Abstract
Cell growth can be suppressed by stressful environments, but the role of stress pathways in this process is largely unknown. Here we show that a cascade of p38β mitogen activated protein kinase and p38 regulated/activated kinase (PRAK) plays a role in energy starvation-induced suppression of mammalian target of rapamycin (mTOR), that energy starvation activates the p38β-PRAK cascade, and that p38β- or PRAK-deletion diminishes energy depletion-induced suppression of mTORC1 and reduction of cell size. We show that p38β-PRAK operates independent from the known mTORC1 inactivation pathways – phosphorylation of tuberous sclerosis protein 2 (TSC2) and raptor by AMP activated protein kinase (AMPK), and surprisingly, PRAK directly regulates Ras homolog enriched in brain (Rheb), a key component of the mTORC1 pathway by phosphorylation. Phosphorylation of Rheb at serine 130 by PRAK impairs Rheb’s nucleotide-binding ability and inhibits Rheb-mediated mTORC1 activation. The direct regulation of Rheb by PRAK integrates a stress pathway with the mTORC1 pathway in response to energy depletion.
The p38 mitogen activated protein kinase (MAPK) signaling pathway is evolutionarily conserved from yeast to human and participates in a variety of cellular responses1–4. There are four mammalian members in the p38 group of MAPK: p38α, p38β, p38γ, and p38δ5–8. While similarities in activation and function have been observed, each p38 isoform also has specific functions9. Although activation of p38 MAPKs by different stimuli is cell type-dependent, various stress stimuli, including energy stress, activate the p38 pathway in all cells, and thus the p38 pathway is considered to be a major stress-activated signaling pathway10. A number of substrates of p38 group MAPKs have been identified, including transcription factors and protein kinases, and the p38 pathway not only regulates gene expression but also some other cellular responses to stress.
mTOR is a highly conserved protein kinase that plays a critical role in controlling cell growth and metabolism11–13. mTOR exists in two distinct complexes called mTORC1 and mTORC214. The immunosuppression drug rapamycin inhibits mTORC115 but not mTORC216. The two complexes catalyze phosphorylation of different substrates and thus execute different functions17. mTORC1 integrates signals from growth factors, nutrients, stress, and energy in regulating cell growth. mTORC2 phosphorylates the hydrophobic motif of AKT (also known as protein kinase B) and modulates cytoskeleton organization18. mTORC1 contains regulatory-associated protein of mTOR (raptor)19, mammalian lethal with Sec13 protein 8 (mLST8, also known as GβL)20, praline-rich AKT substrate 40 kDa (PRAS40)21, and DEP-domain-containing mTOR interacting protein (Deptor)22. AMPK is upstream of mTORC1 in energy starvation-induced cellular response23,24. Energy depletion activates AMPK, which increases TSC2’s GTPase-activation protein (GAP) activity by phosphorylation of TSC225. TSC2 is a GAP of a small G-protein Rheb26, and Rheb is a key regulator of mTORC127,28. Since GTP form of Rheb activates mTORC1, TSC2 negatively regulates mTORC126,29,30. Direct phosphorylation of raptor by AMPK has been found and raptor phosphorylation also negatively regulates the activity of mTORC131. The p70 S6 kinase (S6K1) and eIF4E binding protein 1 (4EBP1) are key regulators of translation, and are the most well characterized targets of mTORC132. Phosphorylation of S6K and 4EBP1 by mTORC1 leads to increased levels of translation of specific mRNAs, which is part of the mechanism used by mTORC1 to regulate cell growth.
Crosstalk between the p38 and mTOR pathways has been reported. Phosphorylation of serine 1210 of TSC2 by MK2, a downstream kinase of p38α, creates a 14-3-3 binding site, which prevents TSC2 from inhibition of mTORC133. An involvement of the p38 pathway in H2O2- and other stimuli-induced mTORC1 activation was also reported recently34. Since both mTOR and p38 pathways are evolutionally conserved signal pathways, we are interested in their relationship during the cellular response to energy stress, and found that the p38β-PRAK cascade is essential for energy starvation-induced inactivation of mTORC1.
RESULTS
p38β is essential for energy depletion-induced inhibition of mTORC1
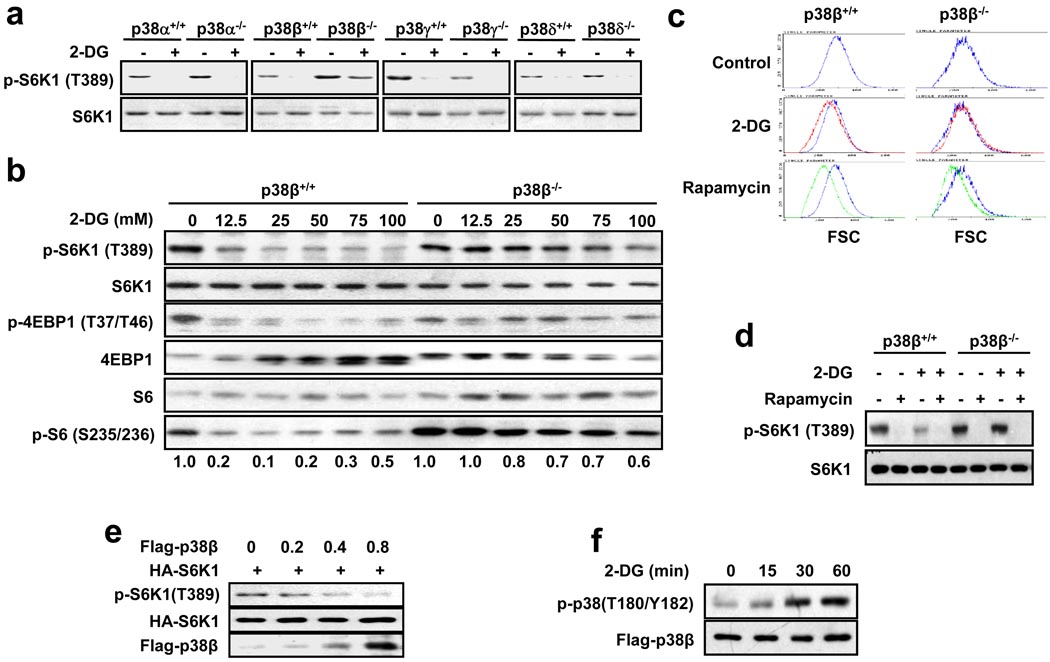
To determine the involvement of the p38 pathway in energy depletion-induced inactivation of mTORC1, we analyzed whether knockout of p38α, p38β, p38γ, or p38δ could affect 2-deoxy-glucose (2-DG)-induced dephosphorylation of p70 S6K1 (S6K1), an in vitro model for measuring energy depletion-induced inactivation of mTORC125. 2-DG-induced dephosphorylation of S6K1 in p38α, p38γ, and p38δ knockout MEF cells is similar to that in their corresponding p38α+/+, p38γ+/+, and p38δ+/+ MEF cells, but the dephosphorylation of S6K1 was blocked in p38β−/− MEF cells in comparison with p38β+/+ MEF cells (Fig. 1a), indicating that p38β is important for S6K1 dephosphorylation. 25 mM of 2-DG can effectively inhibit the phosphorylation of S6K1, 4EBP1, and S6 in wildtype MEF cells, but not in p38β−/− cells, even at higher 2-DG doses (Fig. 1b). Dynamic analysis also shows the impaired 2-DG response in p38β−/− cells (Supplementary Information, Fig. S1a). The p38β-specific effect on 2-DG-induced dephosphorylation of S6K1 was also observed in HEK293 cells (Supplementary Information, Fig. S1b). Since mTORC1 controls cell size35,36, we analyzed the size of wildtype and p38β−/− MEFs before and after 2-DG and rapamycin treatment. Interestingly, 2-DG did not reduce the size of p38β−/− cells as it did in wildtype MEF cells, while inhibition of mTORC1 by rapamycin reduced cell size in both cells (Fig. 1c). Consistent with the cell size data, rapamycin eliminated phospho-S6K1 in both cells, while 2-DG’s effect on phospho-S6K1 was impaired in p38β−/− cells (Fig. 1d). In support of the role of p38β in 2-DG-induced inactivation of mTORC1, we found that overexpression of p38β, but not other p38 group members, suppressed S6K1 phosphorylation (Fig. 1e, Supplemental Information, Fig. S1c), though 2-DG activated not only p38β but also some other p38 group members (Fig. 1f, Supplementary Information, Fig. S1d).
