Abstract
In response to DNA double-strand breaks (DSBs), BRCA1 forms biochemically distinct complexes with certain other DNA damage response proteins. These structures, some of which are required for homologous recombination (HR)-type DSB repair, concentrate at distinct nuclear foci that demarcate sites of genome breakage. Polyubiquitin binding by one of these structures, the RAP80/BRCA1 complex, is required for efficient BRCA1 focal recruitment, but the relationship of this process to the execution of HR has been unclear. We found that this complex actively suppresses otherwise exaggerated, BRCA1-driven HR. By controlling the kinetics by which other BRCA1-interacting proteins that promote HR concentrate together with BRCA1 in nuclear foci, RAP80/BRCA1 complexes suppress excessive DSB end processing, HR-type DSB repair, and overt chromosomal instability. Since chromosomal instability emerges when BRCA1 HR function is either unbridled or absent, active tuning of BRCA1 activity, executed in nuclear foci, is important to genome integrity maintenance.
Keywords: BRCA1, RAP80, homologous recombination, ionizing radiation-induced foci
BRCA1 p220 is a tumor-suppressing protein that, when functionally inactivated, triggers the development of familial breast and ovarian cancer. At least a fraction of its tumor suppression function is linked to its ability to participate in genome integrity control. In particular, it supports the repair of double-strand DNA breaks (DSBs) by error-free homologous recombination (HR), and there are genetic links between its HR and its breast cancer suppression functions (Chen et al. 1999; Moynahan et al. 1999; Scully and Livingston 2000; Walsh and King 2007). Thus, the ability of BRCA1 to support HR appears to be of high clinical significance, although why a breakdown in this function would translate into a markedly increased risk of breast and ovarian as opposed to other cancers is unknown.
In cells exposed to ionizing radiation (IR), BRCA1 concentrates in distinctive nuclear foci (i.e., IR-induced foci [IRIF]). These structures, in part, demarcate sites of genome breakage (Scully et al. 1997a; Chen et al. 1998; Rogakou et al. 1999; Paull et al. 2000; Stewart et al. 2003). BRCA1 colocalizes in them together with phospho-H2AX (γH2AX), MDC1, and several other DNA damage response (DDR) proteins, some of which interact with BRCA1 to execute key steps in the HR process (Scully et al. 1997a; Chen et al. 1998; Cortez et al. 1999; Zhong et al. 1999; Paull et al. 2000; Goldberg et al. 2003; Xie et al. 2010). IRIF are composed, in part, of chromatin, and they are suspected of playing a critical role in a proper IR response (Bekker-Jensen et al. 2006; Stucki and Jackson 2006). However, how they operate in this regard is largely unclear.
Multiple groups, including our own, have reported that a ubiquitin-binding nuclear protein, RAP80/UIMC1, facilitates the recruitment of BRCA1 to IRIF, at least in part, by binding to specific K63- or K6-linked polyubiquitin structures at or near sites of DNA DSBs (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007; Yan et al. 2007). RAP80 depletion leads to a loss of most, albeit not all, BRCA1 at IRIF, a defective G2/M checkpoint, and increased cellular sensitivity to IR (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007; Yan et al. 2007). Why not all BRCA1 is concentrated at IRIF as a result of RAP80 function has also been unclear.
BRCA1/BARD1 heterodimers and RAP80 interact, and these complexes contain additional subunits, including Abraxas/CCDC98 (which contributes to the tethering of BRCA1 to RAP80), BRCC36 (a deubiquitinase), BRCC45/BRE, and MERIT40/NBA1 (Kim et al. 2007b; Liu et al. 2007; Sobhian et al. 2007; Wang and Elledge 2007; Wang et al. 2007, 2009; Feng et al. 2009; Shao et al. 2009). Although this multisubunit complex is clearly engaged in securing much of the ambient BRCA1 in IRIF, and thereby in maintaining certain aspects of a proper response to DSB, how, if at all, it operates with respect to BRCA1-driven DSB repair (DSBR) has also been unknown.
In this study, we describe the results of experiments aimed at exploring whether endogenous RAP80 influences endogenous BRCA1 HR function. Surprisingly, the data reveal that BRCA1 can elicit normal or excessive levels of HR function in cells, depending, respectively, on whether it is a normal component of an intact RAP80 complex, and whether or not this complex has been rendered functionally defective. Indeed, there is a strong correlation between the integrity of these complexes; their localization in IRIF; and the prevention of excessive, BRCA1-driven HR function. This implies, for the first time, that RAP80 complexes, likely docked at IRIF, contribute to a process that down-modulates BRCA1 HR function. Furthermore, overt genomic instability resulted when cells that had undergone a breakdown in the integrity of their RAP80 complexes acquired DSBs and expressed a hyperrecombination phenotype. Thus, either insufficient BRCA1-dependent HR (Moynahan et al. 1999, 2001; Scully et al. 2000; Thompson and Schild 2001) or, paradoxically, the emergence of excessive, BRCA1-dependent HR activity—each arising in an abnormal physiological setting—is associated with the development of overt chromosomal instability.
Results
Effect of RAP80 depletion on the relative concentration of various BRCA1 partner proteins in IRIF
Besides its interaction with RAP80-Abraxas, BRCA1 forms distinctive biochemical complexes upon interacting with other key proteins; i.e., BACH1 (also known as BRIP1 or FANCJ), CtIP, and PalB2-BRCA2 (Wong et al. 1998; Yu et al. 1998; Cantor et al. 2001; Greenberg et al. 2006; Xia et al. 2006; Sy et al. 2009; Zhang et al. 2009). In response to DNA damage, these three BRCA1-containing complexes join forces with several other proteins—including TopBP1, MRE11–RAD50–NBS1, and RAD51—to participate in the development of HR-type DSBR (Chen et al. 1999, 2008; Greenberg et al. 2006; Sartori et al. 2007). Like BRCA1/RAP80 complexes, these three sets of structures (i.e., complexes) also concentrate in IRIF (Greenberg et al. 2006; Sobhian et al. 2007; Wang et al. 2007). Thus far, it has not been possible to dissociate the IRIF localization of these complexes from their checkpoint/HR-associated functions, although which comes first after a DSB develops is unknown (Bekker-Jensen et al. 2006).
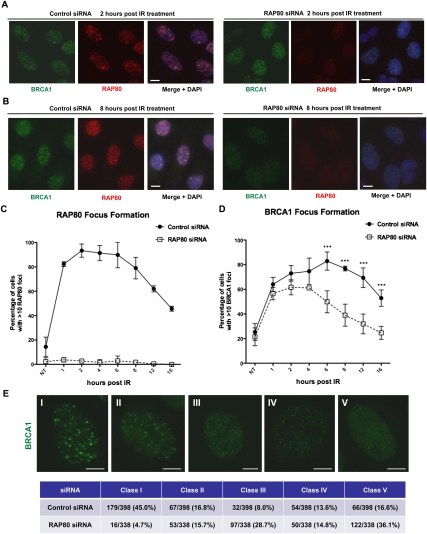
We examined the dynamics of nuclear focus (IRIF) formation by several BRCA1-interacting proteins in response to IR in the presence and absence of RAP80. IRIF were analyzed in cells before and at seven different time points after exposing them to 5 Gy IR (Fig. 1). Cell cycle analysis showed that the general distribution of the various cell cycle populations was not significantly different between cells exposed to control siRNA and those exposed to RAP80 siRNA within the 16-h experimental period (Supplemental Fig. S1A). Surprisingly, although RAP80 was effectively depleted during the entire time course (Supplemental Fig. S1B), and very few cells were positive for RAP80 IRIF by immunofluorescence staining (Fig. 1A–C), we observed comparable fractions of cells that displayed abundant BRCA1 IRIF 1 h after IR treatment in both control and RAP80-depleted cells (Fig. 1D). On the other hand, while the percentage of control cells with >10 BRCA1 IRIF per cell continued to increase and peaked ∼6 h post-IR, in RAP80-depleted cells, a leveling off followed by significant decreases of BRCA1 IRIF-positive cells were observed after the first hour (Fig. 1D). Conceivably, the BRCA1 IRIF detected at 1 h reflect the initial recognition of/interaction with DNA lesions by BRCA1 (Celeste et al. 2003; Bekker-Jensen et al. 2006). Indeed, consistent with data in our previous report (Sobhian et al. 2007), at 1 h after DSB formation, BRCA1 remained weakly associated with the center (as opposed to the entire γH2AX-staining structure) of laser-induced DNA damage stripes generated in RAP80-depleted cells (Supplemental Fig. S1C). This segment is where, as others have suggested, the damaged DNA is concentrated (Celeste et al. 2003; Bekker-Jensen et al. 2006; Greenberg et al. 2006; Mailand et al. 2007). This result would imply that RAP80 function does not include a major contribution to the recognition of damaged DNA by BRCA1. We noted that several earlier studies reported more dramatic effects on BRCA1 IRIF in cells depleted of RAP80 (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007). However, no detailed time-course analysis was reported in those studies, and IRIF data were analyzed using images with lower resolution than those employed here.
