Summary
The cullin-RING ubiquitin ligases (CRL) are the largest family of multi-subunit E3 ligases in eukaryotes, which ubiquitinate protein substrates in numerous cellular pathways. CRLs share a common arched scaffold and a RING domain catalytic subunit, but use different adaptors and substrate receptors to assemble unique E3 machineries. In comparison to the first CRL structure, recent findings have revealed increased complexity in the overall architecture and assembly mode of CRLs, including multi-domain organization, inter-domain flexibility, and subunit dimerization. These features highlight the capacity of CRLs to catalyze protein ubiquitination under distinct cellular contexts and in response to diverse signals. As the first installment of a two-review series, this article will focus on recent advances in our understanding of CRL assembly mechanisms.
Introduction
Protein ubiquitination is a post-translational modification with such diverse targets, effects, and regulatory mechanisms that it rivals protein phosphorylation in its scope. Ubiquitin conjugation not only controls protein degradation by the 26S proteasome, but also affects protein sorting, enzymatic activity, and protein-protein interactions [1]. These ubiquitination-dependent changes in protein function regulate biological processes including cell cycle progression, gene transcription, signal transduction, DNA replication, DNA repair, and even viral and bacterial infections. The 76 amino acid ubiquitin polypeptide is covalently linked to target proteins through an ordered relay of conjugation reactions catalyzed by a three-enzyme cascade (E1-E2-E3) [2]. The E3 ubiquitin ligases catalyze the final step, mediating the formation of an isopeptide bond between a lysine residue on the substrate protein and the C-terminal carboxyl group of ubiquitin. Since E3s dictate the specificity toward a variety of substrates, they are the most structurally complex and diverse enzymes in the pathway. Based on sequence motifs and the mechanism of ubiquitin conjugation, ubiquitin ligases can be divided into two major classes: the HECT (Homologous to E6-AP C-Terminus) and the RING (Really Interesting New Gene) E3s. The HECT E3s pass ubiquitin from a E2 to a substrate via an E3-ubiquitin intermediate [3], whereas the RING E3s promote direct ubiquitin transfer from an E2 to a target protein [4].
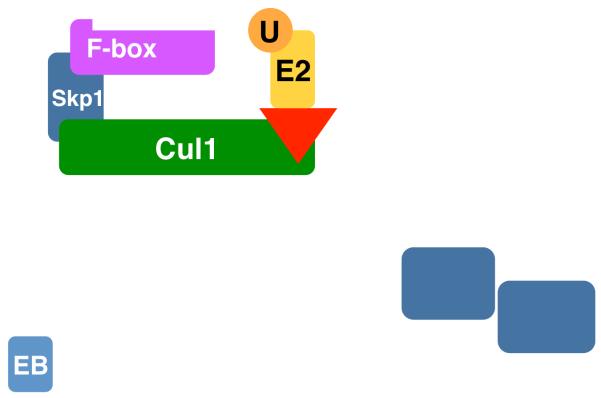
The cullin-RING ubiquitin ligases (CRLs) represent a superfamily of multi-component RING-class E3 complexes, and are organized by a cullin scaffold protein and a catalytic RING subunit, Rbx1 or Rbx2 [5]. The human genome codes for six closely related cullin proteins, Cul1, Cul2, Cul3, Cul4A, Cul4B, and Cul5. In complex with Rbx1 or Rbx2, these cullin proteins nucleate different subfamilies of CRLs, CRL1–CRL5 (Figure 1). Built on the conserved cullin-RING enzymatic core, CRLs adopt a modular architecture to achieve exquisite specificity toward a large number of substrates. To recognize specific target proteins, each cullin-RING complex can assemble with its own dedicated family of substrate receptor (SR) proteins, which are individually bridged to the cullin scaffold by an adaptor protein. These SR proteins feature various protein-protein interaction domains to bind specific sequences of substrates, known as “degrons”. The six cullin-RING complexes, together with their numerous adaptor-SR partners, yield an estimated 500 CRL family members in total. In addition, besides the canonical cullin proteins, the human genome has three additional proteins, APC2, PARC, and Cul7, which exhibit homology to the cullin domain of Cul1–5 but diverge elsewhere, and are considered atypical cullins [6,7].
