Abstract
During a study of the gene coding for alpha-galactosidase (EC 3.2.1.22), the lysosomal enzyme deficient in Fabry's disease, RT-PCR amplification of alpha-galactosidase mRNAs obtained from three different tissues isolated from males revealed a substantial number of clones with a U to A conversion at the nucleotide position 1187. Such a modification of the coding sequence would result in an amino acid substitution in the C-terminal region (Phe396Tyr) of the enzyme. Neither PCR analysis of the genomic sequence nor the RT-PCR amplification of RNA obtained by in vitro transcription of the wild-type cDNA showed this change in the sequence. Multiple genes, pseudogenes are allelic variants were excluded. Hence, we propose RNA editing as a mechanism responsible for this base change in the alpha-galactosidase RNA.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backus J. W., Smith H. C. Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res. 1992 Nov 25;20(22):6007–6014. doi: 10.1093/nar/20.22.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier H., Lee M. C., Sekiya T., Kuchino Y., Nishimura S. Two nucleotides next to the anticodon of cytoplasmic rat tRNA(Asp) are likely generated by RNA editing. Nucleic Acids Res. 1992 Jun 11;20(11):2679–2683. doi: 10.1093/nar/20.11.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R., Van den Burg J., Brakenhoff J. P., Sloof P., Van Boom J. H., Tromp M. C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986 Sep 12;46(6):819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Beutler E., Gelbart T. Gaucher disease associated with a unique KpnI restriction site: identification of the amino-acid substitution. Ann Hum Genet. 1990 May;54(Pt 2):149–153. doi: 10.1111/j.1469-1809.1990.tb00371.x. [DOI] [PubMed] [Google Scholar]
- Bishop D. F., Calhoun D. H., Bernstein H. S., Hantzopoulos P., Quinn M., Desnick R. J. Human alpha-galactosidase A: nucleotide sequence of a cDNA clone encoding the mature enzyme. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4859–4863. doi: 10.1073/pnas.83.13.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boström K., Garcia Z., Poksay K. S., Johnson D. F., Lusis A. J., Innerarity T. L. Apolipoprotein B mRNA editing. Direct determination of the edited base and occurrence in non-apolipoprotein B-producing cell lines. J Biol Chem. 1990 Dec 25;265(36):22446–22452. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll D. M., Lakhe-Reddy S., Oleksa L. M., Martinez D. Induction of RNA editing at heterologous sites by sequences in apolipoprotein B mRNA. Mol Cell Biol. 1993 Dec;13(12):7288–7294. doi: 10.1128/mcb.13.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egebjerg J., Kukekov V., Heinemann S. F. Intron sequence directs RNA editing of the glutamate receptor subunit GluR2 coding sequence. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10270–10274. doi: 10.1073/pnas.91.22.10270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C. M., Desnick R. J. Molecular basis of Fabry disease: mutations and polymorphisms in the human alpha-galactosidase A gene. Hum Mutat. 1994;3(2):103–111. doi: 10.1002/humu.1380030204. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R., Devereux J. PEPPLOT, a protein secondary structure analysis program for the UWGCG sequence analysis software package. Nucleic Acids Res. 1986 Jan 10;14(1):327–334. doi: 10.1093/nar/14.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., Single F. N., Köhler M., Sommer B., Sprengel R., Seeburg P. H. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993 Dec 31;75(7):1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- Hodges P., Scott J. Apolipoprotein B mRNA editing: a new tier for the control of gene expression. Trends Biochem Sci. 1992 Feb;17(2):77–81. doi: 10.1016/0968-0004(92)90506-5. [DOI] [PubMed] [Google Scholar]