Figure 1.
p38β is essential for the 2-DG-induced cellular response. (a) 2-DG-induced dephosphorylation of S6K1 is compromised in p38β−/− MEF cells. p38α−/−, p38β−/−, p38γ−/−, p38δ−/−, and their corresponding wildtype (+/+) MEF cells were treated with or without 25 mM 2-DG for 60 minutes. Cell lysates were analyzed by immunoblotting for the levels of the indicated proteins and phosphorylation states. (b) p38β+/+ and p38β−/− MEF cells were treated with different doses of 2-DG for 30 minutes and analyzed as in a. The relative levels of phosphorylated S6 were determined by Image J software (Wayne Rasband, NIH, USA). (c) The 2-DG-induced but not rapamycin-induced cell size decrease is compromised in p38β−/− MEF cells. p38β+/+ and p38β−/− MEF cells were treated with nothing (control), 6.25 mM 2-DG, or 20 nM rapamycin for 60 hours. FACS analysis was performed to determine cell size. The X-axis indicates relative cell size. Cell size distribution of the control, as indicated by the blue line, is included in each image for comparison. (d) 2-DG-induced but not rapamycin-induced S6K1 dephosphorylation is compromised in p38β−/− MEF cells. The cells treated as in c were analyzed as in a to determine the phosphorylation levels of S6K1. (e) p38β overexpression suppresses S6K1 phosphorylation. The HA-S6K1 expression plasmid (0.1 µg) was co-transfected with 0, 0.2, 0.4, and 0.8 µg of the Flag-p38β expression plasmid into HEK293 cells. Cell lysates were analyzed as in a. (f) 2-DG induces phosphorylation of p38β. MEF cells were transfected with Flag-p38β and treated with 25 mM 2-DG for 0, 15, 30, and 60 minutes. Flag-p38β phosphorylation levels and Flag-p38β levels in Flag-immunoprecipitates were determined by Western blotting using anti-phospho-p38 and anti-Flag antibodies, respectively. (Note: Due to the similar size of p38 family members, there is currently no method available to specifically detect p38β phosphorylation. Therefore, we used transiently expressed Flag-p38β to determine whether 2-DG can activate p38β.)
PRAK is essential for energy depletion-induced inhibition of mTORC1
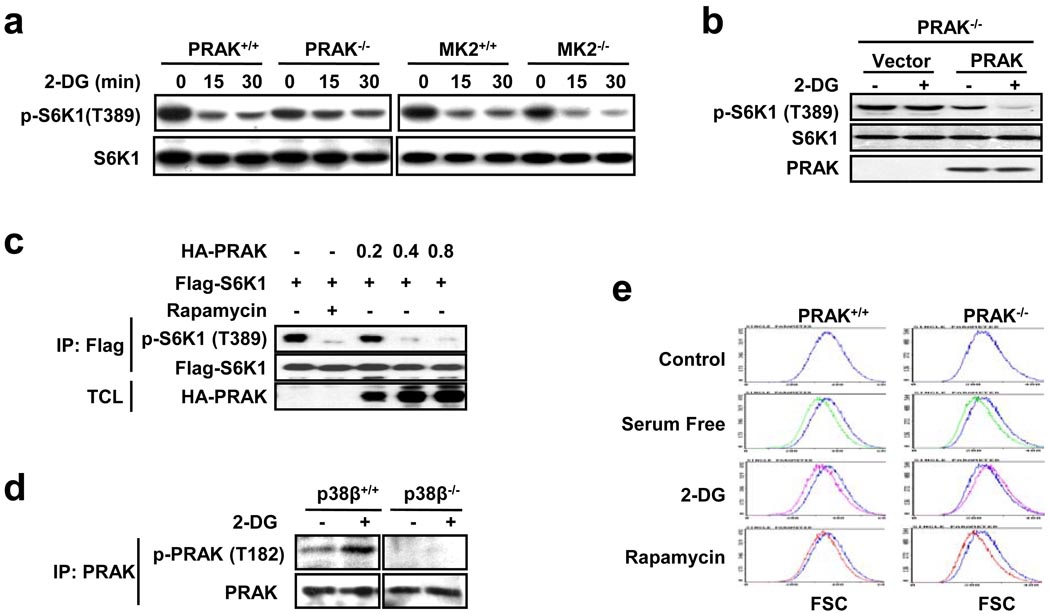
One means by which the p38 group kinases execute their function is through their downstream kinases. To evaluate the possibility that p38β regulates mTORC1 via activation of its downstream kinase, we analyzed whether deletion of PRAK37 or MK238,39, the kinases that can be activated by p38β40,41, affects 2-DG-induced dephosphorylation of S6K1 in MEF cells. Deletion of PRAK but not MK2 impaired 2-DG-induced dephosphorylation of S6K1 (Fig. 2a). In support of the role of PRAK in mTORC1 inactivation, reconstitution of PRAK expression in PRAK−/− MEFs restored 2-DG-induced dephosphorylation of S6K1 in PRAK−/− cells (Fig. 2b), and overexpression of PRAK inhibited S6K1 phosphorylation (Fig. 2c). Knockdown of PRAK but not MK2 in HEK293 cells also impaired 2-DG-induced dephosphorylation of S6K1 (Supplementary Information, Fig. S2a, S2b). 2-DG can activate PRAK, as demonstrated by T182 phosphorylation, and this activation is eliminated by p38β knockout (Fig. 2d), indicating that the p38β-mediated regulation of mTORC1 activity occurs at least partly through PRAK. As with p38β, PRAK is also required for the 2-DG-induced decrease in cell size and has no role in the serum deprivation- or rapamycin-induced change in cell size (Fig. 2e).
Figure 2.
PRAK is essential for the 2-DG-induced cellular response. (a) 2-DG-induced dephosphorylation of S6K1 is compromised in PRAK−/− MEF cells. PRAK+/+, PRAK−/−, MK2+/+, and MK2−/− MEF cells were treated with 25 mM 2-DG for 0, 15, or 30 minutes, then analyzed by immunoblotting for the levels of the indicated proteins and phosphorylation states. (b) Reconstitution of PRAK restores 2-DG-induced dephosphorylation of S6K1 in PRAK−/− MEF cells. PRAK−/− MEF cells were infected with an empty viral vector or a PRAK-expressing lentivirus 36 hours prior to a treatment with nothing or 25 mM 2-DG for 30 minutes. Phosphorylation of S6K1, S6K1 protein levels, and PRAK protein levels were determined by immunoblotting with corresponding antibodies. (c) PRAK overexpression suppresses S6K1 phosphorylation in a dose-dependent manner. The Flag-S6K1 expression plasmid (0.1 µg) was co-transfected with 0, 0.2, 0.4, and 0.8 µg of the HA-PRAK expression plasmid. Cells treated with 20 nM rapamycin were included as a control. Flag-S6K1 was immunoprecipitated by anti-Flag antibody-conjugated protein A/G-sepharose beads and phosphorylation of Flag-S6K1 was determined by immunoblotting with corresponding antibodies. IP: immunoprecipitates; TCL: total cell lysates. (d) Activation of PRAK by 2-DG is defective in p38β−/− MEF cells. p38β+/+ and p38β−/− MEF cells were treated with nothing or 25 mM 2-DG for 30 minutes. PRAK was immunoprecipitated with mouse anti-PRAK antibodies, then the phosphorylation of PRAK and PRAK protein levels were determined by immunoblotting with corresponding antibodies. (e) The 2-DG-induced, but not serum starvation- and rapamycin-induced cell size decrease is compromised in PRAK−/− MEF cells. PRAK+/+ and PRAK−/− MEF cells were treated with nothing (control), 6.25 mM 2-DG, or 20 nM rapamycin for 60 hours, or they were serum-starved for 36 hours. Cells were harvested for FACS analysis to determine cell size. The X-axis indicates relative cell size. The cell size distribution curve of the control, as indicated by the blue line, is included in each image for comparison purposes.
