Abstract
Uncovering conserved alternative splicing (AS) events can identify AS events that perform important functions. This is especially useful for identifying premature stop codon containing (PTC) AS isoforms that may regulate protein expression by being targets for nonsense mediated decay. This report discusses the identification of a PTC containing splice isoform of the TFIIIA gene that is highly conserved in land plants. TFIIIA is essential for RNA Polymerase III-based transcription of 5S rRNA in eukaryotes. Two independent groups have determined that the PTC containing alternative exon is ultraconserved and is coupled with nonsense-mediated mRNA decay. The alternative exon appears to have been derived by the exonization of 5S ribosomal RNA (5S rRNA) within the gene of its own transcription regulator, TFIIIA. This provides the first evidence of ancient exaptation of 5S rRNA in plants, suggesting a novel gene regulation model mediated by the AS of an anciently exonized non-coding element.
Key words: alternative splicing, TFIIIA, cis-element, 5S rRNA, exonization, exaptation
Alternative splicing (AS) creates multiple mRNA transcripts or isoforms, from a single gene. Alternative splicing is common in metazoans and, to a lesser extent, in plants.1 Upwards of 80% of human genes have been demonstrated to undergo AS,2,3 while 20–40% of plant genes produce two or more transcript isoforms.1,4 While the proportion of plant genes that generate alternative transcripts is far less than that of animal genes, another intriguing difference is the predominance of certain alternative splicing mechanisms. The basic classes of alternative transcripts (exon-skipping, alternative acceptor/donor sites, etc.) are present in both plants and animals; however, the most abundant human AS event is exon skipping and the most abundant plant AS event is intron retention. Cases of exon skipping and intron retention are relatively rare in plants and animals, respectively. This bias may reflect differences in the molecular mechanism of splice site recognition between plants and animals.1
Approximately 60–75% of all AS events occur within the translated regions of mRNAs,5,6 which can effect protein function (reviewed in ref. 6) and influence protein diversity. It is also quite common for alternative splicing to result in isoforms that contain a premature termination codon (PTC) which, subsequently, become targets for nonsense mediated decay (NMD).7 NMD is an RNA surveillance system that recognizes mRNAs containing premature termination codons (PTCs) and targets them for degradation.8 Lewis and Brenner9 proposed that a process of regulated unproductive splicing and translation (RUST) may function to regulate protein expression by generating NMD-targeted isoforms. Premature termination codon containing isoforms that result from alternative splicing are common in plants1,4,10 and animals.9 However, it is not clear to what extent PTC containing isoforms are indeed coupled to NMD as a mode of post-transcriptional gene regulation versus simply being the by-products of noisy splicing. Recent studies suggest that a large fraction of AS events are a result of noise in the splicing machinery and lead to unstable protein products if translated11,12 rather than being an effector of an NMD mediated expression regulation process.13
While many PTC containing transcript isoforms may result from imprecise splicing, there remain several examples of genes that are truly regulated by coupling PTC containing alternatively spliced transcript isoforms with NMD. The majority of these encode components of the ribosome14,15 and splicing machinery.14–18 One interesting feature associated with some genes regulated by AS-coupled NMD is the presence of ultraconserved sequence elements. Lareau et al.18 identified ultraconserved elements associated with cassette exons within some mammalian serine/arginine (SR) rich proteins that result in PTC containing transcript isoforms when spliced in. Evidence suggests that the PTC containing isoforms are subject to NMD and that when abundant, these SR proteins affect the splicing of their own mRNA to increase the proportion of NMD target isoforms, which are degraded by the NMD pathway. Thus, the regulation of unproductive splicing of SR protein transcripts is proposed to play a role in the auto-regulation or homeostatic control of these splicing regulators.18 Consistent with this, Zhang et al.13 examined human and mouse orthologues of well annotated protein coding genes for PTC containing transcript isoforms and determined that only the small fraction (2%) that exhibited strong evolutionary conservation of the PTC containing transcript isoforms were likely to represent bona fide examples of regulated unproductive splicing coupled to NMD. These observations suggest that the portions of the gene directly influencing production of the PTC containing isoform are under purifying selection and will remain conserved between organisms with conserved PTC containing splice isoforms, assuming that the mechanisms of alternative splice determination and PTC recognition were also conserved. Therefore, most genes that truly utilize AS-coupled NMD to regulate protein expression post-transcriptionally should having evolutionarily conserved PTC containing isoforms that can be identified during comparative genomics searches.