Figure 1.
Modular assembly of cullin-RING ubiquitin ligases (CRLs). Cullin scaffold proteins (Cul1-5, green) in complex with Rbx1/2 (R, red) form the catalytic cores of CRLs. Ubiquitin (U)-charged E2 enzymes are recruited to CRLs by Rbx1/2. Skp1, Elongin C, and DDB1 serve as the adaptor (light blue) of CRL1(SCF), CRL2/5, and CLR4, respectively. F-box proteins, BC-box proteins, BTB-domain proteins, and DCAF proteins function as the substrate receptors (magenta) of CLR1(SCF), CRL2/5, CLR3, and CLR4, respectively. Adaptor binding regions of all cullins are colored in cyan.
The canonical CRL architecture: rigid scaffold and analogous adaptors
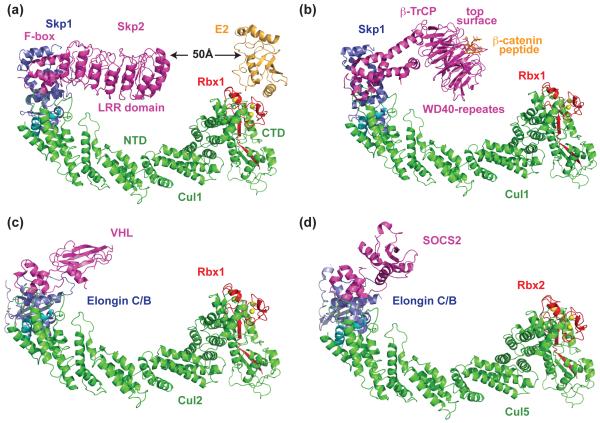
Our understanding of the architecture and assembly logic of CRLs began with a structural model of a complete CRL1 complex, SCFSkp2, which was derived from the crystal structures of two overlapping sub-complexes (Figure 2a) [8,9]. As the founding member of all CRLs, the SCF complex consists of the Cul1 scaffold, the Rbx1 RING subunit, the adaptor protein Skp1, and a substrate receptor F-box protein. Skp2, in this case, is an F-box protein that mediates cell cycle progression by recruiting the Cdk inhibitor p27 to the SCF for ubiquitination [10,11]. The SCFSkp2 structural model revealed several key structural features underlying its ubiquitin ligase functions. First, the central scaffold protein Cul1 adopts an overall elongated crescent-shaped structure consisting of a stalk-like N-terminal domain (NTD) and a globular C-terminal domain (CTD). Based on sequence homology, Cul2–5 are expected to form the same structure. Second, Skp1 is docked to Cul1 through two conserved α-helices near the Cul1 N-terminus, while Rbx1 is held at the other end by the Cul1 CTD. Third, tightly packed interfaces are found throughout the entire SCF, suggesting that parts of the E3 machinery act like a rigid platform to promote ubiquitin transfer from the E2 to the substrate protein. Finally, the structural configuration of SCF brings the substrate-binding LRR domain of Skp2 to the same side of the E3 scaffold where an E2 molecule is modeled. Upon modeling a bound E2 [12], a 50 Å gap, however, is found in between the substrate-binding site of the E3 and the active site of the E2 (Figure 2a) [9]. This rigid yet open arrangement is also supported by all subsequently reported Skp1-F box protein structures (Figure 2b) [13,14].
Figure 2.
Structural models of CRL1 and CRL2 showing a common assembly mode. (a) Structural model of SCFSkp2 obtained by superimposing the Skp1-Skp2 complex and the Cul1-Rbx1-Skp1-Skp2 F-box domain structure [8,9]. The E2 (orange) is modeled by superimposing Rbx1 RING domain with the UbcH7-bound c-Cbl RING domain [12]. Zinc atoms are shown as yellow spheres. The two Cul1 α-helices used to bind Skp1 and Skp2 F-box motif are colored in cyan. (b) Structural model of SCFβ-TrCP in complex with phosphorylated β-catenin degron peptide. The model is obtained by superimposing Skp1 in the structures of Skp1–β-TrCP and Cul1-Rbx1-Skp1-Skp2 F-box domain complexes [63]. β-TrCP uses the top surface of its WD40-repeat β-propeller domain to recognize and present its substrate β-cateinin. (c) Structural model of CRL2VHL obtained by superimposing Elongin C in the Elongin BC-VHL complex and Skp1 in the Cul1-Rbx1-Skp1-Skp2 F-box domain complex [17]. (d) Structural model of CRL5SOCS2 obtained by superimposing Elongin C in the Elogin BC-SOCS2 complex and Skp1 in the Cul1-Rbx1-Skp1-Skp2 F-box domain complex [18].