Figure 1.
BRCA1 IRIF formation in the presence and absence of RAP80. U2OS cells treated with control or RAP80 siRNA were irradiated with 5 Gy IR or mock-treated (designated as NT) and released for different periods of time. (A,B) Representative images showing BRCA1 and RAP80 IRIF at 2 h (A) and 8 h (B) post-IR. Nuclei were counterstained with DAPI. (C,D) Percentages of cells, noted above, that contain RAP80 (C) and BRCA1 (D) IRIF. At the indicated intervals, cells containing at least 10 distinct foci, as recognized by relevant antibodies in immunofluorescence experiments, were denoted and counted. At least 100 cells were counted for each category of foci at each time point. The experiment was repeated three times and error bars indicate standard deviation. Data in D were analyzed using two-way ANOVA and, when a significant difference between control and RAP80-depleted cells was present, the actual significance values were marked. (***) P < 0.001. (E) Distribution of control and RAP80-depleted cells in five distinct categories based on intensity of BRCA1 IRIF (class I–class V) at 4 h after 5 Gy IR. A representative cell is shown for each class. All images were obtained at the same magnification and exposure time. During the analysis, each nucleus in a given culture was characterized as belonging to one of the above-noted cell classes. At the completion of the experiment, the distribution of nuclei among these classes in control and RAP80-depleted cultures was assessed and then compared. Bars, 10 μm. Numbers of cells in each class and total numbers of cells counted, as well as their corresponding percentages in control or RAP80-depleted cells, are summarized in the table. All images were analyzed in parallel for each experiment, and the data represented in the table are that obtained, collectively, from five identical experiments.
To better compare the BRCA1 immunostaining of IRIF that arise in the presence and absence of RAP80, we performed detailed examinations of BRCA1 IRIF after 5 Gy IR and assigned cells to five different classes based on the BRCA1 immunostaining intensity of their IRIF (Fig. 1E). The results showed that RAP80 depletion was associated with a major loss of cells that carried larger, more strongly staining BRCA1 IRIF (class I and II cells, reduced from 61.8% to 20.4%), and equally significant increases in cells carrying smaller, more weakly staining IRIF (class III and IV cells, increased from 21.6% to 43.5%). Therefore, our results show that RAP80 depletion leads to a shift in an irradiated culture from a predominance of cells containing more intensely staining BRCA1 foci to a predominance of cells that contained less intensely staining structures.
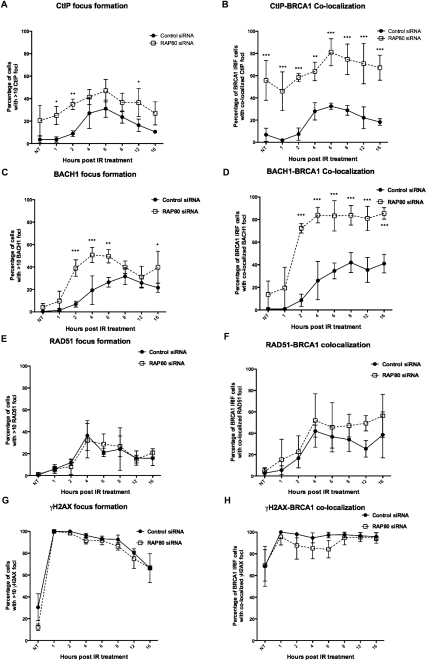
We then examined the dynamics of BRCA1-containing IRIF that also contained CtIP and BACH1—two BRCA1-interacting proteins, each of which is known to engage in the performance of HR (Litman et al. 2005; Limbo et al. 2007; Sartori et al. 2007; Takeda et al. 2007). In control cells, both CtIP and BACH1 were first observed to form IRIF in significant numbers (∼40%) of cells 4 h after IR (Fig. 2A,C; see also Supplemental Fig. S2A,B). Consistent with the recently identified role of CtIP in promoting DSB end resection during early stages of HR-mediated DSBR (Sartori et al. 2007; Takeda et al. 2007; Mimitou and Symington 2008; You et al. 2009), the percentage of CtIP IRIF-positive cells gradually decreased after the initial increases (Fig. 2A; Supplemental Fig. S2A). In contrast, in RAP80-depleted cells, significant numbers of cells were already positive for CtIP-containing and BACH1-containing IRIF by 1 and 2 h post-IR, respectively; i.e., earlier than normal (Fig. 2A,C; see also Supplemental Fig. S2A,B). Furthermore, in RAP80-depleted cells, many more cells contained significant numbers of BRCA1 IRIF that costained with CtIP (58.4% vs. 7.3%) or BACH1 (72.3% vs. 8.6%) antibody within 2 h, and this result persisted for up to 16 h post-IR (Fig. 2B,D; see also Supplemental Fig. S2A,B). At later time points, although BRCA1 IRIF in RAP80-depleted cells diminished in abundance and persisted in fewer of these than in control cells (Fig. 1D), most of them continued to costain with CtIP or BACH1 antibody (Fig. 2B,D; see also Supplemental Fig. S2A,B). The colocalization of CtIP with BRCA1 foci appeared to occur earlier than was observed for BACH1 foci. Even in nonirradiated cells, CtIP foci were seen in ∼19% of RAP80-depleted cells (Fig. 2A; Supplemental S2A, NT), and most of them colocalized with sporadic BRCA1 foci in these cells (Fig. 2B; Supplemental S2A, NT). One possible interpretation of the increased colocalization of sporadic CtIP and BRCA1 foci in the absence of RAP80 is that more extensive end processing of spontaneous single-strand breaks and DSBs is ongoing in these cells (please see below). Such a significant increase was not observed for sporadic BACH1 foci, which remained low in both control and RAP80-depleted cells without IR (Fig. 2C,D; Supplemental S2B, NT). Conceivably, BACH1 is not involved in repairing spontaneous DNA breaks in this setting, and further studies would be needed to clarify the particular role of CtIP. Moreover, in each irradiated cell that contained >10 BRCA1 foci, it appeared that more of these foci in RAP80-depleted cells costained with CtIP or BACH1 antibodies than was the case in control cells (Supplemental Fig. S2A,B). We also examined the dynamics of RAD51 IRIF in the same setting. Although the development of RAD51 IRIF followed comparable trends in both control and RAP80- depleted cells (Fig. 2E), RAP80 depletion led to a slightly higher percentage of BRCA1 IRIF-containing cells in which RAD51 colocalized in these IRIF at later times in the experiment (Fig. 2F; see also Supplemental Fig. S2C).
Figure 2.
Dynamics of IRIF formation. U2OS cells were treated as described in Figure 1. At the indicated intervals, cells containing at least 10 distinct foci, as recognized by relevant antibodies in immunofluorescence assays, were identified and counted. Experiments were repeated three times, and 80–150 cells were counted for each category of foci at each time point for each experiment. (A) Percentages of cells containing CtIP foci. (B) Percentages of cells containing BRCA1 foci with colocalized CtIP. (C) Percentages of cells containing BACH1 foci. (D) Percentages of cells containing BRCA1 foci with colocalized BACH1. (E) Percentages of cells containing RAD51 foci. (F) Percentages of cells containing BRCA1 foci with colocalized RAD51. (G) Percentages of cells containing γH2AX foci. (H) Percentages of cells containing BRCA1 foci with colocalized γH2AX foci. Data were analyzed using two-way ANOVA and, when a significant difference between control and RAP80-depleted cells was present, the actual significance values were marked. (***) P < 0.001; (**) P < 0.01; (*) P < 0.5. Please see also Supplemental Figure S2.
Notably, in RAP80-depleted cells, no significant differences were observed in the dynamics of γH2AX IRIF formation (Fig. 2G) and, as expected, nearly all remaining BRCA1 IRIF still colocalized with γH2AX foci (Fig. 2H), suggesting that the observed differences in BRCA1/CtIP/BACH1 IRIF dynamics are not caused by a gross change in γH2AX-containing chromatin structure at and adjoining sites of DNA breakage. Moreover, RAP80 depletion did not affect the abundance of BRCA1, CtIP, BACH1, or RAD51 as individual proteins in cell extracts (Supplemental Fig. S1B), arguing against possible changes in the availability of these proteins in RAP80-depleted cells.