p38β-PRAK cascade is selectively involved in certain energy depletion-induced mTORC1 inactivation
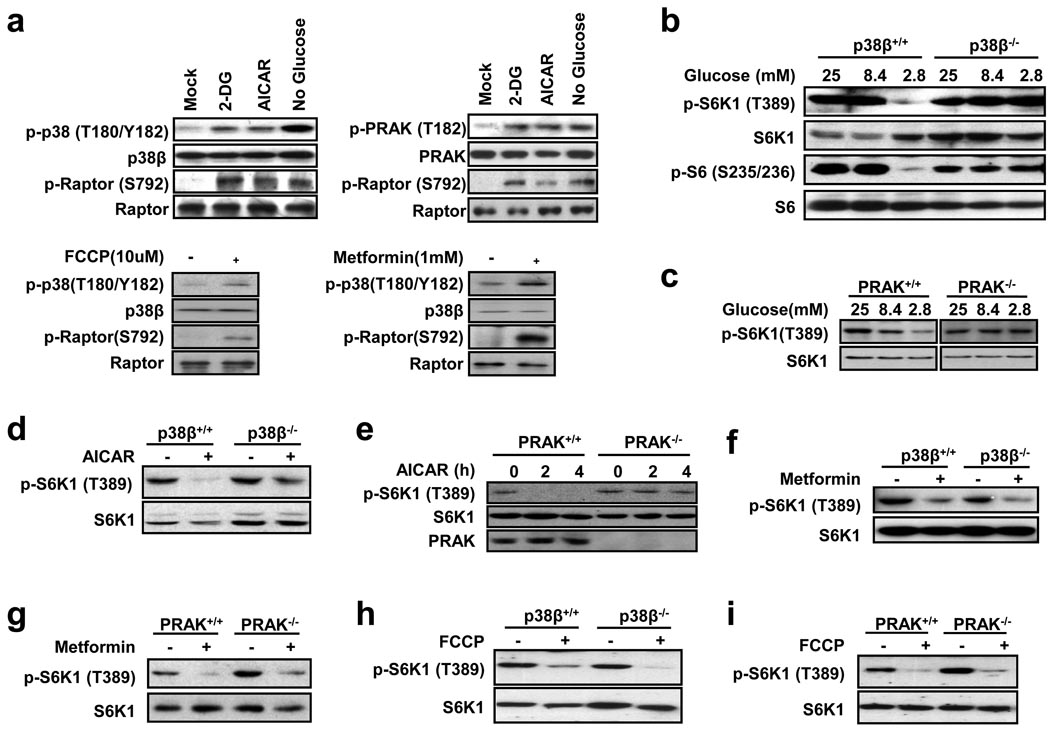
Since 2-DG functions via inhibition of glycolysis to cause energy depletion in cells, glucose deprivation in culture medium should induce similar energy starvation. As anticipated, glucose deprivation induced activation of p38β and PRAK, as indicated by the increase in phosphorylation levels of these proteins (Fig. 3a), and low glucose-induced dephosphorylation of S6K1 was impaired in p38β−/− and PRAK−/− cells (Fig. 3b, 3c). PRAK deletion sensitized MEF cells to glucose deprivation-induced death, and rapamycin treatment was able to rescue this phenotype (Supplementary Information, Fig. S2c). AMPK plays a key role in cellular energy homeostasis by sensing changes in the AMP:ATP ratio42. Its activity can be determined by phosphorylation of its substrate raptor. We treated cells with 5-aminoimidazole-4-carboxamide 1-β-D-ribofuranoside (AICAR) and metformin24, the compounds that can activate AMPK, and found that both can activate p38β and AMPK (Fig. 3a), but only AICAR-induced dephosphorylation of S6K1 was impaired in p38β−/− and PRAK−/− cells (Fig. 3d, 3e, 3f, 3g). We also examined carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP), an uncoupler of oxidative phosphorylation, and found that it activated p38β and AMPK (Fig. 3a), but its mediated dephosphorylation of S6K1 was not p38β-PRAK dependent (Fig. 3h, 3i). It is unclear why p38β-PRAK has no role in metformin- and FCCP-induced inactivation of mTORC1. A recent report showed that the metformin-induced inactivation of mTORC1 is independent from AMPK43, which is consistent with our observation that the mechanism used by metformin to induce mTORC1 inactivation is different from that used by AICAR.
Figure 3.
p38β and PRAK selectively participate in certain forms of energy depletion-induced mTORC1 inactivation. (a) MEF cells were infected with a lentivirus expressing p38β (top left and bottom panels) or PRAK (top right panel), and treated with 50 mM 2-DG for 30 minutes, 2 mM AICAR for 4 hours, glucose free medium for 24 hours, 10 µM FCCP for 15 minutes, or 1 mM metformin for 24 hours. Phosphorylation and protein levels of p38β, PRAK, and raptor were determined by immunoblotting with corresponding antibodies. (b) Glucose starvation-induced dephosphorylation of S6K1 is compromised in p38β-deficient MEF cells. p38β+/+ and p38β−/− MEF cells were cultured in DMEM containing 25, 8.4, or 2.8 mM glucose for 24 hours. Phosphorylation and protein levels of S6K1 or S6 were determined by immunoblotting with corresponding antibodies. (c) Glucose starvation-induced dephosphorylation of S6K1 is compromised in PRAK-deficient MEF cells. The same as in b except PRAK+/+ and PRAK−/− MEF cells were used. (d) p38β+/+ and p38β−/− MEF cells were stimulated with 0.5 mM AICAR for 4 hours. Cell lysates were analyzed by immunoblotting for protein and phosphorylation levels of S6K1. (e) The same as in d except PRAK+/+ and PRAK−/− MEF cells were used. (f) Metformin-induced dephosphorylation of S6K1 is not affected by p38β deficiency in MEF cells. p38β+/+ and p38β−/− MEF cells were stimulated with 1 mM metformin for 24 hours. Cell lysates were analyzed by immunoblotting for protein and phosphorylation levels of S6K1. (g) Metformin-induced dephosphorylation of S6K1 is not affected by PRAK deficiency in MEF cells. The same as in f except PRAK+/+ and PRAK−/− MEF cells were used. (h) FCCP-induced dephosphorylation of S6K1 is not affected by p38β deficiency in MEF cells. p38β+/+ and p38β−/− MEF cells were stimulated with 10 µM FCCP for 15 minutes. Phosphorylation and protein levels of S6K1 were determined by immunoblotting with corresponding antibodies. (i) FCCP-induced dephosphorylation of S6K1 is not affected by PRAK deficiency in MEF cells. The same as in h except PRAK+/+ and PRAK−/− MEF cells were used.
We also tested other stimuli that can affect mTORC1 activity and found that PRAK deletion has little or no effect on the insulin-induced phosphorylation of S6K1 (Supplementary Information, Fig. S3a), or on the serum or amino acid depletion-mediated dephosphorylation of S6K1 (Supplementary Information, Fig. S3b, S3c). Rapamycin- and high osmolarity (sorbitol)-mediated dephosphorylation of S6K1 were also not affected by PRAK knockout (Supplementary Information, Fig. S3c). Collectively, the p38β-PRAK pathway selectively participates in 2-DG-, AICAR-, and glucose deprivation-induced inactivation of mTORC1.