Recently, two independently conducted comparative genomic analysis identified a highly conserved sequence element associated with an alternatively spliced cassette exon in the gene encoding the TFIIIA transcription factor in rice (Oryza sativa) and Arabidopsis thaliana19,20 that diverged 140–150 Mya.21 Hammond et al.20 searched for structured cis-regulatory RNA elements based on nucleotide and secondary structure conservation between noncoding regions in rice and Arabidopsis orthologous genes. Fu et al.19 identified the same sequence element while performing a comparative genome-wide search for AS events conserved between A. thaliana and O. sativa. TFIIIA (Transcription Factor for polymerase III A) was the first zinc finger transcription factor characterized, and is essential for the Pol III-based transcription of 5S ribosomal RNA (5S RNA) that is required for the assembly of the large ribosomal subunit.22 TFIIIA can bind to both 5S rRNA and 5S rDNA,23,24 and these binding properties have lead to the proposal of a negative feedback auto-regulation model for 5S rRNA transcription in metazoans.25 In contrast to the regulation of 5S RNA by TFIIIA, and its interaction with 5S rRNA, the transcriptional regulation of TFIIIA is not well understood; however, post-transcriptional regulation of TFIIIA had never been reported previously in any species.
The Arabidopsis TFIIIA (AtTFIIIA) gene consists of seven exons, the third of which is alternatively skipped (ES: Exon-Skipped isoform) or included (EI: Exon-Inclusive isoform) to produce either of two transcript isoforms. The AtTFIIIA ES isoform encodes the fully functional peptide containing all nine zinc fingers, while the EI isoform contains a PTC that resides within the cassette exon retained in this isoform. Translation of the EI isoform results in a truncated protein that contains only the first two zinc-fingers and is unable to bind 5S rRNA or initiate transcription of 5S rDNA.19 A dramatic accumulation of the PTC containing transcript in Arabidopsis seedlings following cycloheximide treatment19 or in the presence of a UPF1 mutant background26 demonstrates that the AtTFIIIA EI transcript is a target of NMD.
Fu et al.19 provide further support that the AtTFIIIA EI isoform is a target of NMD by evaluating phenotype and transcript abundance in transgenic Arabidopsis overexpressed in either of the two isoforms. The hypothesis that expression of TFIIIA is regulated by RUST predicts that the EI isoform serves no purpose other than as a ‘dead-end’ to translation. Therefore, exogenous overexpression of the EI transcript isoform should not affect the expression level of the endogenous ES isoform or plant fitness. Indeed, the phenotype of transgenic Arabidopsis plants that overexpress the EI isoform were indistinguishable from non-transgenic (wild-type) Arabidopsis plants. On the other hand, exogenous overexpression of the functional transcript would produce an “overexpression” phenotype that may affect morphology or plant fitness. In addition, if the cell regulates AtTFIIIA expression by RUST, then the cell should respond to rising TFIIIA protein and/or 5S rRNA levels by influencing the alternative splicing decision during processing of the endogenous AtTFIIIA pre-mRNA to increase the ratio of PTC containing EI to ES isoform. Three independent transgenic ES overexpression events were assessed for EI and ES isoform expression levels and their affect on Arabidopsis phenotype. ES isoform overexpression lines exhibit abiotic stress-related phenotypes, specifically sensitivity to salt and osmotic shock, with severities that correlate to the relative level of accumulation of the transgenic AtTFIIIA ES isoform. Additionally, the endogenous AtTFIIIA EI/ES ratios were strongly positively correlated with the levels of ES isoform accumulation in the transgenic lines. These results demonstrated that increasing the expression of AtTFIIIA (and presumably the amount of TFIIIA protein and 5S rRNA) with AtTFIIIA overexpression constructs results in a corresponding increase in the amount of EI isoform produced during processing of the endogenous AtTFIIIA mRNA. This result demonstrated that high levels of AtTFIIIA protein and/or 5S rRNA cause a greater fraction of the AtTFIIIA transcripts to be spliced into the unproductive isoform and then degraded through NMD, thereby downregulating AtTFIIIA production.