A number of these proteins are known to form distinct protein complexes with BRCA1 and BARD1, the heterodimeric, nuclear partner of BRCA1 (Scully et al. 1997b; Wong et al. 1998; Yu et al. 1998; Cantor et al. 2001; Greenberg et al. 2006). Thus, the question of whether RAP80 controls the extent and/or stability of BRCA1 complex formation with CtIP, BACH1, or RAD51 was raised. When anti-BRCA1 coimmunoprecipitates were generated using extracts of control and RAP80 shRNA-expressing cells, it was clear that these coimmunoprecipitates retained endogenous BRCA1, CtIP, and BACH1 in normal amounts compared with control cells (Supplemental Fig. S3). We were not able to determine the amount of RAD51 in these coimmunoprecipitation experiments using antibodies against endogenous proteins due to a high background. But when coimmunoprecipitations were performed using cells expressing epitope-tagged BARD1 (eBARD1), it was apparent that comparable amounts of RAD51 were associated with eBARD1/BRCA1 regardless of the levels of RAP80 (data not shown). Thus, the RAP80 depletion-associated changes in the kinetics and extent of concentration of these proteins in, and the dynamics and extent of their colocalization with, BRCA1 in IRIF were not a product of concomitant alterations in either their intracellular abundance or their ability to form complexes with BRCA1.
Taken together, these analyses revealed that (1) RAP80 is required for the long-term maintenance of BRCA1 IRIF, but not for the initial concentration of BRCA1 at sites of DNA damage; (2) loss of RAP80 leads to a shift in the predominance of cells, from those containing more strongly staining BRCA1 IRIF to those containing weakly staining BRCA1 IRIF; and (3) RAP80 actively regulates the timing and extent of the accumulation of CtIP, BACH1, and RAD51 in BRCA1-containing IRIF through a mechanism affecting neither the intracellular concentrations of these proteins nor their ability to interact with BRCA1.
Effect of RAP80 and its associated proteins on homology-mediated DSBR
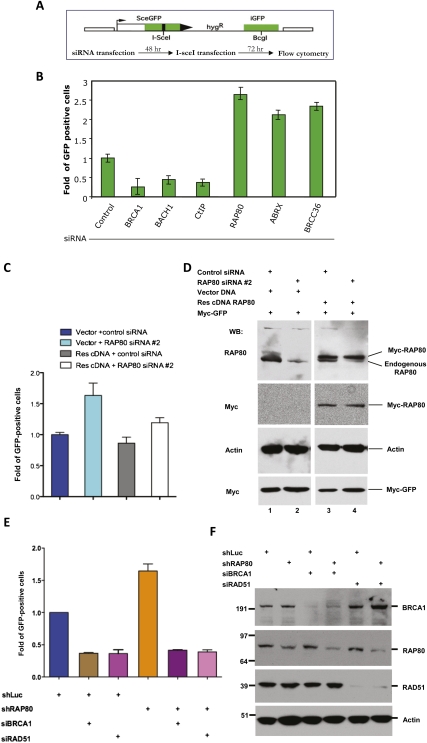
Since RAP80 appeared to suppress premature concentrations of CtIP and BACH1—and, to a less extent, RAD51—in BRCA1-containing IRIF, all of which are involved in HR-mediated DSBR (HR), we asked whether RAP80 plays a role in regulating HR. U2OS-DR cells containing an integrated, single-copy HR reporter with an I-SceI recognition site were employed to measure HR activity (Pierce et al. 1999; Xia et al. 2006). HR-mediated repair of the single DSB induced by I-SceI endonuclease restores a functional eGFP gene. The cells that have undergone HR are green and can be scored by flow cytometry (Fig. 3A). As expected, treating cells with siRNAs that target BRCA1, BACH1, or CtIP led to a marked decrease of GFP-positive cells compared with the control (Fig. 3B). However, RAP80 depletion using four different, nonoverlapping siRNAs or a lentiviral shRNA consistently led to a significant increase in GFP-positive cells, implying that there had been an increased frequency of HR in this setting (Fig. 3B; see also Supplemental Fig. S4). siRNA directed at another member of the RAP80–BRCA1 complex—i.e., Abraxas or BRCC36—gave similar results (Fig. 3B), and analogous results were obtained in mouse embryonic stem (ES) cells following Rap80 depletion (data not shown). Transient expression of exogenous RAP80 from an siRNA-resistant cDNA restored near-normal levels of HR in cells depleted of endogenous RAP80 (Fig. 3C,D), reinforcing the notion that the hyperrecombination phenotype was specifically due to RAP80 depletion. These data suggest that the integrity of RAP80 complexes normally contributes to measurable suppression of HR function. Since the enhanced HR effect that was observed in this setting proved to be BRCA1- and RAD51-dependent (Fig. 3E,F), one can further argue that these RAP80-containing structures normally suppress excessive, BRCA1-dependent HR activity.
Figure 3.
RAP80 depletion leads to an increased frequency of homology-mediated DSBR. (A) A schematic diagram summarizing the I-SceI-inducible DSBR reporter in U2OS-DR cells and the relevant HR measurement procedure. (B) A histogram showing the relative change in GFP-positive cells after I-SceI induction in cells transfected with control siRNA or siRNA targeting BRCA1, BACH1, CtIP, RAP80 (siRAP80-1), Abraxas, or BRCC36. The percentage of GFP-positive cells after I-SceI expression in control siRNA-treated samples was normalized to 1. Data were collected from three independent experiments. Error bars indicate standard deviation. (C,D) Rescue of increased HR in RAP80-depleted cells by expression of an siRNA-resistant RAP80 cDNA. (C) Histogram showing the relative change of I-SceI-induced HR in cells treated with control or RAP80 siRNA and cotransfected with an siRNA-resistant RAP80 cDNA (Res cDNA) or the vector DNA. The levels of GFP-positive cells in vector + control siRNA cotransfected cells were normalized to 1. Experiments were performed in triplicate, and error bars indicate standard deviation. (D) Western blots showing the efficiency of endogenous RAP80 depletion by siRNA and expression of a Myc-tagged RAP80 cDNA resistant to siRNA. Actin served as a loading control for Western blots, and the expression of a Myc-tagged GFP cDNA (cloned in the same vector) in parallel cell cultures served as controls for transfection efficiency. Please note the total amounts of protein loaded in lanes 3 and 4 (cf. actin bands) were less than those in lanes 1 and 2 in order to distinguish endogenous and tagged RAP80 using the same RAP80 antibody. (E,F) The excessive HR-mediated DSBR in RAP80-depleted cells is BRCA1- and RAD51-dependent. (E) HR-mediated repair of I-SceI-induced DSB. Experiments were performed as described in A. Specifically, U2OS-DR cells expressing an shRNA that targets luciferase (shLuc) or RAP80 (shRAP80) were transfected with control siRNA or an siRNA directed at BRCA1 or RAD51. The histogram shows the relative increase in GFP-positive cells of each category. The levels of GFP-positive cells in shLuc-expressing cells were normalized to 1. Experiments were performed in triplicate, and error bars indicate standard deviation. (F) Western blots reveal the efficiency of protein depletion by RAP80 shRNA and/or siRNA directed at BRCA1 or RAD51. Fifteen micrograms of protein was loaded in each lane, and actin (detected by blotting) served as a loading control.
To test whether cell proliferation or viability effects underlie the apparent enhancement in HR activity, cell cycle and apoptosis assays were performed on control and RAP80-depleted cells. RAP80 depletion failed to elicit a change in overall cell cycle distribution or an altered apoptotic cell population within the experimental time frame (Supplemental Fig. S1A; data not shown). These results argue against an increase in the S/G2 cell population or a change in cell death frequency indirectly contributing to the increased HR activity, described above. Moreover, in keeping with the notion that the RAP80-enhanced DSBR effect is executed primarily at the level of HR, RAP80 depletion did not appear to affect the abundance of a nonhomologous end-joining (NHEJ)-derived product that was assessed using a PCR-based assay (Supplemental Fig. S5; Nakanishi et al. 2005). These results aside, it is conceivable that other forms of error-prone DSBR, even some forms of NHEJ, could have been perturbed in this setting (please see the Discussion).