Sharing sequence homology to Skp1, Elongin C, together with Elongin B, serves as the common adaptor for CRL2 and CRL5. A family substrate receptors called BC-box containing proteins, include the von Hippel-Lindau (VHL) tumor suppressor and suppressors of cytokine signaling (SOCS) proteins, are recruited to Cul2 or Cul5 by interacting with Elongin BC [15,16]. Crystal structures of the Elongin BC-VHL and Elongin BC-SOCS2 complexes revealed a Bric-a-brac, Tramtrack, and Broad complex (BTB) fold of Elongin C, which is also adopted by the Cul1 adaptor Skp1 (Figure 2c) [8,17,18]. Interactions between Elongin BC and the BC-box motif closely resemble those for Skp1 binding of the F-box motif. Moreover, Elongin BC contacts to Cul2 or Cul5 likely parallel those for Skp1 with Cul1 [15]. Overall, all available crystal structures support a similar architecture and a common assembly mode shared by CRL1, 2, and 5 (Figure 2c, 2d).
CRL4A E3 machinery: mobile propellers and cryptic helices
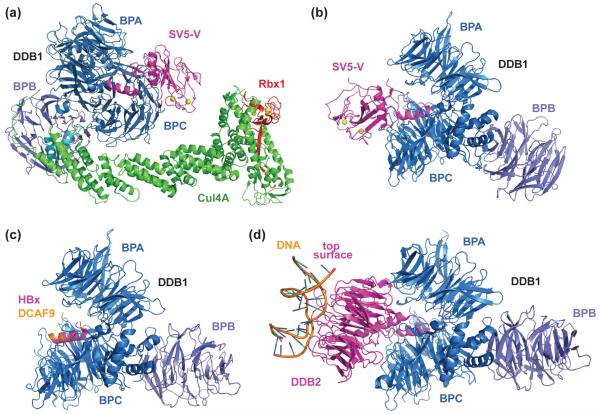
In contrast to the common structural features of CRL1, 2, and 5, a series of recent studies have revealed an unexpected and far more complex architecture for CRL4A. Conserved from yeast to humans, Cul4A adopts an elongated structure, analogous to other cullins [19]. Unlike other cullins, however, Cul4A utilizes as its adaptor the 127 kDa damaged DNA binding protein 1 (DDB1) as its adaptor, which lacks a BTB fold [20,21]. Rather, DDB1 contains three distinct WD40-like β-propeller domains, designated BPA, BPB, and BPC, and a C-terminal helical domain [22]. The BPB domain tethers DDB1 to Cul4A by interacting with the Cul4A NTD, while BPA and BPC form a structurally coupled double propeller, pointing away from the cullin scaffold (Figure 3a). Intriguingly, Cul4A interacts with the DDB1 BPB domain, again, via the same two α-helices corresponding to those that Cul1 uses to bind Skp1, even though DDB1 and Skp1 share no sequence homology or structural similarity. As a result, the relative positions of adaptor, SR, and Rbx1 on the cullin scaffold remain conserved among CRLs.
Figure 3.