p38β-PRAK cascade is independent from AMPK and TSC2
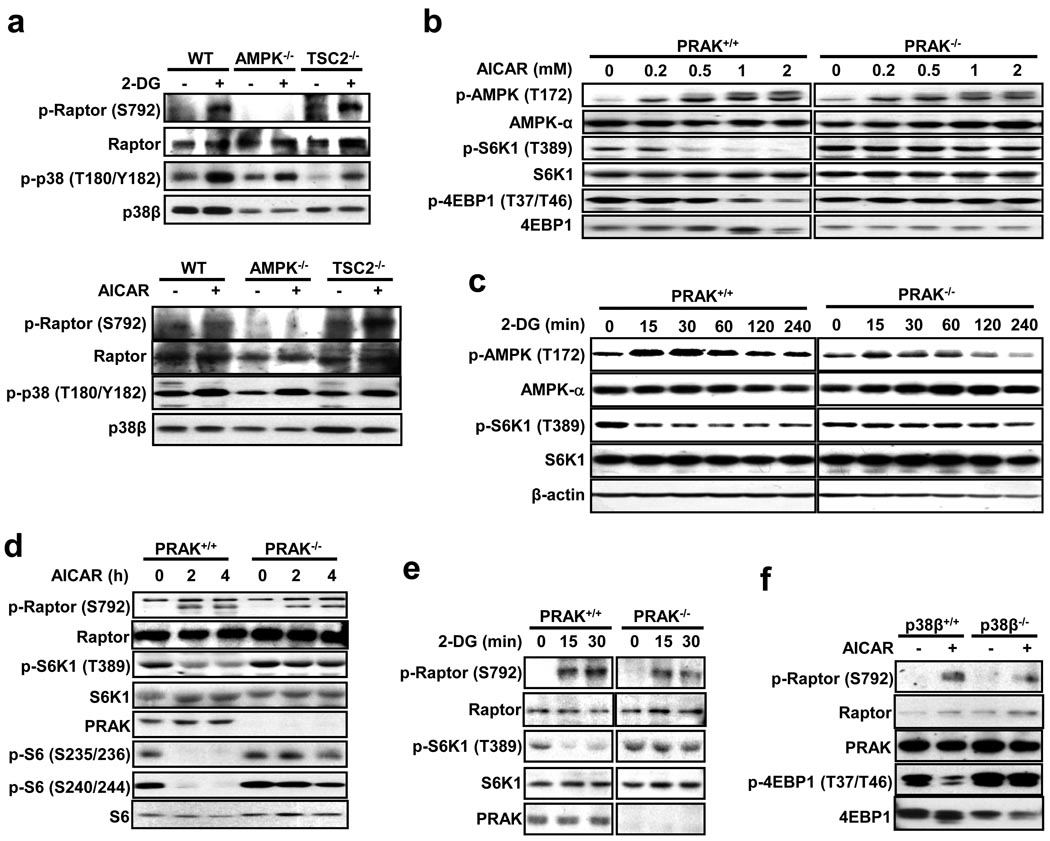
The p38β-PRAK cascade should function upstream of mTORC1, since deletion of p38β or PRAK does not affect rapamycin-mediated dephosphorylation of S6K1 and cell size reduction (Fig. 1c, 1d, 2e). To determine whether p38β is downstream of AMPK in suppressing mTORC1, we used AMPKα1/α2 double knockout MEFs. Consistent with published data31, 2-DG and AICAR were not able to induce raptor phosphorylation in AMPK-deficient cells (Fig. 4a). Since 2-DG- and AICAR-induced p38β phosphorylation was not affected by AMPK deletion (Fig. 4a), AMPK is not upstream of p38β activation. Consistently, p38β phosphorylation was also not affected by deletion of TSC2, a downstream target of AMPK (Fig. 4a). Because treatment with AICAR or 2-DG induced similar T172 phosphorylation of AMPK in wildtype and PRAK−/− MEF cells (Fig. 4b, 4c), p38β-PRAK should have no role in AMPK activation. The activation of AMPK by AICAR and 2-DG in wildtype and PRAK−/− cells was confirmed by the phosphorylation of the AMPK substrate raptor (Fig. 4d, 4e). Similar data was obtained using p38β−/− cells (Fig. 4f). Thus, AMPK and p38β-PRAK are parallel pathways.
Figure 4.
PRAK's function in suppressing mTORC1 is independent from AMPK. (a) MEF cells with the indicated gene deletions were infected with a lentivirus expressing p38β and stimulated with or without 25 mM 2-DG for 30 minutes or 2 mM AICAR for 4 hours. Cell lysates were analyzed by immunoblotting for the protein and phosphorylation levels of raptor and p38β. (b) AICAR-induced AMPK phosphorylation is comparable in PRAK+/+ and PRAK−/− cells. PRAK+/+ and PRAK−/− MEF cells were stimulated with different doses of AICAR for 2 hours. Phosphorylation and protein levels of AMPK, S6K1, and 4EBP1 were determined by immunoblotting with corresponding antibodies. (c) 2-DG-induced AMPK activation in MEF cells is not affected by PRAK knockout. PRAK+/+ and PRAK−/− MEF cells were treated with 25 mM 2-DG for 0, 15, 30, 60, 120, or 240 minutes. Cell lysates were analyzed by immunoblotting for the levels of the indicated proteins and their phosphorylation states. (d) PRAK+/+ and PRAK−/− MEF cells were treated with 0.5 mM AICAR for 0, 2, and 4 hours. Cell lysates were analyzed by immunoblotting for the levels of the indicated proteins and their phosphorylation states. (e) 2-DG-induced raptor phosphorylation is not compromised in PRAK-deficient MEF cells. PRAK+/+ and PRAK−/− MEF cells were treated with 25 mM 2-DG for 0, 15, or 30 minutes. Cell lysates were analyzed by immunoblotting for the levels of the indicated proteins and their phosphorylation states. (f) AICAR-induced raptor phosphorylation is not compromised in p38β-deficient MEF cells. p38β+/+ and p38β−/− MEF cells were stimulated with 0.5 mM AICAR for 4 hours. Cell lysates were analyzed by immunoblotting for the levels of the indicated proteins and their phosphorylation states.
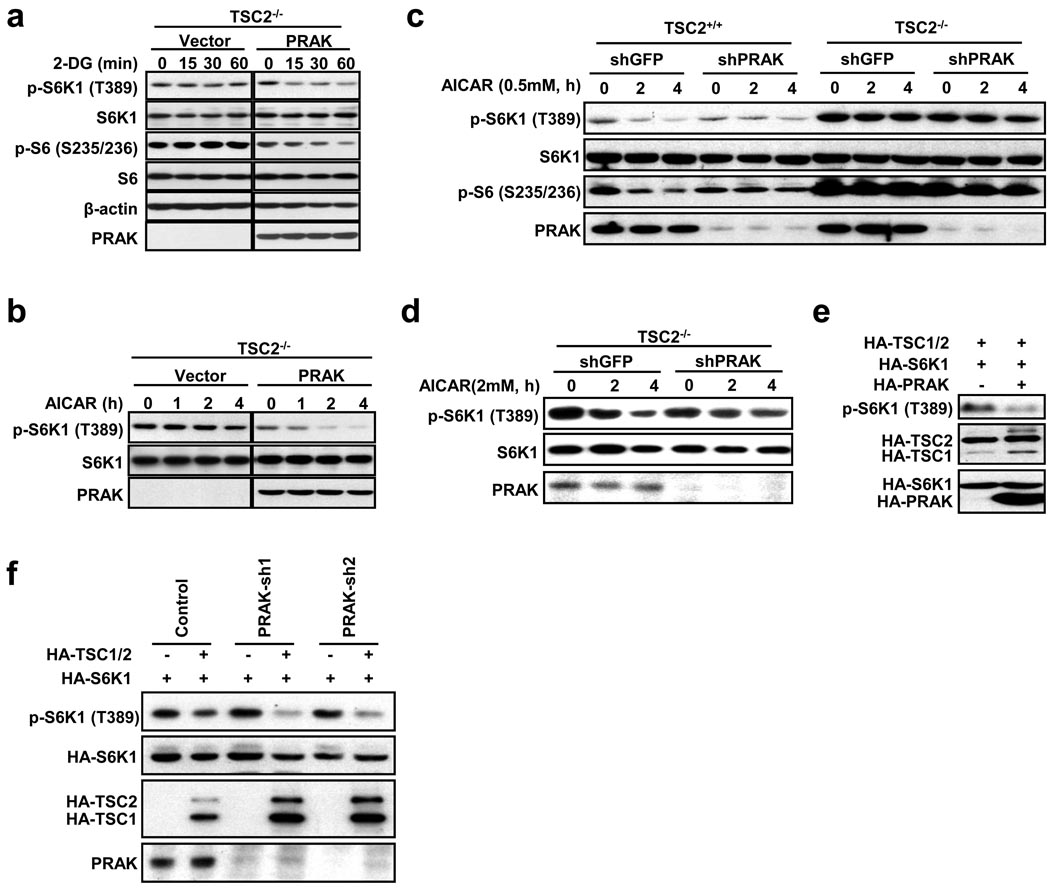
TSC2 is a suppressor of mTORC1 and acts downstream of AMPK25. It is known that 2-DG- or AICAR-induced dephosphorylation of S6K1 is impaired in TSC2−/− cells25. We overexpressed PRAK in TSC2−/− cells and found that both 2-DG and AICAR can downregulate S6K1 phosphorylation in PRAK-overexpressing TSC2−/− cells, but not in the vector-transfected TSC2−/− cells (Fig. 5a, 5b), indicating that overexpressed PRAK can compensate for the loss of TSC2 function in TSC2−/− cells. Since a high dose (2 mM) of AICAR can suppress S6K1 phosphorylation in TSC2−/− cells31, we tested whether PRAK knockdown could make TSC2−/− cells more resistant to AICAR. TSC2−/− cells were resistant to low dose (0.5 mM) AICAR-induced inactivation of mTORC1 and, as anticipated, we were not able to see any effect of PRAK knockdown (Fig. 5c). We observed that knockdown of PRAK in TSC2−/− cells enhances the resistance to 2 mM AICAR in these cells (Fig. 5d), consistent with the conclusion that PRAK is in parallel with TSC2 in regulating mTORC1. In addition, we observed that expression of PRAK further suppresses S6K1 phosphorylation in the presence of overexpressed TSC2 in wildtype cells (Fig. 5e), and that TSC2 overexpression-mediated inhibition of S6K1 phosphorylation does not require PRAK (Fig. 5f). Collectively, our data show that TSC2 and PRAK are independent from each other and can compensate for each other in the inactivation of mTORC1.