RAP80 suppresses excessive DSB end processing
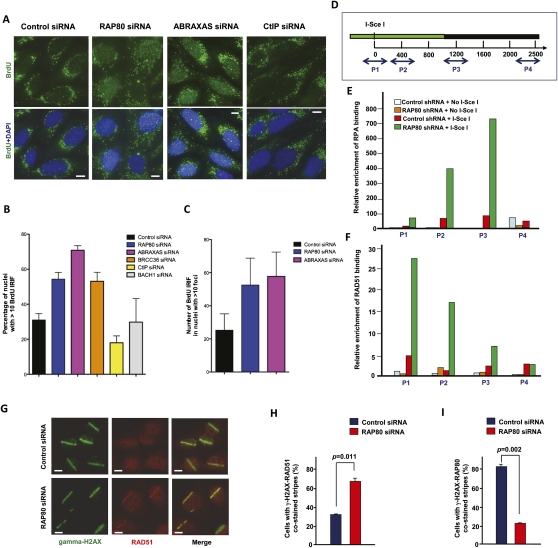
Several recent studies showed that CtIP, in conjunction with the MRE11 complex, is required for the DSB end processing reactions that generate ssDNA regions to support subsequent HR-mediated DSBR (Limbo et al. 2007; Sartori et al. 2007). Since we observed that CtIP formed IRIF earlier in the time course of a DSB response in the absence of RAP80 than in control cells (Fig. 2; Supplemental Fig. S2), one of the possible mechanisms by which RAP80 suppresses HR is to limit the extent of DSB end processing that generates substrates for homology-dependent DNA repair. To test whether there was ssDNA generated by DSB end processing, we searched for 5-bromo-2-deoxyuridine (BrdU)-containing IRIF that were detected under nondenaturing conditions. This method detects only BrdU that is incorporated into ssDNA (Raderschall et al. 1999; Sartori et al. 2007). Indeed, RAP80 depletion led to a significantly higher percentage of nuclei, each containing >10 BrdU foci, at 2 h after 5 Gy IR (Fig. 4A,B; see also Supplemental Fig. S6). Depletion of Abraxas or BRCC36 (a component of the RAP80 complex) led to a similar increase in BrdU foci-positive nuclei after IR (Fig. 4A,B). There were also significantly more BrdU IRIF in these nuclei in RAP80- or Abraxas-depleted cells than were observed in controls (Fig. 4A,C). In contrast, in parallel experiments, CtIP siRNA resulted in suppression of such BrdU foci (Fig. 4A,B; see also Supplemental Fig. S6). As negative controls, few or no foci were seen in the nuclei of nonirradiated cells either before or after RAP80, Abraxas, BRCC36, or CtIP depletion (data not shown).
Figure 4.
RAP80/BRCA1 complexes are required to suppress excessive DSB end processing. (A) Representative images showing IR-induced ssDNA foci in control, RAP80, Abraxas, or CtIP siRNA-treated U2OS cells. Cells were immunostained with an anti-BrdU antibody under nondenaturing conditions 2 h after 5 Gy IR treatment. Nuclei were counterstained with DAPI. Bars, 10 μm. (B) A histogram showing the percentage of cells containing >10 nuclear BrdU/ssDNA foci. Experiments were performed in triplicate, and at least 100 cells were counted in each experiment for each treatment. Error bars indicate standard deviation. (C) A histogram showing the average number of nuclear BrdU/ssDNA foci in cells containing 10 or more such foci. At least 120 nuclei were counted for each treatment. Error bars indicate standard deviation. (D) A schematic diagram showing the locations of four sets of primers used in the ChIP assays (marked as P1, P2, P3, or P4). The associated numbers correspond to the numbers of base pairs (bp) from the I-SceI recognition site to the position of a relevant 5′ primer. The genomic region that contains the sequences in which HR is expected is marked in green. (E) Results of RPA ChIP assays. (F) Results of RAD51 ChIP assays. The extent of RPA or RAD51 antibody binding at each location was calculated by comparing each antibody binding results with binding results obtained at the same DNA location with irrelevant IgG and normalized with the amounts of input DNA. Levels of RPA or RAD51 binding were all normalized to that obtained at position P1 in control cells (i.e., in the absence of RAP80 knockdown and I-SceI expression). (G) Representative images showing the recruitment/accumulation of RAD51 to UV laser-induced DSB stripes in cells treated with control or RAP80 siRNA. Bars, 10 μm. (H,I) Histograms showing the percentage of γ-H2AX-positive stripes in control siRNA or RAP80 siRNA-treated cells that were stained with RAD51 (H) or RAP80 (I) antibody, respectively. Data were collected from two independent experiments with at least 60 stripes scored in each experiment. Error bars indicate standard deviation.
As an independent assay to investigate the role of RAP80 in regulating DSB end processing, chromatin immunoprecipitation (ChIP) was used to determine whether RAP80 influences the association of RPA or RAD51 with chromatin at and near a DSB that is actively generated at a unique I-SceI site (Fig. 4D). According to the current view of HR-mediated DSBR, RPA binds to ssDNA generated by end processing complexes (i.e., CtIP/MRE11-containing complexes, etc.) at DSBs, and this is an early event in the activation of homology-dependent DNA repair (Raderschall et al. 1999; Limbo et al. 2007; Sartori et al. 2007; Takeda et al. 2007; Mimitou and Symington 2009; You et al. 2009). RAD51 then replaces RPA on ssDNA to promote recombination with homologous DNA (Sung and Robberson 1995; Sugiyama et al. 1997; Golub et al. 1998; McIlwraith et al. 2000; Sigurdsson et al. 2001). Thus, localization of RPA and RAD51 at and near such a DSB would reflect the performance of two major early steps in the HR process. Results of a ChIP assay performed on the GFP HR reporter after I-SceI expression showed that RAP80 depletion (by siRNA or shRNA) was accompanied by a dramatic increase in the abundance of RPA (Fig. 4E) and RAD51 (Fig. 4F) localized at and near the I-SceI cutting site compared with control cells. The controls here included cells stably transduced with control shRNA (directed against luciferase) that were or were not treated with I-SceI. Of note, the levels of RPA or RAD51 binding to this DNA region were comparably low at position P4, which is ∼2.3 kb away from the DSB and ∼1.2 kb distal to the homology region (Fig. 4D, green section), suggesting that RPA or RAD51 coating did not spread beyond a certain distance from the DSB in RAP80-depleted cells. This is in keeping with what happens at DSBs in both yeast and mammalian cells (Sugawara et al. 2003; Rodrigue et al. 2006).
In addition, ∼63% of UV laser-induced DSB-containing stripes were strongly positive for RAD51 immunostaining in cells treated with RAP80 siRNA, compared with ∼37% of stripes in control cells (Fig. 4G–I). The laser stripe results are also consistent with the BrdU-labeled ssDNA foci and the ChIP results, and, together, these data are likely reflections of the existence of more robust DSB end processing/resection and HR activity than that observed in control cells.
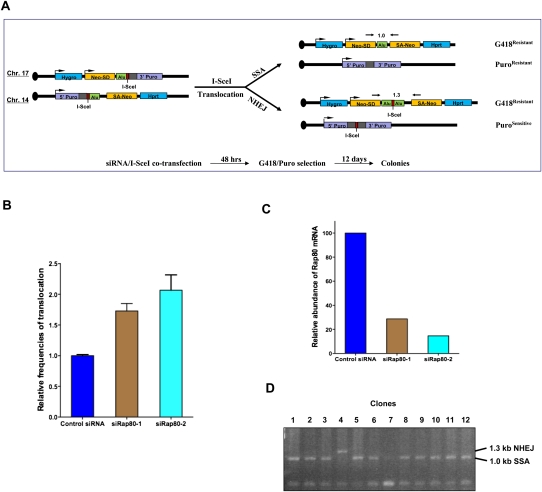
End resection to generate ssDNA is also required for other types of homology-dependent DSBR, such as single-strand annealing (SSA) (Ivanov et al. 1996; Paques and Haber 1999; Stark et al. 2004; Mimitou and Symington 2009). We tested whether increased DSB end processing observed in RAP80-depleted cells might have an effect on SSA using a previously described chromosome translocation assay (Elliott et al. 2005; Weinstock et al. 2006). The reporter murine ES cells that were used here contain two I-SceI sites located on chromosomes 17 and 14, each of which was flanked by two short, homologous sequences (290 base pairs [bp] and 265 bp, respectively, one of which was derived from a human Alu repeat). Repair of I-SceI-induced DSBs may lead to translocation events between the two chromosomes, which renders cells harboring such events resistant to G418 and/or puromycin (Fig. 5A). We found that knockdown of Rap80 using two different siRNA reagents led, in each case, to an approximately twofold increase in drug-resistant colonies recovered after repair of I-SceI-induced DSBs (Fig. 5B,C). Consistent with a previous study (Elliott et al. 2005), SSA was the predominant repair mechanism that generated drug-resistant clones (Fig. 5D; data not shown). These observations suggest that RAP80 may suppress end processing not only during DSBR for canonical HR, but also during a different, homology-dependent repair mechanism (i.e., SSA).
Figure 5.