Crystal structures of CRL4SV5-V and DDB1 in complex with viral hijackers, DCAF H-box motif, and DDB2. (a) Crystal structure of the Cul4A-Rbx1-DDB1-SV5-V complex with all subunits in (near) full-length forms [19]. The two α-helices used by Cul4A to bind DDB1 are colored in cyan. The BPB domain of DDB1 is colored (slate) differently from the BPA-BPC double propeller (marine) for clarity. (b) Bipartite interaction of SV5-V with DDB1 [22]. The SV5-V N-terminal α-helix is inserted into the pocket created in between the DDB1 BPA and BPC β-propeller domains. The SV5-V C-terminal domain containing two zinc ions (yellow) forms an additional interface with the DDB1 BPC domain. (c) Superimposed crystal structures of DDB1 in complex with the H-box motif of either HBx (magenta) or DCAF9 (orange) [27•]. Superimposable structures of DDB1 bound to the H-box motif of other DCAFs are not shown. (d) Crystal structure of DDB1 in complex with the DCAF protein DDB2 and UV-damaged DNA duplex [28••]. DDB2 binds DDB1 via a bipartite interface, similar to SV5-V. The N-terminal α-helix of DDB2 sits at the same position as that of SV5-V and the H-box motif of HBx and other DCAFs. DDB2 uses its narrower top surface, which faces away from DDB1, to recognize DNA lesions. Note that the position and orientation of the DDB1 BPB domain relative to the BPA-BPC double propeller is different in all structures.
The endogenous CRL4 substrate receptors, termed DCAFs (DDB1 and Cul4A associated factors) or DWDs (DDB1-binding WD40 proteins), were identified in several independent analyses of DDB1-interacting proteins [19,23-25]. The majority of these DCAFs contain six or more WD40 motifs, which fold as propeller blades and pack radially around a central axis to form a complete β-propeller domain.
The first clue toward the binding mode of DCAFs on DDB1 came from the co-crystal structure of DDB1 and a viral hijacker protein [22]. In order to block antiviral interferon signaling, the paramyxovirus V protein exploits the ubiquitin ligase activity of Cul4A-DDB1 by recruiting a non-cognate target, the STAT1-STAT2 heterodimer, to DDB1 for ubiquitination and subsequent degradation [26]. The crystal structure of DDB1 in complex with the simian virus 5 (SV5) V protein shows that the viral hijacker captures DDB1 through a bipartite interaction (Figure 3b). The N-terminal region of SV5-V forms an α-helix and inserts itself into a deep pocket sandwiched by the BPA and BPC domains of DDB. The C-terminal region of SV5-V adopts a zinc-stabilized globular domain and makes additional contacts with the DDB1 double propeller. Since SV5-V is able to recruit an unnatural target to CRL4, it likely does so by mimicking the endogenous DDB1 substrate receptors. Supporting this notion, the SV5-V protein can compete with endogenous DCAF proteins for DDB1 interaction, even though it does not contain a WD40 β-propeller domain [22].
Structural analysis of two additional viral hijackers of Cul4A-DDB1, the hepatitis B virus X (HBx) protein and the woodchuck hepatitis protein X (WHx), provided additional insights into the mechanism by which DCAFs anchor themselves to DDB1 [27•]. The crystal structure of DDB1 in complex with an HBx or WHx peptide demonstrated that both viral proteins use an α-helix to bind the DDB1 double propeller groove, like SV5-V (Figure 3c). Competitive DDB1 binding experiments further implied that DCAFs might dock to DDB1 via a similar helical motif. In fact, a combination of bioinformatic and crystallographic approaches helped identify such a motif in at least seven cellular DCAFs [27•]. Strikingly though, this 13 amino acid helical motif, termed the H-box, is unusually promiscuous in terms of its sequence, having few stringent sequence requirements for DDB1 binding. On the one hand, the poorly conserved H-box motif imposes a challenge for its identification in other DCAF proteins. On the other hand, its structurally promiscuous binding mode may explain why Cul4A-DDB1 is a frequent target of highly variable and compact viral proteins.
The crystal structure of a DDB1-DDB2 complex painted a complete picture of how a cellular DCAF protein is anchored to the CRL4 adaptor [28••]. Capable of specifically recognizing UV-induced DNA lesions, the DCAF protein DDB2 acts as a damage sensor and forms a CRL4 complex with Cul4A-DDB1 to ubiquitinate histones and DNA repair proteins [29,30]. At the structural level, DDB2 engages DDB1 in a fashion reminiscent of the SV5-V-DDB1 interaction (Figure 3d). The N-terminal region of DDB2 folds into a helix-loop-helix structure that nestles in between the DDB1 BPA-BPC cleft. The first helix of DDB2 plays an identical role as the H-box motif found in viral hijackers and other DCAF proteins. The C-terminal region of DDB2 adopts a canonical WD40 β-propeller fold with two characteristic opposite-facing surfaces. This β-propeller substantiates the formation of the DDB1-DDB2 complex by packing extensively against DDB1 through its “bottom” surface. In doing so, the DDB2 β-propeller presents its “top” surface to bind damage-containing DNA duplex (Figure 3d). Although the crystal structures of additional DDB1-DCAF complexes remain to be determined, it is tempting to speculate that most H-box and WD40 repeats-containing DCAFs will assume the same binding mode upon docking to DDB1. Supporting this model, most if not all β-propeller-containing CRL SR proteins use their “top” surface to bind and present substrates (Figure 2b) [13,14,31-33].