Figure 5.
PRAK's function in suppressing mTORC1 is independent from TSC2. (a) TSC2−/− MEF cells were infected with vector or PRAK-expressing lentivirus. 48 hours post-infection, cells were treated with 2-DG for 0, 15, 30, or 60 minutes and analyzed by immunoblotting for the levels of the indicated proteins and their phosphorylation states. (b) TSC2−/− MEF cells were infected with an empty vector or PRAK-expressing lentivirus. Cells were treated with 0.5 mM AICAR for 0, 1, 2, or 4 hours. Cell lysates were analyzed as in a. (c) TSC2+/+ and TSC2−/− MEF cells were infected with lentiviruses expressing GFP shRNA or PRAK shRNA. Cells were then stimulated with 0.5 mM AICAR for 0, 2, or 4 hours. Cell lysates were analyzed as in a. (d) TSC2−/− MEF cells were infected with lentiviruses expressing GFP shRNA or PRAK shRNA. Cells were then stimulated with 2 mM AICAR for 0, 2, or 4 hours. Cell lysates were analyzed as in a. (e) PRAK further enhances S6K1 dephosphorylation in TSC1/2 overexpressing cells. HEK293 cells were transfected with plasmids as indicated. The level of phospho-S6K1 and HA-TSC1/2, HA-S6K1, and HA-PRAK were determined by Western blotting using anti-phospho-S6K1 and anti-HA antibodies, respectively. (f) TSC2 suppresses S6K1 phosphorylation in both PRAK wildtype and PRAK knockdown cells. HEK293 cells were infected with lentiviruses expressing control shRNA, PRAK-sh1, or PRAK-sh2. 48 hours post-infection, HA-S6K1 was co-transfected with an empty vector or HA-TSC1 plus HA-TSC2 (HA-TSC1/2). The cell lysates were analyzed as in e.
PRAK phosphorylates Rheb at serine 130 and reduces Rheb’s ability to activate mTORC1
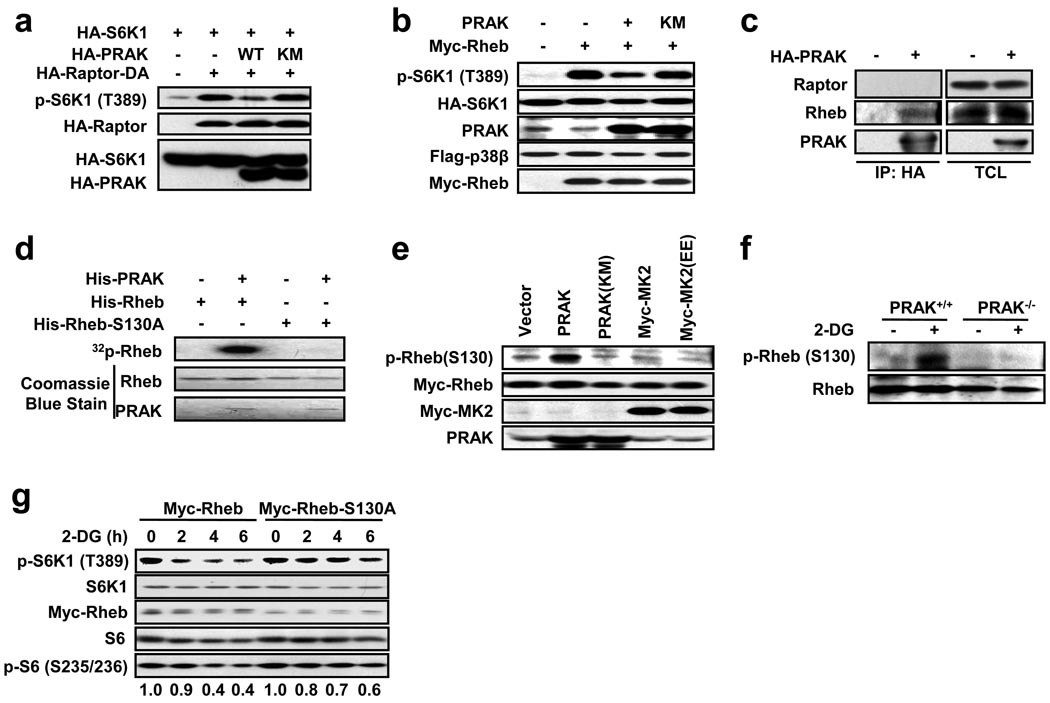
Raptor is a component of mTORC1 and is a target of AMPK in suppressing mTORC1 activity31. Neither a knockout cell line nor a dominant negative mutant of raptor is available; however, a fusion of raptor with the c-terminal 20 amino acid portion of Rheb can create a dominant active raptor (Raptor-DA)44 (K-L Guan, unpublished data). Raptor-DA expression activated mTOR, as determined by increased phosphorylation of S6K1, and overexpression of wildtype PRAK, but not the inactive PRAK(KM), inhibited Raptor-DA-induced increase of S6K1 phosphorylation (Fig. 6a). Raptor-DA-induced S6K1 phosphorylation requires Rheb44. Expression of Rheb enhanced S6K1 phosphorylation, and overexpression of PRAK, but not PRAK(KM), inhibited Rheb-induced S6K1 phosphorylation (Fig. 6b), suggesting that PRAK is in parallel with Raptor-Rheb or directly regulates Rheb. We then found that PRAK can directly interact with Rheb but not Raptor, as immunoprecipitation of HA-PRAK in HEK293 cells pulls down endogenous Rheb protein (Fig. 6c). As with other kinase-substrate interactions, a kinase-dead mutant of PRAK (PRAK(KM)) was able to more efficiently pull down Rheb when these two proteins were coexpressed (Supplementary Information, Fig. S4a). An in vitro kinase assay showed that PRAK phosphorylates Rheb (Fig. 6d), suggesting that Rheb is a substrate of PRAK. To identify the phosphorylation site on Rheb, PRAK-phosphorylated Rheb was subjected to phosphopeptide mapping by mass spectrometry. Spectrum searches revealed two potential phosphorylation sites on Rheb (Supplementary Information, Fig. S4b). We mutated these sites in Rheb to alanine (A) and found that only the mutation of serine 130 (S130) abolished the phosphorylation of Rheb by PRAK (Fig. 6d, Supplementary Information, Fig. S4c), demonstrating that PRAK phosphorylates S130 on Rheb. In order to determine whether S130 phosphorylation occurs in cells, we generated anti-S130-phospho-Rheb antibodies. Coexpression of Rheb with PRAK, but not PRAK(KM), MK2, or an active MK2 mutant (MK2(EE)), increased Rheb S130 phosphorylation (Fig. 6e). S130 phosphorylation of endogenous Rheb by PRAK overexpression was also observed (Supplementary Information, Fig. S4d). These data indicate that S130 is an in vivo phosphorylation site of PRAK. S130 in Rheb is the only in vivo phosphorylation site targeted by PRAK, as coexpression of PRAK but not PRAK(KM) with Rheb led to a single band shift on the Phos-tag gel (Supplementary Information, Fig. S4e). No band shift was observed in S130A (A) Rheb and S130A/T44A (AA) Rheb (Supplementary Information, Fig. S4e), confirming the phosphorylation at S130. Therefore, PRAK phosphorylates a single S130 site on Rheb.