Because genes whose protein expression levels are truly modulated by regulated unproductive splicing coupled to NMD are expected to exhibit strong evolutionary sequence conservation of the PTC containing transcript isoforms, and given the presence of PTC-containing AS isoforms conserved between A. thaliana and O. sativa, it is not unexpected that this regulatory mechanism is associated with TFIIIA. What was surprising was the degree of sequence conservation observed across a very wide evolutionary time scale. Available plant EST and genomic sequences provide the means to examine the extent of conservation of the alternative exon within the TFIIIA gene of many plant species. Analysis of available and experimentally isolated sequences identified 52 TFIIIA orthologues from a diverse group of land plants that include many flowering plants, the bryophyte moss (Physcomitrella patens), and an early vascular plant (Selaginella moellendorffii) that last shared a common ancestor with angiosperms over 400 Mya. No evidence for the alternative exon was found in the green alga Chlamydomonas or other non plant genomes.19 The pervasiveness of the alternative exon throughout a large and evolutionary diverse collection of land plants suggests it performs an important functional role, which is underscored by the finding that the alternative exon was the most conserved feature of the TFIIIA orthologues (Fig. 1). Most significant was the obvious sequence similarity between the alternative exon and 5S rRNA. Multiple sequence analysis identifies highly conserved sequence blocks between all 5S-like alternative exons and 5S rRNA sequences, suggesting that the 5S rRNA-like element may adopt a similar secondary structure to that of 5S rRNA.19,20 Computational19,20 and experimental20 evidence suggests that the 5S rRNA-like element associated with the alternative exon adopts many 5S rRNA secondary structure elements including Helix III and loop D that are required for ribosomal protein L5 binding, and helix IV and loop E that can interact with TFIIIA protein19,22 (Fig. 2). The invariance throughout all land plants examined for nucleotides that may maintain these specific 5S rRNA-like secondary structural elements within the 5S rRNA-like alternative exon suggests that this element is under strong purifying selection to maintain these structural elements and that regulation of TFIIIA alternative splicing may occur via protein factors that normally interact with 5S rRNA.
Figure 1.
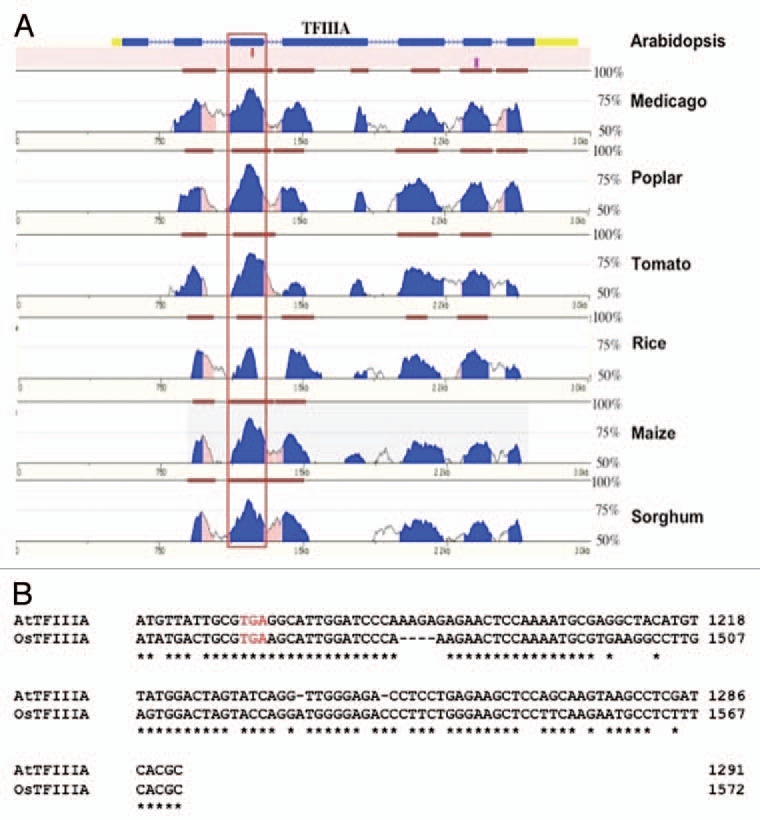
An example of sequence conservation between the alternative exons in a selection of plant TFIIIA genes. (A) Vista plots of pair-wise alignments between TFIIIA from Arabidopsis and its orthologue in medicago, poplar, tomato, rice maize and sorghum. The alternative exon (boxed) is the most conserved feature between Arabidopsis TFIIIA and its orthologues in other plant species. Levels of sequence identity between Arabidopsis TFIIIA and its orthologues in the six other plants species displayed are depicted as blue peaks. Brown bars signify segments that pass the alignment criteria of 70% identity over a window of 100 bp. (B) Detailed sequence alignment between the PTC containing alternative exon of Arabidopsis TFIIIA and one of the PTC containing alternative exons of rice TFIIIA—rice contains a duplication of the alternatively spliced exon (see text). Sequence identity across the alternatively splice exon is 74%. The in-frame stop codon introduced when this exon is included in the transcript is represented by red text.