Besides a sophisticated multi-domain organization, DDB1 distinguishes itself from other CRL adaptors by displaying four distinct domain arrangements in the currently documented structures (Figure 3b, 3c, 3d) [19,22,28••]. In the crystals, a large degree of rotational and torsional flexibility has been observed between the DDB1 BPA-BPC double propeller and the BPB domain. It is conceivable that such flexibility might allow CRL4 to accommodate DCAF-substrate complexes of various sizes and shapes, and to support the assembly of unique polyubiquitin chains. Alternatively, it has been suggested that the structural plasticity might enable a CRL4 complex to sample a large area centered around the DCAF-substrate complex [28••]. Such a feature might be essential for the functions of various CRL4 complexes, many of which are specialized for promoting protein ubiquitination in the context of the ever-changing chromatin landscape [29].
CRL3 substrate adaptors: All-in-one and two-for-one
Distinct from all other CRLs, CRL3s have integrated the functions of cullin adaptor and substrate receptor into a single polypeptide, which also forms an obligate dimer. BTB domain proteins were identified by several independent studies as the substrate receptors of Cul3 [34-37]. Members of the BTB domain protein family use a BTB domain to bind the cullin scaffold and a Kelch, MATH, ZnF, or other protein-protein interaction domain to recruit substrate [38]. As earlier structural studies have defined a general role of the BTB domain in protein dimerization [39,40], BTB-containing CRL3 adaptor proteins can induce dimerization of CRL3 with two juxtaposed substrate-binding sites and two catalytic cores.
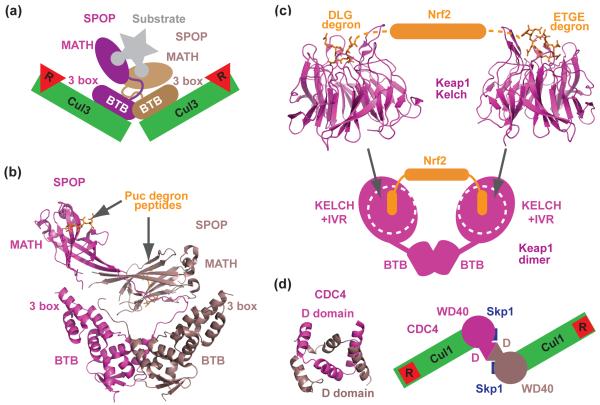
A recent comprehensive study of CRL3SPOP indeed led to a model describing how a BTB-mediated CRL3 dimer can ubiquitinate a single target protein recognized by two flexibly orientated substrate-binding sites [41••]. The Cul3 substrate receptor SPOP uses a MATH domain to recruit substrates such as MAPK phosphatase Puc, the X chromosome regulator MacroH2A, and the transcription factor Ci/Gli for polyubiquitination [24,42,43]. Based on a series of biochemical analyses, multiple copies of a newly mapped degron sequence were identified in most CRL3SPOP substrates; and a preferred 1:2 complex between SPOP and Puc was detected in the presence of excess Puc. The BTB domain of SPOP was shown to dimerize and form a complex with Cul3 at a 2:2 ratio, whereas a dimerization-defective mutant of SPOP was unable to ubiquitinate substrate in vitro, despite retaining competence for Cul3 binding. Together, these results suggest a model in which a SPOP dimer nucleates two copies of Cul3-Rbx1 on one side and can tether one copy of a substrate on the other (Figure 4a). In support of this model, the crystal structure of a nearly full-length SPOP dimer bound to the substrate degron peptide revealed asymmetric positioning of the MATH domains relative to the symmetrical BTB dimerization module (Figure 4b). This striking feature, together with additional biophysical evidence, suggests that the MATH domains are flexibly tethered to the BTB dimer and are capable of simultaneously engaging two degrons of the same polypeptide substrate. Such a binding mode might allow protein degradation to be fine-tuned through avidity upon degron optimization on a single substrate. It further suggests that regulated SPOP-dimerization or induced rigidity of the mobile linker could plausibly affect the efficiency of substrate ubiquitination.