Figure 6.
PRAK regulates Rheb by phosphorylation of Rheb on serine 130. (a) PRAK suppresses dominant active raptor (Raptor-DA)-mediated S6K1 phosphorylation. HEK293 cells were transfected with plasmids as indicated. The levels of phospho-S6K1, HA-raptor, HA-S6K1, and HA-PRAK were determined by immunoblotting with corresponding antibodies. (b) PRAK inhibits Rheb-induced phosphorylation of S6K1. HEK293 cells were transfected with expression plasmids of Flag-p38β, HA-S6K1, PRAK, and Myc-Rheb as indicated. Cell lysates were analyzed as in a. (c) HA-PRAK interacts with endogenous Rheb. The HA-PRAK expression vector was transfected into HEK293 cells. HA-PRAK was immunoprecipitated with anti-HA conjugated protein A/G-sepharose beads. The immunoprecipitates (IP) and total cell lysate (TCL) were analyzed by immunoblotting for HA-PRAK, Rheb, and raptor. (d) PRAK phosphorylates Rheb on S130 in vitro. His-Rheb or His-Rheb-S130A was incubated without or with PRAK in a kinase buffer containing γ-32P-ATP. Phosphorylation of Rheb was detected by autoradiography. Proteins of Rheb and PRAK were determined by Coomassie blue staining. (e) PRAK, but not MK2, phosphorylates Rheb in mammalian cells. The expression plasmids for PRAK, PRAK(KM), Myc-MK2, or Myc-MK2(EE) were co-transfected with the Myc-Rheb expression plasmid into HEK293 cells, as indicated. Cell lysates were analyzed by immunoblotting with corresponding antibodies. (f) PRAK is required for 2-DG-induced Rheb phosphorylation. PRAK+/+ and PRAK−/− MEF cells were stimulated with or without 25 mM 2-DG for 30 minutes. Phosphorylation of Rheb and Rheb protein levels were determined by immunoblotting using anti-phospho-Rheb and anti-Rheb antibodies, respectively. (g) Rheb-S130A inhibits 2-DG-induced dephosphorylation of S6K1. HEK293 cells were transfected with wildtype Myc-Rheb or Myc-Rheb-S130A 24 hours prior to treatment with 25 mM 2-DG for 0, 2, 4, or 6 hours. The levels of phospho-S6K1, S6K1, Myc-Rheb, phospho-S6, and S6 were determined by immunoblotting with their corresponding antibodies. The relative levels of phosphorylated S6 were determined by Image J software.
We then analyzed whether 2-DG or AICAR induces Rheb phosphorylation and found that 2-DG and AICAR induced endogenous Rheb phosphorylation in wildtype but not PRAK−/− MEF cells (Fig. 6f, Supplementary Information, Fig. S4f). Overexpression of wildtype Rheb in HEK 293 cells did not affect 2-DG-induced S6K1 dephosphorylation, while overexpression of Rheb-S130A inhibited 2-DG-induced S6K1 dephosphorylation (Fig. 6g), suggesting that the S130A mutant has a dominant negative effect on 2-DG-induced response. Consistently, overexpression of PRAK can suppress Rheb-mediated S6K1 phosphorylation, but has no effect on Rheb-S130A-mediated S6K1 phosphorylation (Supplementary Information, Fig. S4g). The phosphorylated form of Rheb should be less capable of inducing S6K1 phosphorylation, since the phospho-mimic mutant Rheb S130E had less effect on S6K1 phosphorylation (Supplementary Information, Fig. S4h). These data indicate that phosphorylation of S130 on Rheb reduces its ability to activate mTORC1.
Phosphorylation of Rheb by PRAK decreases Rheb’s ability in guanine nucleotides binding
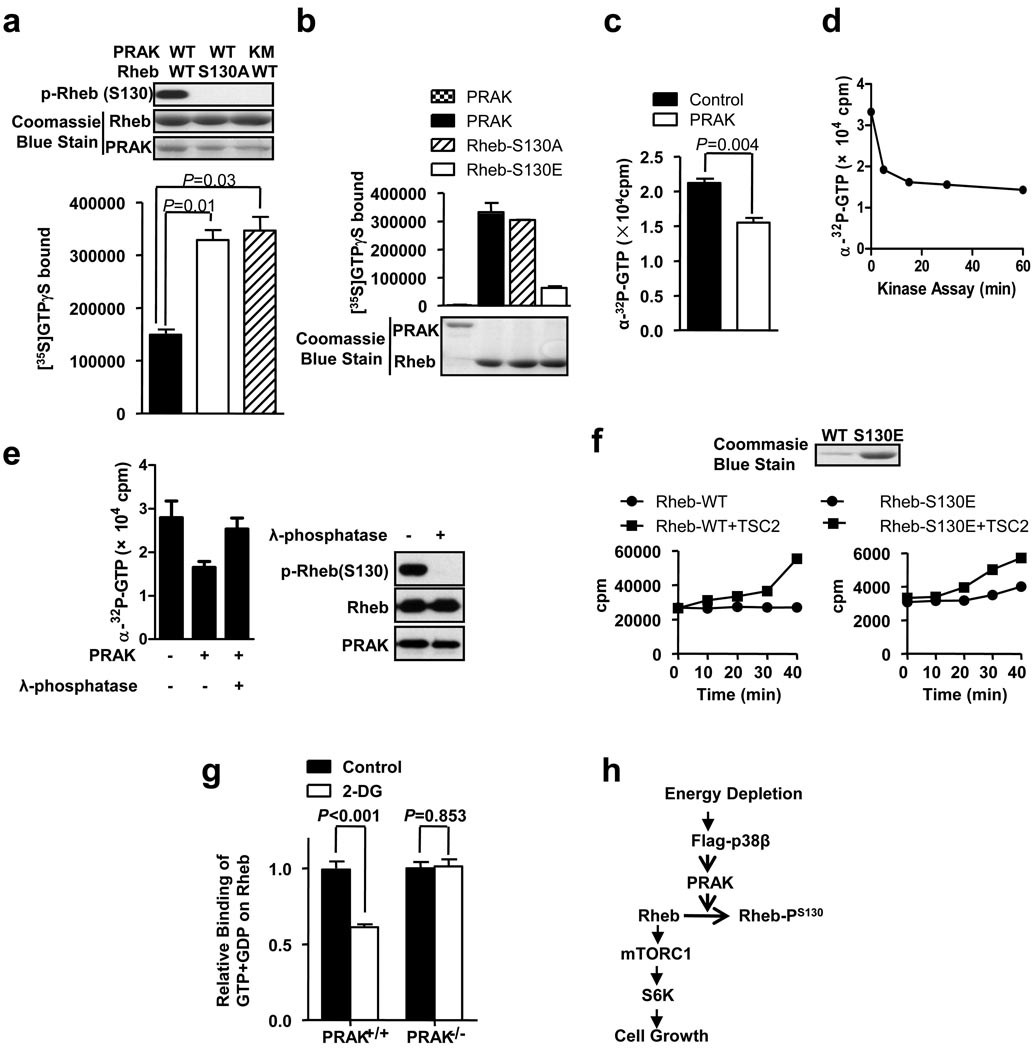
In attempts to understand why S130 phosphorylation inactivates Rheb, we analyzed the amount of nonhydrolyzable GTP (GTPγS) loading on recombinant Rheb or Rheb-S130A in the presence of wildtype PRAK or KM mutant of PRAK in vitro. The amount of GTPγS that is bound to the wildtype PRAK-treated Rheb is about half of that which is bound to the nonphosphorylatable mutant Rheb-S130A or inactive PRAK-treated sample (Fig. 7a). Because Rheb cannot hydrolyze GTP in the absence of its GAP TSC230, a similar result was obtained when α-32P-GTP was used (Supplementary Information, Fig. S5a). Given the fact that only about half of Rheb was phosphorylated by PRAK in vitro (Supplementary Information, Fig. S5b), S130 phosphorylation should significantly impair GTP binding to Rheb. Because we already reached our limit to increase the activity of recombinant PRAK (see supplemental materials), we were unable to phosphorylate all Rheb in vitro, and therefore cannot obtain a precise percentage of inhibition. We also analyzed the GTPγS binding of Rheb mutants. GTPγS loading in S130A was about the same as that in wildtype Rheb, GTPγS loading in S130E was much less than that in wildtype, and as a negative control, PRAK protein had no GTPγS binding (Fig. 7b). The GDP loading was also less in S130E (Supplementary Information, Fig. S5c). Thus, the phosphorylation of S130 decreases guanine nucleotides binding to Rheb.