Figure 2.
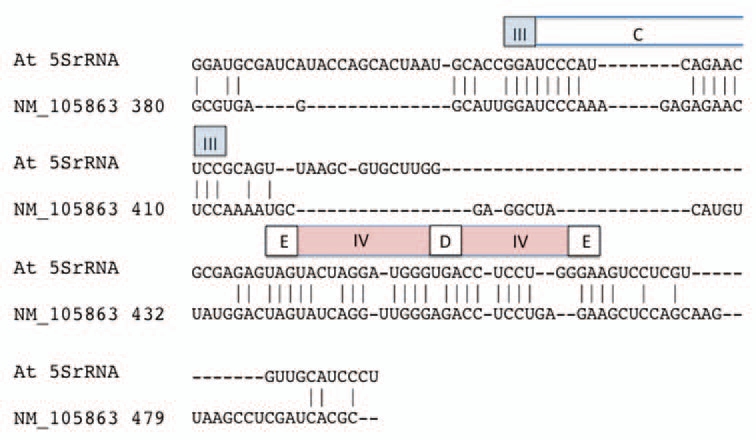
Alignment of the alternative exon of Arabidopsis TFIIIA to Arabidopsis 5SrRNA. The alternative exon sequence is represented within Gen-Bank accession NM_105863 from nucleotide position 380 to 479. Regions corresponding to 5SrRNA secondary structure features are depicted above the sequence segment in which they are found. Elements are named according to the standard 5S rRNA nomenclature: helices are designated by roman numerals (III–V) in colored boxes; loop regions are designated by letters (C–E) in open boxes.
What is the molecular mechanism responsible for regulating the splicing decision? The SC35 SR protein in mammals is thought to regulate its own expression through a negative feedback loop mediated by alternative splicing and NMD.18 Likewise, since TFIIIA is known to bind 5S rRNA, Fu et al.19 proposed a simple model by which TFIIIA protein binds the 5S rRNA-like element associated with the alternative exon and affects splicing by either acting as a splicing factor or competing with another splice factor by occupying splice sites or cis-splicing regulatory regions such as exon splice enhancers or suppressors. Under this model, and consistent with experimental observations, TFIIIA would interact with its own pre-mRNA via the 5S rRNA-like structural elements helix IV and loop E. This would influence inclusion of the PTC containing exon and trigger NMD. Conversely, when TFIIIA levels are low, the PTC containing exon is skipped during mRNA processing so that the ES isoform is produced, which leads to an increase in TFIIIA protein levels. This model suggests that TFIIIA interacts directly with TFIIIA pre-mRNA; however, Fu et al. did not rule out the possibility that other factors in addition to or in place of, TFIIIA are involved in the regulation model.19 Since TFIIIA is essential for the transcription of 5S rRNA, it is reasonable to expect that the mechanism regulating expression of TFIIIA would be sensitive to 5S rRNA levels. One glaring deficiency of the TFIIIA auto-regulation model is that it is not clear how regulation of TFIIIA is influenced by the level of 5S rRNA.
Elegant biochemical analysis by Hammond et al.20 demonstrated that ribosomal protein L5 directly binds the conserved helix III-like structure of the 5S rRNA like element associated with the alternative exon, which parallels a known binding interaction between L5 and 5S rRNA.22 Hammond et al. further determined that the binding of L5 to the 5srRNA like element is responsible for splicing control. Binding of L5 protein to the 5S rRNA element associated with the alternative exon in the AtTFIIIA premRNA results in skipping of this exon during mRNA production, while absence of L5 binding results in production of the EI isoform that is subject to NMD. When free levels of L5 are low due to low production or to complex formation with 5S rRNA, L5 is less available to bind to the 5S rRNA-like element within the AtTFIIIA pre-mRNA and splicing results in the PTC containing EI form. Conversely, the binding of L5 to AtTFIIIA pre-mRNA influences the splicing machinery to favor production of the ES isoform. This regulatory logic coordinates the levels of 5S rRNA with L5 ribosomal protein and makes sense in light of the observation that 5S rRNA must be bound with L5 prior to its incorporation into the ribosome22 Hammond et al.20 suggest that L5 binding may displace an exonic splice factor from the pre-mRNA, thus effecting the splicing decision. However, it may be the secondary structure of the 5S rRNA-like element that actually influences the splicing decision. There is evidence that secondary structure can influence alternative splicing27,28 potentially by masking splice sites.28 Schwartz et al.27 determined that secondary structure plays a role in exon recognition and that increased stability of secondary structure is inversely correlated to exon inclusion levels. Perhaps L5 binding further stabilizes the secondary structure of the 5S rRNA-like element containing alternate exon leading to its exclusion during AtTFIIIA pre-mRNA processing. Whether it is secondary structure or presence/absence of protein factors that further influence the splicing decision remains to be determined.