Rap80 depletion results in increased frequencies of chromosomal translocations. (A) A schematic diagram showing the Alu translocation reporter in HomAlu mouse ES cells and the relevant experimental procedure. The centromere of mouse chromosome 17 or 14 is indicated as a black dot. (Hygro) Hygromycin resistance gene; (Hprt) hypoxanthine phosphoribosyltransferase gene; (Alu) two identical Alu elements derived from the MLL gene; (Neo-SD and SA-Neo) two portions of the neomycin/G418 resistance gene with splice donor and splice acceptor; (3′ Puro and 5′ Puro) 3′ and 5′ portions of the puromycin resistance gene. Gray boxes adjacent to 5′ and 3′ Puro indicate homologous sequences from the puromycin cassette. Locations of PCR primers for analyzing translocation products and the predicted sizes of such PCR products are also indicated. (B) A histogram showing the results of Alu recombination/translocation assays. Relative frequencies of translocations after I-SceI expression were compared with that of control siRNA-treated samples (normalized to 1). Translocations in vector transfected (i.e., no I-SceI) samples were undetectable (i.e., frequency <5 × 10−7) and are not shown in the histogram. Data were collected from two independent experiments in which each assay was performed in triplicate. (C) Results of quantitative RT–PCR showing the efficiency of Rap80 siRNA treatment. The amount of Rap80 mRNA in control siRNA-treated cells was normalized to 100. (D) A partial gel image of PCR analysis on translocation products in genomic DNA derived from G418-resistant clones. The size of PCR products correspondent to NHEJ- or SSA-mediated translocation is indicated. At least 100 clones were analyzed each for cells treated with control, siRap80-1, or siRap80-2 siRNA. Only 12 PCR results are shown for control (lanes 1–6) and siRap80-1-treated (lanes 7–12) cells.
Thus, RAP80 contributes to the suppression of exaggerated, BRCA1-dependent HR activity, and its failure to do so is accompanied by marked enrichment at DSBs of an end processing factor, CtIP; of ssDNA binding of RPA; and of a vital HR recombinase, RAD51.
Effect of RAP80 depletion on chromosome structure
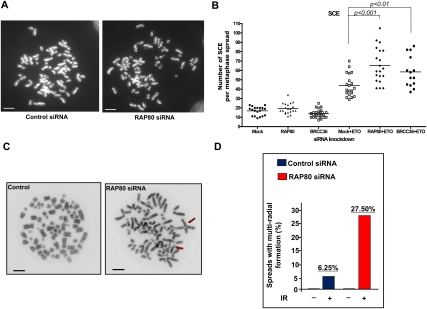
To investigate whether the RAP80 depletion/hyperrecombination phenotype is accompanied by gross chromosomal abnormalities, rates of sister chromatid exchange (SCE) were examined before and after treating cells with etoposide, a potent DSB inducer and anti-cancer drug (Nitiss 2002). Although the rates of SCE were comparably low before DNA damage in control and RAP80-depleted cells, etoposide treatment led to higher rates of SCEs in control siRNA-treated cells and even higher rates in RAP80 (P < 0.001) or BRCC36 (P < 0.01) knockdown cells (Fig. 6A,B).
Figure 6.
Increased chromosomal instability after DNA damage in RAP80-depleted cells. (A) Representative metaphase spreads showing SCEs from U2OS cells transfected with control or RAP80 siRNA. Cells were treated with 200 nM etoposide (ETO) or DMSO for 24 h. Bars, 5 μm. (B) A dot graph depicting the number of SCEs in cells treated with control, RAP80, or BRCC36 siRNA. Each data point represents the total number of SCEs in an individual cell. Horizontal lines indicate the mean number of SCEs in each category of cells. (C) Representative metaphase spreads from cells treated with control or RAP80 siRNA. Arrows point to multiradial chromosomal structures. (D) A histogram summarizing percentages of spreads containing multiradial chromosomal structures 24 h after cells were exposed to 2 Gy IR or mock-treated.
HR is a primary mechanism for repairing DSBs in mammalian cells, and it typically repairs DSBs with high fidelity, since the undamaged sister chromatid is preferentially used as the repair template (Johnson and Jasin 2000). However, recombination between inappropriate homologous sequences may occur if HR fidelity is compromised. This could result in interhomolog recombination and even deleterious genome rearrangements (Moynahan and Jasin 1997; Richardson et al. 1998; Vilenchik and Knudson 2003). One manifestation of illegitimate recombination is the formation of multiradial chromosome structures. Indeed, treating cells with RAP80 siRNA followed by 2 Gy IR resulted in a 4.4-fold increase over control cells in the number of spreads containing multiradial chromosome formations (Fig. 6C,D). Elevated SSA activity, as became apparent in RAP80-depleted cells (cf. Fig. 5), is known to contribute to interchromosome recombination and/or translocations (Deininger et al. 2003; Elliott et al. 2005; Weinstock et al. 2006), and seems a likely contributor to the multiradial chromosome development described here (Fig. 6C,D).
We also considered the possibility that RAP80–BRCA1 complexes are more abundant in G1 as part of a mechanism aimed at suppressing BRCA1-driven HR in G1 when illegitimate recombination might well lead to chromosome rearrangement. We analyzed endogenous BRCA1 complexes in synchronized T98G cells by coimmunoprecipitation. T98G cells were arrested in G0/G1 by serum starvation (since other synchronization protocols can induce significant DNA damage) and then released into media containing 10% serum. After release, a large percentage of cells progressed through the cell cycle in a synchronous manner for ∼28 h (Supplemental Fig. S7A). The results derived from endogenous/endogenous coimmunoprecipitation analysis showed that, although there was indeed less CtIP and BACH1 associated with BRCA1 in G0/G1 than in S/G2, the levels of BRCA1-interacting RAP80 remained constant throughout the cell cycle (Supplemental Fig. S7B). These results argue against the notion that RAP80–BRCA1 complexes are more prominent in G1 as part of a process aimed at suppressing G1-based illegitimate recombination. Instead, a possible mechanism that might limit end resection and recombination in G1 is that CtIP–BRCA1 and BACH1–BRCA1 complexes, both of which promote HR, are less abundant in G0/G1 than in S/G2 (Supplemental Fig. S7B). Indeed, others have suggested the existence of cell cycle-dependent regulation of CtIP activity (Chen et al. 2008; Huertas and Jackson 2009; Yun and Hiom 2009). It remains unclear whether a similar mechanism also regulates BACH1 activity. Parenthetically, it was shown recently that an interaction between BACH1 and BRCA1 is important for BACH1 pro-HR function (Xie et al. 2010).
Taken together, these data suggest that RAP80 plays an unexpected role in suppressing not only exaggerated, but even illegitimate recombination in response to DSBs. They also imply that a functional deficit of RAP80 in cells that have sustained DSBs and manifest an excessive, BRCA1-dependent HR phenotype also renders these cells more likely to develop certain gross chromosomal rearrangements than cells that exhibit normal control of HR activity.
Discussion
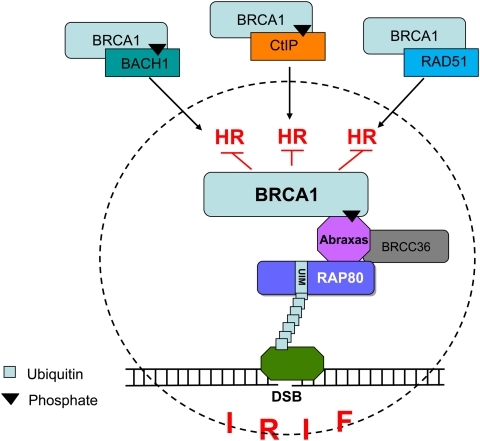
The data reported here point to unexpected conclusions. Intact RAP80 complexes support the localization in IRIF of much of the BRCA1 seen in these structures in a time-dependent manner. They operate in this regard without altering cell cycle progression. However, during the course of an IR-driven DDR, RAP80 complexes appear to suppress the premature and prolonged colocalization at IRIF of BRCA1 and three known partner proteins (CtIP, BACH1, and RAD51), each of which is a component of a distinct, BRCA1-containing complex engaged in supporting HR (Chen et al. 1998, 1999; Cantor et al. 2001; Litman et al. 2005; Greenberg et al. 2006; Sartori et al. 2007; Yun and Hiom 2009). CtIP is known to be involved in end processing at DSBs that are subsequently repaired by HR (Sartori et al. 2007; Takeda et al. 2007; Mimitou and Symington 2008; You et al. 2009). BACH1-containing complexes likely also contribute to the excessive HR response in RAP80-depleted cells, but how they do so is unclear (Litman et al. 2005; Peng et al. 2006; Xie et al. 2010). RAD51, as the mammalian recombinase, plays a central role in supporting HR-mediated DSBR. Since the overall abundance in cells of BRCA1/BARD1–CtIP, BRCA1/BARD1–BACH1, and BRCA1/BARD1–RAD51 complexes, or of these specific protein subunits, per se, was not altered before or after RAP80 depletion, the relative representation of the various complexes in IRIF is not a result of a RAP80 effect on their overall intracellular concentration. Perhaps these foci can accommodate only a limited amount of complex protein, and, in that context, BRCA1–RAP80 complexes gain preferred access to these structures through recognition of K6/K63-linked polyubiquitin chains by RAP80's UIM motifs. If this were the case—and if, in this setting, the foci lack sufficient BRCA1-containing, HR-promoting complexes to trigger HR at excessive amplitude—this might favor suppression of otherwise unbridled BRCA1-dependent HR. Viewed in this manner, RAP80 complexes perform a BRCA1 HR-tuning function that, when properly executed, contributes to a physiologically measured response to DSBs (Fig. 7).