Figure 7.
Rheb phosphorylation by PRAK reduces Rheb’s nucleotide binding ability. (a) Phosphorylation of Rheb in vitro reduces its GTP binding. GST-PRAK or GST-PRAK(KM) was treated with His-p38β plus His-MKK6b(E), and then was purified by GST pull-down. GST-Rheb or GST-Rheb(S130A) was incubated with or without activated GST-PRAK or GST-PRAK(KM) in a kinase buffer containing [35S]GTPγS. The samples were then applied to a GTP loading assay. Error bars indicate mean +/− s.e.m., n=3 samples, Student’s t-test. (b) The phospho-Rheb mimic mutant GST-Rheb-S130E bound much less GTP. GST-Rheb, GST-Rheb-S130A, and GST-Rheb-S130E were applied to a GTP loading assay (upper panel). The amounts of proteins are shown in the lower panel. (c) Phosphorylation of Rheb releases bound GTP. GST-Rheb was loaded with α-32P-GTP first, and was then treated with or without activated PRAK for 60 min at 30 °C. The bound GTP was determined. (d) The reduction of bound α-32P-GTP during Rheb phosphorylation. α-32P-GTP-loaded GST-Rheb was incubated with or without activated GST-PRAK for 0, 5, 15, 30, or 60 min at 30 °C. The bound GTP was determined. (e) Dephosphorylation of phospho-Rheb recovers Rheb’s α-32P-GTP binding ability. α-32P-GTP-loaded GST-Rheb was treated with or without PRAK and then treated with or without λ-phosphatase, as indicated. The bound GTP was determined (left panel). The levels of phospho-Rheb, Rheb, and PRAK were shown in right panel. (f) Rheb-wt or Rheb-S130E was applied in a GTP hydrolysis assay in the absence and presence of HA-TSC2 (left two panels). The Coomassie blue staining of Rheb-wt and Rheb-S130E protein is shown in right panel. (g) PRAK+/+ and PRAK−/− MEF cells were metabolically labeled with 32P-phosphate, then treated with or without 25mM 2-DG for 30 min. Endogenous Rheb was immunoprecipitated. GTP and GDP bound to Rheb was analyzed by thin layer chromatography and quantitated by a Phosphorimager. Data are represented as the mean +/− s.e.m., n=4 samples, Student’s t-test. The relative binding of GTP+GDP on Rheb was calculated by setting the binding of GTP+GDP on Rheb in control groups to 1.0. (h) A proposed model of the p38β-PRAK cascade in regulating the mTORC1 pathway following energy starvation.
Since Rheb may already bind with GTP before being phosphorylated by PRAK in cells, we loaded GTP onto Rheb first and then analyzed the effect of PRAK phosphorylation of Rheb on the amount of GTP bound to Rheb. Phosphorylation of Rheb by PRAK released GTP that was bound to Rheb (Fig. 7c). Most of the bound GTP was released within 5 minutes (Fig. 7d). Thus, S130 phosphorylation of Rheb not only made Rheb less capable of accepting GTP loading, but also reduced its ability to retain the already-bound GTP. To further evaluate this notion, we used λ phosphatase to remove the phosphorylation from in vitro phosphorylated Rheb and found that dephosphorylation of phospho-Rheb increased GTP binding (Fig. 7e). To determine whether S130 phosphorylation affects Rheb’s GTP hydrolysis activity, we analyzed GTP hydrolysis in wildtype Rheb and Rheb-S130E in the presence or absence of TSC2. Because of the reduced GTP binding capacity by Rheb S130E, we used an excess of Rheb-S130E in comparison with wildtype Rheb in the experiment (Right panel of Fig. 7f). Both Rheb and Rheb-S130E alone had little or no GTP hydrolyzing activity, but both can hydrolyze GTP in the presence of TSC2 (Fig. 7f). Because of the difference of GTP binding activity, we cannot determine whether S130 phosphorylation affects the hydrolysis activity of Rheb. To determine whether PRAK can regulate Rheb in culture cells, we metabolically labeled wildtype and PRAK−/− cells with 32P-phosphate. The cells were treated with or without 2-DG for 30 min, Rheb molecules were immunoprecipitated, and bound nucleotides were analyzed. Ras was used as a control. The ratio between GTP and GDP is about the same as in wildtype and PRAK−/− cells, and is slightly increased after 2-DG treatment in both cells (Supplementary Information, Fig. S5d). 2-DG reduced Rheb’s guanine nucleotides binding in wildtype cells but not in PRAK−/− cells (Fig. 7g, Supplementary Information, Fig. S5e), while 2-DG had no effect on Ras’s GTP binding (Supplementary Information, Fig. S5f), confirming the selective regulation of Rheb by PRAK.
Discussion
Previously published studies and our data described above indicate that the routes of AMPK-TSC2, AMPK-raptor, and p38β-PRAK are independent events that mediate the suppression of mTORC1. The kinetics of their activation seems to have some differences. For example, the activation of p38β-PRAK occurs later than the initial reduction of mTORC1 activity in energy-starved cells (Fig. S1a, 1f), suggesting that TSC2 or raptor suppresses mTORC1 during the early time points. It is important to note that deletion of either TSC2 or PRAK is sufficient to impair energy depletion-induced inhibition of mTORC1 (no data for raptor, due to raptor−/− cells not being available), suggesting that these parallel signaling events are not simply additive. In addition, the overexpression experiments showed that any of these three genes by itself can modulate mTORC1 activity. Overactivation of one pathway can even compensate for the insufficiency of the others, as we observed that overexpression of PRAK is sufficient to suppress mTORC1 in the absence of TSC2 (Fig. 5a, 5b), overexpression of TSC2 is enough to inhibit mTORC1 regardless of the presence or absence of PRAK (Fig. 5f), and overexpression of TSC2 in PRAK−/− cells can partially rescue the AICAR-induced decrease of S6K1 phosphorylation (Supplementary Information, Fig. S6). It is possible that in order to inactivate mTORC1, the total activity of these pathways needs to reach a threshold. These three pathways could also operate in a network of offsetting inputs that amplify each other in inactivating mTORC1. It is clear that this is a complicated regulatory system and many other possibilities remain.
mTOR receives input from multiple signaling pathways, including those that regulate growth factors, nutrients, and cellular stresses. Recently, Cully et al. reported that the p38 pathway participates in H2O2 and other stimuli induced activation of mTORC1, and Drosophila p38b plays a role in mTORC1 activation34. We tested the role of p38β in H2O2-mediated changes of mTORC1. We observed suppression of mTORC1 by H2O2 (Supplementary Information, Fig. S7a), which is consistent with previous reports45. We found that p38β had no role in this response (Supplementary Information, Fig. S7b). On the other hand, we observed that serum starvation reduced basal levels of phospho-S6K1, and under this condition, H2O2 can increase the phosphorylation of S6K1 (Supplementary Information, Fig. S7c), which is similar to the observation by Cully et al.34. We determined that H2O2-induced mTORC1 activation is not dependent on p38β (Supplementary Information, Fig. S7d). Since Drosophila p38b has the same level of homology to mammalian p38α and p38β, we then analyzed involvement of p38α. H2O2-induced reduction of S6K1 phosphorylation in the cells cultured in serum-containing medium was not affected by p38α deletion (Supplementary Information, Fig. S7e). Under serum-starved conditions, the basal level of phospho-S6K1 in p38α−/− MEF cells was higher than that in wildtype cells, and H2O2-induced increase of S6K1 phosphorylation was impaired in p38α−/− cells (Supplementary Information, Fig. S7f). Thus, it is possible that p38α is responsible for the H2O2-induced mTORC1 activation observed by Cully et al.34.