- Jameson B. A., Wolf H. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput Appl Biosci. 1988 Mar;4(1):181–186. doi: 10.1093/bioinformatics/4.1.181. [DOI] [PubMed] [Google Scholar]
- Janke A., Päbo S. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res. 1993 Apr 11;21(7):1523–1525. doi: 10.1093/nar/21.7.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T., Ishimaru S., Higashijima S., Takayama E., Akimaru H., Sone M., Emori Y., Saigo K. Identification of a different-type homeobox gene, BarH1, possibly causing Bar (B) and Om(1D) mutations in Drosophila. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4343–4347. doi: 10.1073/pnas.88.10.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornreich R., Desnick R. J., Bishop D. F. Nucleotide sequence of the human alpha-galactosidase A gene. Nucleic Acids Res. 1989 Apr 25;17(8):3301–3302. doi: 10.1093/nar/17.8.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan K. M., Gray M. W. Editing of transfer RNAs in Acanthamoeba castellanii mitochondria. Science. 1993 Feb 5;259(5096):812–816. doi: 10.1126/science.8430334. [DOI] [PubMed] [Google Scholar]
- Nutt S. L., Hoo K. H., Rampersad V., Deverill R. M., Elliott C. E., Fletcher E. J., Adams S. L., Korczak B., Foldes R. L., Kamboj R. K. Molecular characterization of the human EAA5 (GluR7) receptor: a high-affinity kainate receptor with novel potential RNA editing sites. Receptors Channels. 1994;2(4):315–326. [PubMed] [Google Scholar]
- Nutt S. L., Kamboj R. K. RNA editing of human kainate receptor subunits. Neuroreport. 1994 Dec 20;5(18):2625–2629. doi: 10.1097/00001756-199412000-00055. [DOI] [PubMed] [Google Scholar]
- Powell L. M., Wallis S. C., Pease R. J., Edwards Y. H., Knott T. J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987 Sep 11;50(6):831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994 May;19(1):55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Schmieden V., Kuhse J., Betz H. Mutation of glycine receptor subunit creates beta-alanine receptor responsive to GABA. Science. 1993 Oct 8;262(5131):256–258. doi: 10.1126/science.8211147. [DOI] [PubMed] [Google Scholar]
- Shah R. R., Knott T. J., Legros J. E., Navaratnam N., Greeve J. C., Scott J. Sequence requirements for the editing of apolipoprotein B mRNA. J Biol Chem. 1991 Sep 5;266(25):16301–16304. [PubMed] [Google Scholar]
- Sharma P. M., Bowman M., Madden S. L., Rauscher F. J., 3rd, Sukumar S. RNA editing in the Wilms' tumor susceptibility gene, WT1. Genes Dev. 1994 Mar 15;8(6):720–731. doi: 10.1101/gad.8.6.720. [DOI] [PubMed] [Google Scholar]
- Sommer B., Köhler M., Sprengel R., Seeburg P. H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991 Oct 4;67(1):11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Teng B., Burant C. F., Davidson N. O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993 Jun 18;260(5115):1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- Tsuji S., Martin B. M., Kaslow D. C., Migeon B. R., Choudary P. V., Stubbleflied B. K., Mayor J. A., Murray G. J., Barranger J. A., Ginns E. I. Signal sequence and DNA-mediated expression of human lysosomal alpha-galactosidase A. Eur J Biochem. 1987 Jun 1;165(2):275–280. doi: 10.1111/j.1432-1033.1987.tb11438.x. [DOI] [PubMed] [Google Scholar]
- Vetrie D., Kendall E., Coffey A., Hassock S., Collins J., Todd C., Lehrach H., Bobrow M., Bentley D. R., Harris A. A 6.5-Mb yeast artificial chromosome contig incorporating 33 DNA markers on the human X chromosome at Xq22. Genomics. 1994 Jan 1;19(1):42–47. doi: 10.1006/geno.1994.1010. [DOI] [PubMed] [Google Scholar]
- Yang J. H., Sklar P., Axel R., Maniatis T. Editing of glutamate receptor subunit B pre-mRNA in vitro by site-specific deamination of adenosine. Nature. 1995 Mar 2;374(6517):77–81. doi: 10.1038/374077a0. [DOI] [PubMed] [Google Scholar]
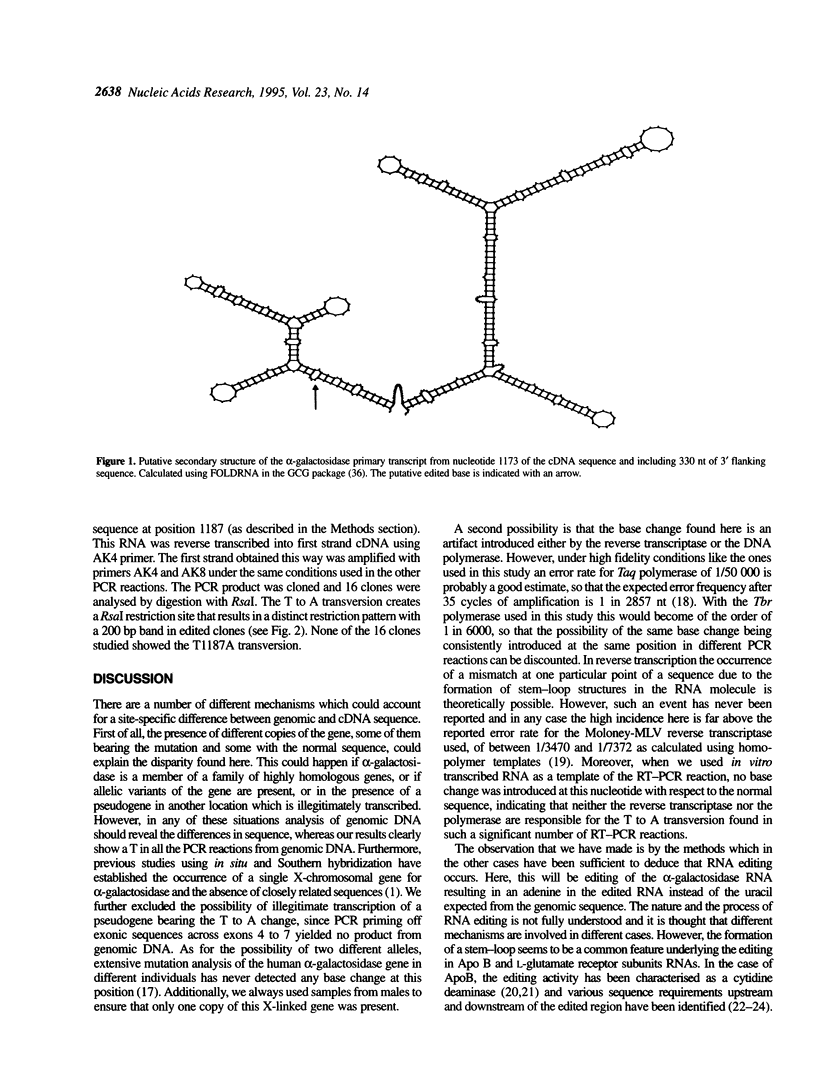
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]