Figure 4.
Dimerization of CRL substrate receptors. (a) A schematic model showing that the asymmetric SPOP dimer recruits two copies of Cul3-Rbx1 and can bind a single substrate polypeptide by recognizing two degrons via two MATH domains [41••]. Degrons are represented by the rounded ends of substrate. (b) Crystal structure of the asymmetric SPOP dimer [41••]. Each MATH domain in SPOP is in complex with a Puc degron peptide. The BTB domains of SPOP mediate dimer formation, while the 3 box domains promote Cul3 binding. (c) Crystal structures of Keap1 Kelch β-propeller in complexes with two distinct degrons of Nrf2 [32,33,54]. A schematic drawing of the full-length Keap1 dimer is shown at the bottom based on the two-site recognition model and EM reconstruction results. Each globular lobe of the Keap1 dimer contains a Kelch β-propeller and the IVR region. (d) Crystal structure of the dimeric Cdc4 D-domain and a top view of a schematic model of a dimeric SCFCdc4 proposed in [62]. The WD40-repeats domains of the Cdc4 dimer are represented by two circles.
Despite the lack of a full-length protein crystal structure, studies on another well-characterized BTB domain protein, Keap1, provides yet another example on how a BTB protein dimer actively controls substrate ubiquitination. Keap1 is a Kelch-repeat-containing BTB domain protein that mediates the constitutive turnover of the transcription factor Nrf2 by CRL3 [44-48]. Under oxidative stress, reactive cysteine residues in Keap1 are oxidized, resulting in a yet-to-be-understood structural shift that hinders Nrf2 ubiquitination. Nrf2 protein then accumulates and initiates a stress-responsive cytoprotective transcriptional program [49-51]. Keap1 is organized into 3 major domains: an N-terminal BTB domain, a C-terminal Kelch repeat domain, and an intervening region (IVR). A rich body of evidence, supports a two-site substrate recognition model, in which two copies of Kelch repeat β-propellers in a Keap1 dimer bind simultaneously to two distinct degrons in Nrf2. Following a potential “hinge and latch” mechanism, the Keap1 dimer can only ubiquitinate Nrf2 when both Nrf2 degrons are engaged [52-54]. With the crystal structures of the Keap1 Kelch repeat domain in hand (Figure 4c) [32,33,55], a recent study has reported the overall architecture of the Keap1 homodimer at 24Å resolution by electron microscopy reconstruction [56•]. The major structural population for the Keap1 dimer has the appearance of “a pair of cherries” with two globular lobes, each containing a Kelch propeller packed with an IVR domain, tethered by thin linkers to a forked stem structure formed by dimeric BTB domains (Figure 4c). Since the apparent gap between the two substrate-binding sites matches a calculated distance between the two degrons in a single Nrf2 polypeptide, it has been proposed that the Keap1 dimer is able to simultaneously bind the two degrons to promote Nrf2 ubiquitination. Under electrophilic stress, cysteine modification of Keap1 might induce a major conformational change that alters the distance between the two degron-binding sites, thereby preventing the high-avidity binding, which is important for Nrf2 ubiquitination. This model could explain how environmentally cued structural changes in an SR protein modulate substrate degradation, adding to a growing list of biochemical modifications that regulate interactions between ubiquitin ligases and their targets.