In this paper, we demonstrate that a specific p38 kinase cascade, p38β-PRAK, is essential for energy depletion-induced inactivation of mTORC1 (Fig. 7h). It was not anticipated that PRAK would be able to directly regulate a component of the mTORC1 pathway. It was also surprising that the GTP binding of Rheb was able to be regulated by phosphorylation on S130, as S130 is located between the second 310 helix and helix IV46, a region which is not within or even close to the GTP/GDP binding pocket. It is possible that S130 phosphorylation changes the relative position of the second 310 helix and its flanking loops, profoundly affecting the integrity of the GTP/GDP binding pocket, and leading to a dramatic decrease in nucleotide binding affinity. It is also possible that S130 phosphorylation affects the interaction between Rheb and its regulators. S130 phosphorylation should be specific for Rheb, as this site was not found in other small GTPases (Supplementary Information, Fig. S8a). RhebL1 has a similar function as Rheb, but does not have a corresponding S130 site47. We knocked down Rheb and RhebL1 in MEF cells and found that only Rheb knockdown reduced basal level phosphorylation of S6K1 (Supplementary Information, Fig. S8b). This could be due to the low expression of RhebL1. Since S130 in Rheb is conserved from Drosophila to humans (Supplementary Information, Fig. S8c), the regulation of Rheb by phosphorylation could be an evolutionarily conserved mechanism for regulating mTORC1 activity. It appears that cell growth and cellular energy levels need to be precisely coordinated, and cells have evolved multiple check points to regulate Rheb activity.
Methods
Methods and associated references are provided as supplementary information online.
Supplementary Material
Acknowledgements
We would like to thank Dr. Huiping Jiang (Boehringer Ingelheim Pharmaceuticals, Inc,) for providing p38 conditional knockout mice, Drs Benoit Viollet (Universite Paris Descartes) and Keith R. Laderoute (Stanford University School of Medicine) for AMPKα1/α2−/− cells. This work was supported by grants from NSF China 30830092, 30921005, 30828219, 973 program 2009CB22200, National Institutes of Health AI41637 and AI68896.
Footnotes
Author contributions
M.Z., Y W., X. W., S. W., and J.H. designed and performed the experiments. B. L. and M. D. performed MS analysis. M.Z., H. Z., P. S., S. L., K. G., and J. H. participated in the interpretation of the data. M.Z. and J.H. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Cuenda A, Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta. 2007;1773:1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 2.English JM, Cobb MH. Pharmacological inhibitors of MAPK pathways. Trends Pharmacol. Sci. 2002;23:40–45. doi: 10.1016/s0165-6147(00)01865-4. [DOI] [PubMed] [Google Scholar]
- 3.Nebreda AR, Porras A. p38 MAP kinases: beyond the stress response. Trends Biochem. Sci. 2000;25:257–260. doi: 10.1016/s0968-0004(00)01595-4. [DOI] [PubMed] [Google Scholar]
- 4.Ono K, Han J. The p38 signal transduction pathway: activation and function. Cell Signal. 2000;12:1–13. doi: 10.1016/s0898-6568(99)00071-6. [DOI] [PubMed] [Google Scholar]
- 5.Han J, Lee JD, Bibbs L, Ulevitch RJ. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y, et al. Characterization of the structure and function of a new mitogen-activated protein kinase (p38beta) J. Biol. Chem. 1996;271:17920–17926. doi: 10.1074/jbc.271.30.17920. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Jiang Y, Ulevitch RJ, Han J. The primary structure of p38 gamma: a new member of p38 group of MAP kinases. Biochem. Biophys. Res. Commun. 1996;228:334–340. doi: 10.1006/bbrc.1996.1662. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y, et al. Characterization of the structure and function of the fourth member of p38 group mitogen-activated protein kinases, p38delta. J. Biol. Chem. 1997;272:30122–30128. doi: 10.1074/jbc.272.48.30122. [DOI] [PubMed] [Google Scholar]
- 9.Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- 10.Rincon M, Davis RJ. Regulation of the immune response by stress-activated protein kinases. Immunol. Rev. 2009;228:212–224. doi: 10.1111/j.1600-065X.2008.00744.x. [DOI] [PubMed] [Google Scholar]
- 11.Reiling JH, Sabatini DM. Stress and mTORture signaling. Oncogene. 2006;25:6373–6383. doi: 10.1038/sj.onc.1209889. [DOI] [PubMed] [Google Scholar]
- 12.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat. Rev. Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 15.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 16.Jacinto E, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 17.Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 18.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 19.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 20.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Vander HE, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 22.Peterson TR, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardie DG. Role of AMP-activated protein kinase in the metabolic syndrome and in heart disease. FEBS Lett. 2008;582:81–89. doi: 10.1016/j.febslet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 24.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 26.Garami A, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 27.Aspuria PJ, Tamanoi F. The Rheb family of GTP-binding proteins. Cell Signal. 2004;16:1105–1112. doi: 10.1016/j.cellsig.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 28.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr. Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 30.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Um SH, D'Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Inoki K, Vacratsis P, Guan KL. The p38 and MK2 kinase cascade phosphorylates tuberin, the tuberous sclerosis 2 gene product, and enhances its interaction with 14-3-3. J. Biol. Chem. 2003;278:13663–13671. doi: 10.1074/jbc.M300862200. [DOI] [PubMed] [Google Scholar]
- 34.Cully M, et al. A role for p38 stress-activated protein kinase in regulation of cell growth via TORC1. Mol. Cell Biol. 2010;30:481–495. doi: 10.1128/MCB.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potter CJ, Xu T. Mechanisms of size control. Curr. Opin. Genet. Dev. 2001;11:279–286. doi: 10.1016/s0959-437x(00)00191-x. [DOI] [PubMed] [Google Scholar]
- 37.New L, et al. PRAK, a novel protein kinase regulated by the p38 MAP kinase. EMBO J. 1998;17:3372–3384. doi: 10.1093/emboj/17.12.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freshney NW et al. Interleukin-1 activates a novel protein kinase cascade that results in the phosphorylation of Hsp27. Cell. 1994;78:1039–1049. doi: 10.1016/0092-8674(94)90278-x. [DOI] [PubMed] [Google Scholar]
- 39.Rouse J, et al. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell. 1994;78:1027–1037. doi: 10.1016/0092-8674(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 40.Li Q, et al. Determinants that control the distinct subcellular localization of p38alpha-PRAK and p38beta-PRAK complexes. J. Biol. Chem. 2008;283:11014–11023. doi: 10.1074/jbc.M709682200. [DOI] [PubMed] [Google Scholar]
- 41.New L, Jiang Y, Han J. Regulation of PRAK subcellular location by p38 MAP kinases. Mol. Biol. Cell. 2003;14:2603–2616. doi: 10.1091/mbc.E02-08-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 43.Kalender A, et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 2010;11:390–401. doi: 10.1016/j.cmet.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw RJ, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Y, et al. Structural basis for the unique biological function of small GTPase RHEB. J. Biol. Chem. 2005;280:17093–17100. doi: 10.1074/jbc.M501253200. [DOI] [PubMed] [Google Scholar]
- 47.Yuan J, et al. Identification and characterization of RHEBL1, a novel member of Ras family, which activates transcriptional activities of NF-kappa B. Mol. Biol. Rep. 2005;32:205–214. doi: 10.1007/s11033-005-0984-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.