Figure 7.
A model for HR-tuning function by RAP80/BRCA1-containing complexes present in IRIF. The main subunits of the RAP80 complex (i.e., RAP80, Abraxas, BRCC36, and BRCA1) are designated. Symbols for polyubiquitin and a generic P-Ser/Thr residue bound at the BRCA1 BRCT motifs are also included. The three HR-supporting BRCA1 complexes are also depicted, along with the hypothesized suppression by RAP80 complexes of their concentration in IRIF and their contribution to HR. The hatched circle symbolizes IRIF.
Previous studies showed that IRIF formation of both RAP80 and BRCA1 depends on the upstream DDR factors MDC1 and RNF8 (Sobhian et al. 2007; Wang and Elledge 2007). However, multiple data sets also show that MDC1 and RNF8 promote HR (Zhang et al. 2005; Xie et al. 2007; Huang et al. 2009; data not shown). Therefore, the RAP80-dependent, IRIF-based HR-tuning mechanism likely acts downstream from the above-mentioned factors. Indeed, Xie et al. (2007) showed previously that the HR-supporting function of MDC1 could be separated from its role in supporting BRCA1 or 53BP1 focal recruitment, leaving open the possibility that the role of MDC1 in regulating RAP80 function is a product of a function other than its HR-promoting activity. Whether RNF8 functions in a similar, variegated manner remains to be investigated. In this regard, we noted that it is not uncommon that multiple, sometimes antagonistic, pathways are controlled by the same factor. For example, in response to stalled replication forks, different modes of ubiquitin or SUMO modification of PCNA seem to determine whether to activate translesional synthesis, HR, or other replication bypass pathways (Hoege et al. 2002; Stelter and Ulrich 2003; Papouli et al. 2005; Branzei et al. 2008). Given the presence of multiple functional domains in the key factors MDC1, RNF8, BRCA1, and RAP80—and of several, active, protein-modifying enzymes in the MDC1–RNF8–RAP80 cascade—a similar scenario might exist in this complex pathway.
Irradiation after depletion of RAP80 (or other components of the RAP80–BRCA1 complex) also triggered the development of excessive numbers of ssDNA-containing IRIF (visualized as BrdU foci under nondenaturing conditions). The exaggerated HR effect arising after RAP80 depletion was also accompanied by a marked enhancement in accumulation of RPA and RAD51 at and near a defined DSB. Thus, the failure of RAP80 to elicit a normal level of control of HR activity in this setting, at least in part, is played out at an initial step in the HR process; i.e., DSB end processing. Such an increased end processing activity is correlated with the premature concentration of CtIP in IRIF observed in RAP80-depleted cells. Specifically, this premature concentration effect likely triggered enhanced end processing at DSB that, in turn, amplified the abundance of ssDNA-containing ends at DSB, the enhanced binding at these sites of an ssDNA-binding protein, and the subsequent increase in binding to these regions of a vital strand insertion recombinase (Gupta et al. 2005; Litman et al. 2005; Sartori et al. 2007; Chen et al. 2008). RAP80 depletion also led to a premature accumulation of BACH1 in IRIF. The exact function of BACH1 in HR-mediated DSBR remains unclear, and it is conceivable that RAP80 suppresses HR at a BACH1-dependent step in addition to its effect on DSB end processing. Future studies that elucidate the mechanistic nature of BACH1 functions in HR will lead to a better understanding of such a possibility.
Furthermore, despite an increased accumulation of RAD51 at/near DSBs in RAP80-depleted cells, as measured by ChIP assay or laser stripe analysis, we observed no dramatic changes in the kinetics of formation or the abundance of RAD51-containing IRIF, per se, at various time points in these experiments. Although RAD51 is clearly a central participant in HR, it is currently unclear what particular aspects of RAD51 function are reflected by its concentration in IRIF as opposed to at stripes or on a specific DNA sequence after a vicinal DSB is generated. For example, it is not fully known when during an HR response RAD51 appears in these higher-order structures (i.e., foci) as opposed to on damaged chromatin, as assessed by the two methods noted above. In this regard, others have reported that the formation of RAD51 foci, observed at a given time point after IR, although a phenomenon related to the HR process, does not necessarily provide a quantitative indication of the amplitude or rate of HR-mediated DSBR in response to IR (Haaf et al. 1999; Raderschall et al. 1999; Sy et al. 2009). The fact that RAD51 is regulated by other factors (e.g., BRCA2, RAD51 paralogs, RecQ helicases, etc.) in addition to BRCA1 may contribute to the complex pattern of RAD51 IRIF formation (Chen et al. 1998; Davies et al. 2001; Wu et al. 2001; Yonetani et al. 2005; Bugreev et al. 2007; Hu et al. 2007; Wu 2008).
The results described here also point to a new RAP80-associated cellular function—one that is focused on preventing exaggerated, BRCA1-driven HR and certain manifestations of genome disorder. Whether any elements that contribute to exaggerated, BRCA1-dependent HR, per se, represent direct triggers of the radial chromosome/illegitimate recombination effect that accompanied them is unclear. However, it is well known that recombination between homologous sequences dispersed throughout the genome can lead to deleterious—sometimes cancer-promoting—chromosome rearrangements, and SSA and HR are likely contributors to these events (for review, see Radman et al. 1982; Kolomietz et al. 2002; Thompson and Schild 2002; Griffin and Thacker 2004; Sung and Klein 2006; Reliene et al. 2007; cf. below). A case could also be made for the existence of a link between enhanced SCE that occurs in this setting and enhanced HR, given the sense that SCEs can be a reflection of enhanced recombination activity (Sonoda et al. 1999; Helleday 2003; Wu and Hickson 2003). Nevertheless, the formal possibility remains that RAP80 could also prevent chromosome instability through mechanisms that might not require BRCA1; for example, by suppressing other error-prone repair pathways such as a CtIP-dependent alternative end-joining (Bennardo et al. 2008; Yun and Hiom 2009; Lee-Theilen et al. 2011; Zhang and Jasin 2011), or by removing covalently bound polypeptides at DNA breaks (Iijima et al. 2010).
Paradoxically, markedly enhanced radial formation also occurred within 72 h after Brca1 elimination in mouse cells (Silver et al. 2007), which, based on recent findings (Bunting et al. 2010; Kass et al. 2010), might be the result of aberrant end-joining of spontaneous DSBs that have accumulated in these BRCA1-null cells. In the above-referenced studies, 53BP1, another primary factor involved in DDRs, was shown to inhibit DSB end resection and subsequent HR in the absence of BRCA1. However, whether 53BP1-mediated blockade of end resection imposes some direct measure of control on BRCA1-driven HR in a normal setting is not clear (Xie et al. 2007). One recent study (Mok and Henderson 2010) and our own observations (Y Hu and DM Livingston, unpubl.) showed that 53BP1 and BRCA1 largely exist in nonoverlapping sets of IRIF in wild-type cells. 53BP1 IRIF formation and dynamics were not affected by RAP80 depletion, and 53BP1 was not found in the RAP80/BRCA1-containing nuclear complex (Y Hu and DM Livingston, unpubl.). Therefore, the available data do not yet show that 53BP1 and RAP80 are involved in the same pathway to suppress HR, although this remains a formal possibility. Further studies are needed to address these questions more precisely.
RAP80 depletion from cells is also associated with their hypersensitivity to IR (Kim et al. 2007a; Sobhian et al. 2007; Wang et al. 2007; Yan et al. 2007). IR hypersensitivity, in principle, could arise from the development of either a weak/absent or an exaggerated and inappropriate HR response to DSB formation. In an example of the former case, mammary epithelial cells that have lost BRCA1 function experience defective HR activity and associated genomic instability. This set of events, over time, is suspected of fueling breast carcinoma development (Moynahan et al. 1999; Scully et al. 2000; Moynahan et al. 2001; Thompson and Schild 2001). In an example of the latter case, when RAP80 complex function was rendered defective in cells that retain wild-type BRCA1 expression and experience DSBs, it was associated with exaggerated HR function, increased SCEs, and signs of SSA and illegitimate recombination—again, clear manifestations of genomic instability. One wonders whether such a scenario actually develops in humans that lack a BRCA1 mutation. If so, perhaps over time it is associated with a neoplastic outcome. Viewed in another way, one wonders whether, when BRCA1 function is chronically exaggerated because its ability to operate under the control of the RAP80 complex has been perturbed, its ability to function as a tumor suppressor is also compromised.
Materials and methods
Plasmids, antibodies, and cells
HeLa cells expressing Flag/HA-tagged RAP80 or BARD1 were described previously (Greenberg et al. 2006; Sobhian et al. 2007). Myc-tagged RAP80 was ligated into a pcDNA3-based expression vector. Flag/HA-tagged RAP80 mutants in pOZC vectors were generated using a QuickChange XL mutagenesis kit (Stratagene) or by PCR. Cells overexpressing these mutants were selected magnetically using IL-2Rα antibody-conjugated Dynabeads (Invitrogen). shRNA sequences were cloned into a pLKO.1 lentiviral vector (Moffat et al. 2006).