F-box protein dimerization: alternate and redundant degron recognition
In addition to BTB domain proteins in CRL3, several WD40-repeat-containing F-box proteins, including yeast Cdc4 and its human ortholog Fbw7, have been reported to form functional dimers through a conserved D-domain N-terminal to their F-box motif [57,58]. Cdc4 and Fbw7 are known to promote degradation of Sic1 in yeast and Cyclin E in humans, respectively, and both substrates contain more than one phosphorylation-dependent degron [58-60]. Cdc4 dimerization is important in vivo, as mutations in the substrate recognition domain that leave the D-domain intact yield a dominant-negative phenotype. In human cells, although Fbw7 dimerization is not strictly required for the degradation of Cyclin E, it is necessary for the recognition and ubiquitination of a Cyclin E mutant with sub-optimal degrons. Thus, Fbw7 dimerization may present an alternate route to ubiquitinate substrates in response to a cellular input that does not phosphorylate the target proteins at the optimal degron site [61]. Until the structure of a full-length D-domain-containing F-box protein becomes available, the architecture of a functional CRL1 dimer remains unknown. In a model derived from the crystal structures of separate Cdc4 D- and WD40-repeat domain, as well as small angle X-ray scattering analysis of the Skp1-Cdc4 dimer complex, a side-by-side configuration of a dimeric Skp1-Cdc4 complex has been proposed (Figure 4d) [62]. In the context of the entire SCF, this model places the two substrate-binding sites in the Cdc4 dimer 65 Å apart and both in between two catalytic centers of the CRL1 dimeric complexes. Questions, however, remain as to how such a configuration mediates optimal substrate binding and ubiquitination.
Conclusions
Since the architecture of SCF was elucidated, a number of recent structural studies have revealed new aspects of CRL assembly mechanisms, including diverse domain composition, intrinsic flexibility and polymorphic orientations, as well as dimeric configuration. Upon showcasing the structural complexity of CRLs, these studies have raised many questions toward the functional roles of these new architectural features found in this super-family of E3 machinery. For example, what is the functional advantage of DDB1 in CRL4 and SPOP in CRL3 to have structural plasticity built-in in the SR subunit? Is CRL SR dimerization necessary for high avidity binding of substrate or optimal spatial presentation of substrate, or both? It is conceivable that there might be more, yet-to-be discovered co-factors or conformational rearrangements that mediate stabilization and presentation of bound substrate or release of ubiquitinated product. Our understanding of the CRLs is clearly incomplete. Through continued structural dissection of these fascinating and indispensible enzymes, novel mechanisms regarding their assembly and regulation will surely be uncovered.
Acknowledgement
E.S.Z. and N.Z. are supported by National Institutes of Health (R01 CA107134 and F32 GM093497), Burroughs Wellcome Fund, and National Science Foundation. B.A.S is supported by National Institutes of Health (R01 GM069530) and ALSAC. N.Z. and B.A.S. are investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no competing interest.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM. Annu Rev Biochem. Vol. 70. 2001. Mechanisms underlying ubiquitination; pp. 503–533. [DOI] [PubMed] [Google Scholar]
- 3.Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 4.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 5.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 6.Skaar JR, Florens L, Tsutsumi T, Arai T, Tron A, Swanson SK, Washburn MP, DeCaprio JA. PARC and CUL7 form atypical cullin RING ligase complexes. Cancer Res. 2007;67:2006–2014. doi: 10.1158/0008-5472.CAN-06-3241. [DOI] [PubMed] [Google Scholar]
- 7.Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJ, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 8.Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 9.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 10.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 11.Tsvetkov LM, Yeh KH, Lee SJ, Sun H, Zhang H. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- 12.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 13.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 14.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 15.Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, Staehling-Hampton K, Washburn MP, Conaway RC, Conaway JW. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J Biol Chem. 2008;283:8005–8013. doi: 10.1074/jbc.M706987200. [DOI] [PubMed] [Google Scholar]
- 17.Stebbins CE, Kaelin WG, Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 18.Bullock AN, Debreczeni JE, Edwards AM, Sundstrom M, Knapp S. Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase. Proc Natl Acad Sci U S A. 2006;103:7637–7642. doi: 10.1073/pnas.0601638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 20.Shiyanov P, Nag A, Raychaudhuri P. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J Biol Chem. 1999;274:35309–35312. doi: 10.1074/jbc.274.50.35309. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Zhang Y, Douglas L, Zhou P. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J Biol Chem. 2001;276:48175–48182. doi: 10.1074/jbc.M106808200. [DOI] [PubMed] [Google Scholar]
- 22.Li T, Chen X, Garbutt KC, Zhou P, Zheng N. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell. 2006;124:105–117. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 23.He YJ, McCall CM, Hu J, Zeng Y, Xiong Y. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 2006;20:2949–2954. doi: 10.1101/gad.1483206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 25.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 26.Ulane CM, Horvath CM. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology. 2002;304:160–166. doi: 10.1006/viro.2002.1773. [DOI] [PubMed] [Google Scholar]
- •27.Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol. 2010;17:105–111. doi: 10.1038/nsmb.1719. This study reveals a helical motif, namely the “H-box”, which is conserved in structure but not in sequence and is responsible for docking two viral hijacking proteins and several cellular substrate receptors to the Cul4A-DDB1 E3 complex.