The summation of the experimental analysis by Fu et al.19 and Hammond et al.20 indicates that TFIIIA protein level in Arabidopsis is regulated post-transcriptionally by coupling unproductive splicing with NMD; and, this depends on direct binding of ribosomal protein L5 to an ultra-conserved sequence element with striking similarity to 5SrRNA. Evidence for this 5S rRNA-like alternate exon has been found in all land plants examined (>50), but is absent in metazoans and green algae. Furthermore, phylogentic analyses suggests that the 5S rRNA element originated from insertion of 5S rRNA into an intron of TFIIIA after the separation of land plants and unicellular green alga, but before the divergence of nonvascular and vascular plants, suggesting that this event occurred over 400 Mya.19 A single 5S rRNA-like element is associated with an alternate exon in most plant TFIIIA genes examined. However, the 5S rRNA like element has been duplicated in at least six plant species: 4 monocots from the family Poaceae and 2 Eudicots from the family Asteraceae. Although the origin of the 5S rRNA element is not known, some alternate exons have been shown to originate from retrotransposons insertions such as Alu elements in mammals;29,30 and, the Cassandra retrotranspon carries a 5S rRNA sequence within each of its two long terminal repeats and is present in many plant genomes.31 If the 5S rRNA-like element was derived from an ancient insertion of a Cassandra retrotransposons into an ancestor of land plants it is likely that one copy was lost shortly after insertion, and those species that have two copies of the 5S rRNA-like element acquired this through subsequent duplication.
The discovery of a 5S rRNA-like element acting to regulate TFIIIA protein expression represents the first example of 5S rRNA, a non-coding gene, being exonized into genic sequence in plant genomes. Furthermore, this exonization event is an example of molecular exaptation, where a previously adapted sequence is co-opted to serve a different function. In this instance, the 5S rRNA like element within the plant TFIIIA gene is probably evolved from a 5S rRNA carrying transposon or pseudo-gene that inserted into the TFIIIA locus. Once present in the TFIIIA locus, the 5S rRNA-like structural features of the 5S rRNA-like element have been co-opted to function in a regulatory role that controls TFIIIA expression post transcriptionally in land plants. This represents a particularilly intriguing event because 5S rRNA has thus been co-opted to play a role in regulating the expression of TFIIIA, which is the very transcription factor responsible for activating 5S rRNA transcription. The observation that regulation of TFIIIA by unproductive splicing with NMD is limited to land plants, and overexpression of the AtTFIIIA ES isoform is associated with osmotic stress phenotypes, allows that this regulatory circuit may have contributed to the adaptation of plants to stress when their ancestors invaded land.
While the prevalence of AS in plants is still being determined, and most known plant AS events have not been functionally characterized, evidence suggests that AS participates in important plant functions including development and stress response.1 Using comparative genomics to identify conserved alternative splicing events among evolutionarily distant species can prioritize AS events for functional characterization, help uncover relevant cis- and trans-regulatory factors, and distinguish regulated AS events from noise in the transcription machinery. With more available plant genomes, deeper transcriptome sequencing and the power of comparative genomics, we expect to uncover more conserved AS events and perhaps more cases of coexonization-exaptation in plants.
Acknowledgements
Thanks to Yan Fu for supplying data and analysis included in Figure 1.