The following antibodies were used in immunoprecipitation, Western blot, and immunofluorescence experiments: rabbit polyclonal anti-RAP80 (Bethyl Laboratory; 1:1000 for Western blot and immunofluorescence), rabbit polyclonal anti-RAP80 (1:100 for immunofluorescence) (Sobhian et al. 2007), mouse monoclonal anti-ubiquitin conjugates (Fk2, BioMol; 1:4000 for Western blot), rabbit polyclonal anti-BRCA1 (Millipore; 1:2500 for Western blot and immunofluorescence), mouse monoclonal anti-BRCA1 (MS110; 1:50 for Western blot), mouse monoclonal anti-BRCA1 (D9, Santa Cruz Biotechnology; 1:50 for immunofluorescence and 1:100 for immunoprecipitation), monoclonal anti-mouse BRCA1 (GH118; 1:50 for Western blot) (Ganesan et al. 2002), rabbit polyclonal anti-γH2AX (phospho-S139, Abcam; 1:1000 for Western blot and immunofluorescence), mouse monoclonal anti-γH2AX (JBW301, Millipore; 1:1000 for Western blot and immunofluorescence), rabbit polyclonal anti-RAD51 (H-92, Santa Cruz Biotechnology; 1:2000 for Western blot and 1:250 for immunofluorescence), rabbit polyclonal anti-BACH1 (Sigma; 1:1000 for Western blot), mouse monoclonal anti-BACH1 (1:20 for immunofluorescence) (Cantor et al. 2001), rabbit polyclonal anti-CtIP (Bethyl Laboratory; 1:1000 for Western blot), mouse monoclonal anti-CtIP (1:40 for immunofluorescence) (Yu and Baer 2000), rabbit polyclonal anti-BRCC36 (Bethyl Laboratory; 1:2000 for Western blot), mouse monoclonal anti-BrdU (GE Healthcare; 1:100 for immunofluorescence), mouse monoclonal anti-IL-2Rα (7G7/B6, Millipore), and mouse monoclonal anti-HA (HA.11, Covance; 1:1000 for Western blot). A rabbit polyclonal anti-Abraxas (N2) antibody was raised against a GST-Abraxas fusion protein.
U2OS, U2OS-DR, HeLa, 293T, 293, and T98G cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). All cells were cultivated at 37°C in a humidified incubator in an atmosphere containing 10% CO2. HeLa S3 cells were grown in Joklik's Minimum Essential Medium (Sigma) supplemented with 7% newborn calf serum at 37°C. Mouse ES cells were grown in ES medium (Invitrogen) on either mouse embryonic fibroblast (MEF) feeder cells or gelatinized plates supplemented with leukemia inhibitory factor (LIF), and were cultured at 37°C in a humidified incubator in an atmosphere containing 5% CO2.
RNAi
siRNA or shRNA sequences used in this study (synthesized by Dharmacon or Qiagen) were as follows: siRAP80-1, GUAUUGACUCGGAGACAAA; siRAP80-2, CCAGUUGGAGGUUUAUCAA; siRAP80-3, GCACAAAGACUUCAGAUGCA; siRAP80-4, CUAGUUCAUUGUUGAGCAA; siRAP80-5, GGGUGGGCCAUGUUCAUUA; siRap80-1 (mouse), GAAGCGUAGACUUGAGGAC; siRap80-2 smartpool (mouse), GAAGAUCAGACUAGUGACA, AAGCAGUUGUGUCAGUAUA, GCAUAGGACCACAGGUGUA, and CAAAUUGCUUAGUGGACUU; siBRCA1, AGAUAGUUCUACCAGUAAA; siCtIP, GCUAAAACAGGAACGAAUC; siBACH1, AGCUUACCCGUCACAGCUU; siRAD51, CUAAUCAGGUGGUAGCUCA; siAbraxas, CAGGGUACCUUUAGUGGUU; siBRCC36, GAGGAAGGACCGAGUAGAA; shLuciferase, GTGCGCTGCTGGTGCCAAC; shRAP80-1, GCTCACATATCAGTCAGGGAA; shRAP80-2, AAACTTTTGTTGGAGGAAGAA.
Nontargeting siRNA pools from Dharmacon were used as siRNA controls, and shRNA targeting luciferase was used as an shRNA control in all experiments. siRNA transfections were performed using HiPerFect (Qiagen) or Lipofectamine RNAiMax (Invitrogen) according to the manufacturers' instructions.
I-SceI recombination assay
U2OS cells containing a single copy of the DR-GFP reporter (U2OS-DR) were employed using experimental procedures that were described previously (Xia et al. 2006). Experiments using mouse ES cell lines containing HR/SCR reporter were performed as described in Xie et al. (2004).
For Alu recombination/translocation assays, 6 × 106 HomAlu ES cells (Elliott et al. 2005) were cotransfected with siRNA and pCBASce/vector using Lipofectamine 2000 (Invitrogen) and then selected for G418- or puromycin-resistant colonies. Briefly, cells were trypsinized and resuspended at 1 × 106 cells per milliliter in ES cell culture medium, and then prepared transfection mixtures were added. After incubating for 10 min at 37°C, cells were plated on neomycin–hygromycin–puromycin triple-resistant feeder plates in triplicates. In parallel, 2 × 103 cells from suspension of each treatment were plated to determine plating efficiency, and 2 × 105 cells cotransfected with siRNA + an eGFP expression vector (instead of pCBASce/vector) were plated to determine transfection and knockdown efficiency. Drug selection (G418: 200 μg/mL; puromycin: 2 μg/mL) started 48 h after transfection and was maintained for 8–12 d (no drug selection was imposed in plates used for determining plating or transfection efficiency) until well-defined colonies were observed. Sixty colonies from each treatment were picked and transferred to 96-well feeder plates for further analyses. Plates were then fixed and stained with 0.1% crystal violet/20% ethanol solution for colony counting. Relative frequencies of translocation were calculated based on numbers of colonies after normalization with plating/transfection efficiency of each treatment.
SCE assay and chromosome analysis
The SCE analysis was carried out using a classical BrdU-labeling protocol (German and Alhadeff 2001) with minor modifications: Cells were incubated with 10 μM BrdU for 48 h after treatment with DNA-damaging agents (IR: 0.5–1 Gy; etoposide: 200 nM), collected, and then prepared for analysis of metaphase spreads. Sister chromatids were visualized by staining with acridine orange (Sigma) solution (0.0125 mg/mL). For chromosome analysis, spreads were stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI).
ChIP assay
U2OS cells were incubated with 1% formaldehyde in culture medium for 15 min at room temperature, and then washed with cold PBS/BSA (5 mg/mL) to inactivate residual formaldehyde. Cells were resuspended in lysis buffer (1% SDS, 10 mM EDTA, 0.5 mM EGTA, 50 mM Tris-HCl at pH 7.5, 0.25% Triton X-100, protease inhibitors) and sonicated to fragment chromatin to the desired extent. Cell lysate was diluted in immunoprecipitation dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl at pH 7.5, protease inhibitors), and immunoprecipitations were performed with anti-RPA or anti-RAD51 antibody or IgG control overnight at 4°C. Beads were washed six times with SDS-free RIPA/LiCl buffer (50 mM HEPES at pH 7.5, 1 mM EDTA, 0.7% sodium deoxycholate, 1% NP-40, 0.5 M LiCl, protease inhibitors) and then twice with TE buffer. Immunoprecipitated chromatin was eluted with elution buffer (1% SDS, 0.1 M NaHCO3) and incubated for 16 h at 65°C to reverse the cross-links. DNA was purified using a PCR purification kit (Qiagen) and prepared for quantitative PCRs.
Primers used in ChIP assays were as follow: ChIP-P1F, GAGCAAGGGCGAGGAGCTGT; ChIP-P1R, CCGTAGGTCAGGGTGGTCAC; ChIP-P2F, TCTTCTTCAAGGACGACGGCAACT; ChIP-P2R, TTGTAGTTGTACTCCAGCTTGTGC; ChIP-P3F, ACGAGAGATCTATAGATCTATAGATCATGA; ChIP-P3R, CTCTTTACTGAAGGCTCTTTA; ChIP-P4F, CCGCGACGTCTGTCGAGAAG; ChIP-P4R, GCCGATGCAAAGTGCCGATA.
Detecting BrdU incorporation in ssDNA
The experiment was performed essentially as described (Raderschall et al. 1999). Briefly, U2OS cells on glass coverslips were grown in culture medium containing 10 μM BrdU (Sigma-Aldrich) for 48 h. Cells were irradiated with 5 Gy IR and, after 2 h, coverslips were fixed in absolute methanol for 20 min at −20°C. Coverslips were then rinsed briefly in ice-cold acetone and washed three times with PBS before being exposed to immunofluorescence staining with an anti-BrdU antibody (GE Healthcare).