- ••28.Scrima A, Konícková R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thomä NH. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. The authors determined the crystal structures of the DDB1-DDB2 complex bound to different UV damaged DNA duplexes. This study not only reveals how a DCAF protein (DDB2) is docked to DDB1, but also unveils the structural basis of damaged DNA recognition by DDB2.
- 29.O’Connell BC, Harper JW. Ubiquitin proteasome system (UPS): what can chromatin do for you? Curr Opin Cell Biol. 2007;19:206–214. doi: 10.1016/j.ceb.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Stoyanova T, Roy N, Kopanja D, Raychaudhuri P, Bagchi S. DDB2 (damaged-DNA binding protein 2) in nucleotide excision repair and DNA damage response. Cell Cycle. 2009;8:4067–4071. doi: 10.4161/cc.8.24.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 32.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, Elledge SJ, Harper JW. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 35.Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- 36.Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 37.Pintard L, Willis JH, Willems A, Johnson JL, Srayko M, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- 38.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmad KF, Engel CK, Prive GG. Crystal structure of the BTB domain from PLZF. Proc Natl Acad Sci U S A. 1998;95:12123–12128. doi: 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmad KF, Melnick A, Lax S, Bouchard D, Liu J, Kiang CL, Mayer S, Takahashi S, Licht JD, Prive GG. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12:1551–1564. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- ••41.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. In this comprehensive study, the authors defined regions of SPOP involved in Cul3 binding, mapped the degron of several cellular substrates of the BTB-domain protein SPOP, determined the crystal structures of a SPOP dimer in complex with a substrate degron peptide, and analyzed the molecular composition of the CRL3SPOP-substrate complex. The results support a model that a dimeric CRL3SPOP complex binds a single substrate using two substrate-recruiting domains flexibly linked to the BTB domains.
- 42.Liu J, Ghanim M, Xue L, Brown CD, Iossifov I, Angeletti C, Hua S, Negre N, Ludwig M, Stricker T, et al. Analysis of Drosophila segmentation network identifies a JNK pathway factor overexpressed in kidney cancer. Science. 2009;323:1218–1222. doi: 10.1126/science.1157669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernandez-Munoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci U S A. 2005;102:7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chew EH, Poobalasingam T, Hawkey CJ, Hagen T. Characterization of cullin-based E3 ubiquitin ligases in intact mammalian cells--evidence for cullin dimerization. Cell Signal. 2007;19:1071–1080. doi: 10.1016/j.cellsig.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 54.Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X, Zhang D, Hannink M, Beamer LJ. Crystal structure of the Kelch domain of human Keap1. J Biol Chem. 2004;279:54750–54758. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- •56.Ogura T, Tong KI, Mio K, Maruyama Y, Kurokawa H, Sato C, Yamamoto M. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc Natl Acad Sci U S A. 2010;107:2842–2847. doi: 10.1073/pnas.0914036107. This EM reconstruction study reveals the overall structure of a Keap1 dimer and establishes a spatial relationship among the structural domains of the protein that are important for Nrf2 binding.
- 57.Dixon C, Brunson LE, Roy MM, Smothers D, Sehorn MG, Mathias N. Overproduction of polypeptides corresponding to the amino terminus of the F-box proteins Cdc4p and Met30p inhibits ubiquitin ligase activities of their SCF complexes. Eukaryot Cell. 2003;2:123–133. doi: 10.1128/EC.2.1.123-133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 60.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Molecular Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 61.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 62.Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 63.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Molecular Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]