is a critical regulator of steroid function.
- *25. Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634.. This paper maps cohesin genome-wide in human cells, and together with Parelho et al. [29*], Stedman et al. [30], Rubio et al. [31], shows that cohesin functionally associates with many CTCF binding sites.
- *26. Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119. doi: 10.1371/journal.pbio.1000119.. This study shows that cohesin binds near the transcription start of many active genes, and that hundreds of genes dysregulated in cells from Cornelia de Lange individuals are enriched for genes that bind cohesin.
- *27. Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109.. This paper shows that cohesin co-localizes extensively in a cell-type specific manner with the DNA-bound estrogen receptor, and facilitates the cell’s response to estrogen.
- *28. Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010 doi: 10.1038/nature09380. [Epub ahead of print]. This study shows that Nipbl and cohesin are required for mouse stem cell maintenance through direct regulation of the Oct4 and Nanog pluripotency genes. It also shows that Nipbl and cohesin colocalize at enhancer and promoters of many active genes, and that cohesin facilitates looping between the Nanog enhancer and promoter.
- *29. Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011.. Together with Wendt et al. [25*], Stedman et al. [30] and Rubio et al. [31], this paper shows that cohesin functionally associates with CTCF binding sites.
- 30.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohlsson R, Bartkuhn M, Renkawitz R. CTCF shapes chromatin by multiple mechanisms: the impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma. 2010;119:351–360. doi: 10.1007/s00412-010-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *34. Rhodes JM, Bentley FK, Print CG, Dorsett D, Misulovin Z, Dickinson EJ, Crosier KE, Crosier PS, Horsfield JA. Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev Biol. 2010;344:637–649. doi: 10.1016/j.ydbio.2010.05.493.. This study demonstrates that cohesin regulates the Myc gene in zebrafish, and analyzes genome-wide data from other organisms to show evolutionary conservation of this function.
- 35.Pierce SB, Yost C, Anderson SA, Flynn EM, Delrow J, Eisenman RN. Drosophila growth and development in the absence of dMyc and dMnt. Dev Biol. 2008;315:303–316. doi: 10.1016/j.ydbio.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *36. Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079.. Together with references 37*–39*, this study provides evidence that cohesin functionally contributes to DNA looping between CTCF binding sites.
- *37. Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81.. Together with references 36*, 38* and 39*, this study shows that cohesin functionally contributes to DNA looping between CTCF binding sites.
- *38. Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739.. Together with references 36*, 37* and 39*, this study provides evidence that cohesin stabilizes DNA looping between CTCF sites.
- *39. Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107.. Together with references 36*–38*, this study provides evidence that cohesin functionally contributes to DNA looping between CTCF binding sites.
- 40.Gause M, Schaaf CA, Dorsett D. Cohesin and CTCF: cooperating to control chromosome conformation? Bioessays. 2008;30:715–718. doi: 10.1002/bies.20787. [DOI] [PubMed] [Google Scholar]
- 41.Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132:4743–4753. doi: 10.1242/dev.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melnikova L, Kulikov A, Georgiev P. Interactions between cut wing mutations and mutations in zeste, and the enhancer of yellow and Polycomb group genes of Drosophila melanogaster. Mol Gen Genet. 1996;252:230–236. doi: 10.1007/BF02173768. [DOI] [PubMed] [Google Scholar]
- 43.Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 44.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]