Abbreviations
- 5S rRNA
5S ribosomal RNA
- AS
alternative splicing
- AtTFIIIA
transcription factor IIIA from Arabidopsis thaliana
- EI
exon3 including isoform of TFIIIA mRNA, with premature stop codon
- ES
exon3 skipped isoform of TFIIIA mRNA encoding the functional protein
- PTC
premature termination codon
- NMD
nonsense-mediated mRNA decay
- RUST
regulated unproductive splicing and translation
- Mya
million years ago
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/12684
References
- 1.Barbazuk WB, Fu Y, McGinnis KM. Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 2008;18:14–18. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- 2.Leipzig J, Pevzner P, Heber S. The Alternative Splicing Gallery (ASG): bridging the gap between genome and transcriptome. Nucleic Acids Res. 2004;32:3977–3983. doi: 10.1093/nar/gkh731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 4.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2009;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta S, Zink D, Korn B, Vingron M, Haas SA. Genome wide identification and classification of alternative splicing based on EST data. Bioinformatics. 2004;20:2579–2585. doi: 10.1093/bioinformatics/bth288. [DOI] [PubMed] [Google Scholar]
- 6.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, et al. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Belgrader P, Cheng J, Zhou X, Stephenson LS, Maquat LE. Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol Cell Biol. 1994;14:8219–8228. doi: 10.1128/mcb.14.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 9.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang BB, Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melamud E, Moult J. Stochastic noise in splicing machinery. Nucleic Acids Res. 2009;37:4873–4886. doi: 10.1093/nar/gkp471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melamud E, Moult J. Structural implication of splicing stochastics. Nucleic Acids Res. 2009;37:4862–4872. doi: 10.1093/nar/gkp444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Xin D, Wang P, Zhou L, Hu L, Kong X, et al. Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol. 2009;7:23. doi: 10.1186/1741-7007-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuccurese M, Russo G, Russo A, Pietropaolo C. Alternative splicing and nonsense-mediated mRNA decay regulate mammalian ribosomal gene expression. Nucleic Acids Res. 2005;33:5965–5977. doi: 10.1093/nar/gki905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol. 2007;623:190–211. doi: 10.1007/978-0-387-77374-2_12. [DOI] [PubMed] [Google Scholar]
- 18.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 19.Fu Y, Bannach O, Chen H, Teune JH, Schmitz A, Steger G, et al. Alternative splicing of anciently exonized 5S rRNA regulates plant transcription factor TFIIIA. Genome Res. 2009;19:913–921. doi: 10.1101/gr.086876.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond MC, Wachter A, Breaker RR. A plant 5S ribosomal RNA mimic regulates alternative splicing of transcription factor IIIA pre-mRNAs. Nat Struct Mol Biol. 2009;16:541–549. doi: 10.1038/nsmb.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- 22.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. 5 S rRNA: structure and interactions. Biochem J. 2003;371:641–651. doi: 10.1042/BJ20020872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelke DR, Ng SY, Shastry BS, Roeder RG. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980;19:717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- 24.Pelham HR, Brown DD. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci USA. 1980;77:4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cassiday LA, Lebruska LL, Benson LM, Naylor S, Owen WG, Maher LJ., 3rd Binding stoichiometry of an RNA aptamer and its transcription factor target. Anal Biochem. 2002;306:290–297. doi: 10.1006/abio.2002.5710. [DOI] [PubMed] [Google Scholar]
- 26.Yoine M, Ohto MA, Onai K, Mita S, Nakamura K. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz S, Gal-Mark N, Kfir N, Oren R, Kim E, Ast G. Alu exonization events reveal features required for precise recognition of exons by the splicing machinery. PLoS Comput Biol. 2009;5:1000300. doi: 10.1371/journal.pcbi.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shepard PJ, Hertel KJ. Conserved RNA secondary structures promote alternative splicing. RNA. 2008;14:1463–1469. doi: 10.1261/rna.1069408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lev-Maor G, Ram O, Kim E, Sela N, Goren A, Levanon EY, et al. Intronic Alus influence alternative splicing. PLoS Genet. 2008;4:1000204. doi: 10.1371/journal.pgen.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalendar R, Tanskanen J, Chang W, Antonius K, Sela H, Peleg O, et al. Cassandra retrotransposons carry independently transcribed 5S RNA. Proc Natl Acad Sci USA. 2008;105:5833–5838. doi: 10.1073/pnas.0709698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 33.McGuire AM, Pearson MD, Neafsey DE, Galagan JE. Cross-kingdom patterns of alternative splicing and splice recognition. Genome Biol. 2008;9:50. doi: 10.1186/gb-2008-9-3-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]