Immunofluorescence
Immunofluorescence following irradiation or the generation of UV laser stripes was performed as described previously (Greenberg et al. 2006; Sobhian et al. 2007).
Laser-generated DNA stripes
Laser-induced DNA DSB-containing stripes were generated using a P.A.L.M. MicroBeam laser microdissection system (Carl Zeiss MicroImaging, Inc.) as described previously (Greenberg et al. 2006; Sobhian et al. 2007). Cells grown on coverslips were incubated for 24 h in medium containing 10 μM BrdU (Sigma-Aldrich). Coverslips were transferred to CO2-independent medium (Invitrogen) prior to laser treatment. Laser stripes were generated in 70–100 cells per coverslip with the above-noted laser (λ, 337 nm) using the 40× objective at 63% power. Cells were returned to a cell culture incubator at 37°C and fixed 1 h later following the aforementioned immunofluorescence protocol.
Immunoprecipitation
Immunoprecipitations were performed by incubating cell lysate in NET-N 400 (400 mM NaCl, 0.5 mM EDTA, 20 mM Tris-HCl at pH 7.5, 0.5% NP-40, 1.5 mM MgCl2, 10% glycerol, 5 mM NEM, protease inhibitors) or NET-N150 (the same buffer now containing 150 mM NaCl) in the presence of a primary antibody for 4–16 h at 4°C. This was followed by incubating the reaction mixture with protein A- or G-conjugated beads for an additional hour. Beads were washed four to six times with buffer used in the immunoprecipitation, and immunoprecipitated proteins were eluted with Flag peptide solution (0.2 mg/mL) or 0.1 M glycine (pH 2.5) solution, or by incubating in Laemmli sample buffer for 5 min at 95°C.
Acknowledgments
We thank all of our laboratory colleagues for many helpful discussions. We are also grateful to Dr. Steve Elledge and Brittany Adamson for a critical discussion of our results, and we thank Dr. Junjie Chen for providing an Abraxas/CCDC98 expression vector, Dr. Richard Baer for providing anti-CtIP antibody, Dr. Sharon Cantor for providing anti-BACH1 antibodies, Dr. Maria Jasin for providing the HomAlu ES cell line, and Dr. Peter Sicinski for providing MEFs for feeder cell preparations. This work was supported by grants from the National Cancer Institute and the Breast Cancer Research Foundation to D.M.L. and by a SPORE in breast cancer research to Dana-Farber/Harvard Cancer Center. Y.H. was supported by a U.S. Department of Defense post-doctoral fellowship award (W81XWH-09-1-0632).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.2011011.
Supplemental material is available for this article.
Note added in proof
Findings on the role of RAP80 and its associated proteins in regulating homology-directed DSBR through the suppression of excessive end resection were also reported by Dr. Roger Greenberg's group (Coleman and Greenberg 2011).
References
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol 173: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM 2008. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4: e1000110 doi: 10.1371/journal.pgen.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M 2008. SUMOylation regulates Rad18-mediated template switch. Nature 456: 915–920 [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV 2007. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev 21: 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, et al. 2001. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105: 149–160 [DOI] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5: 675–679 [DOI] [PubMed] [Google Scholar]
- Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, et al. 1998. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell 2: 317–328 [DOI] [PubMed] [Google Scholar]
- Chen JJ, Silver D, Cantor S, Livingston DM, Scully R 1999. BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway. Cancer Res 59: 1752s–1756s [PubMed] [Google Scholar]
- Chen L, Nievera CJ, Lee AY, Wu X 2008. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem 283: 7713–7720 [DOI] [PubMed] [Google Scholar]
- Coleman KA, Greenberg RA 2011. The BRCA1–RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem doi: 10.1074/jbc.M110.213728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Qin J, Elledge SJ 1999. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286: 1162–1166 [DOI] [PubMed] [Google Scholar]
- Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC 2001. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell 7: 273–282 [DOI] [PubMed] [Google Scholar]
- Deininger PL, Moran JV, Batzer MA, Kazazian HH 2003. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev 13: 651–658 [DOI] [PubMed] [Google Scholar]
- Elliott B, Richardson C, Jasin M 2005. Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol Cell 17: 885–894 [DOI] [PubMed] [Google Scholar]
- Feng L, Huang J, Chen J 2009. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev 23: 719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan S, Silver DP, Greenberg RA, Avni D, Drapkin R, Miron A, Mok SC, Randrianarison V, Brodie S, Salstrom J, et al. 2002. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell 111: 393–405 [DOI] [PubMed] [Google Scholar]
- German J, Alhadeff B 2001. Analysis of sister-chromatid exchanges. Curr Protoc Hum Genet 8: 8.6.1–8.6.10 doi: 10.1002/0471142905.hg0806s02 [DOI] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, Bartek J, Jackson SP 2003. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421: 952–956 [DOI] [PubMed] [Google Scholar]
- Golub EI, Gupta RC, Haaf T, Wold MS, Radding CM 1998. Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res 26: 5388–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM 2006. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev 20: 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin CS, Thacker J 2004. The role of homologous recombination repair in the formation of chromosome aberrations. Cytogenet Genome Res 104: 21–27 [DOI] [PubMed] [Google Scholar]
- Gupta R, Sharma S, Sommers JA, Jin Z, Cantor SB, Brosh RM Jr 2005. Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J Biol Chem 280: 25450–25460 [DOI] [PubMed] [Google Scholar]
- Haaf T, Raderschall E, Reddy G, Ward DC, Radding CM, Golub EI 1999. Sequestration of mammalian Rad51-recombination protein into micronuclei. J Cell Biol 144: 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T 2003. Pathways for mitotic homologous recombination in mammalian cells. Mutat Res 532: 103–115 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. 2007. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev 21: 3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J 2009. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol 11: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284: 9558–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima J, Zeng Z, Takeda S, Taniguchi Y 2010. RAP80 acts independently of BRCA1 in repair of topoisomerase II poison-induced DNA damage. Cancer Res 70: 8467–8474 [DOI] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE 1996. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics 142: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RD, Jasin M 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J 19: 3398–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass EM, Moynahan ME, Jasin M 2010. Loss of 53BP1 is a gain for BRCA1 mutant cells. Cancer Cell 17: 423–425 [DOI] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X 2007a. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316: 1202–1205 [DOI] [PubMed] [Google Scholar]
- Kim H, Huang J, Chen J 2007b. CCDC98 is a BRCA1–BRCT domain-binding protein involved in the DNA damage response. Nat Struct Mol Biol 14: 710–715 [DOI] [PubMed] [Google Scholar]
- Kolomietz E, Meyn MS, Pandita A, Squire JA 2002. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer 35: 97–112 [DOI] [PubMed] [Google Scholar]
- Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J 2011. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat Struct Mol Biol 18: 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB 2005. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8: 255–265 [DOI] [PubMed] [Google Scholar]
- Liu Z, Wu J, Yu X 2007. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol 14: 716–720 [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- McIlwraith MJ, Van Dyck E, Masson JY, Stasiak AZ, Stasiak A, West SC 2000. Reconstitution of the strand invasion step of double-strand break repair using human Rad51 Rad52 and RPA proteins. J Mol Biol 304: 151–164 [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2009. DNA end resection: many nucleases make light work. DNA Repair (Amst) 8: 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124: 1283–1298 [DOI] [PubMed] [Google Scholar]
- Mok MT, Henderson BR 2010. A comparison of BRCA1 nuclear localization with 14 DNA damage response proteins and domains: identification of specific differences between BRCA1 and 53BP1 at DNA damage-induced foci. Cell Signal 22: 47–56 [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M 1997. Loss of heterozygosity induced by a chromosomal double-strand break. Proc Natl Acad Sci 94: 8988–8993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M 1999. Brca1 controls homology-directed DNA repair. Mol Cell 4: 511–518 [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Cui TY, Jasin M 2001. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res 61: 4842–4850 [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M 2005. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci 102: 1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss JL 2002. DNA topoisomerases in cancer chemotherapy: using enzymes to generate selective DNA damage. Curr Opin Invest Drugs 3: 1512–1516 [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD 2005. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell 19: 123–133 [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10: 886–895 [DOI] [PubMed] [Google Scholar]
- Peng M, Litman R, Jin Z, Fong G, Cantor SB 2006. BACH1 is a DNA repair protein supporting BRCA1 damage response. Oncogene 25: 2245–2253 [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13: 2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raderschall E, Golub EI, Haaf T 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci 96: 1921–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M, Jeggo P, Wagner R 1982. Chromosomal rearrangement and carcinogenesis. Mutat Res 98: 249–264 [DOI] [PubMed] [Google Scholar]
- Reliene R, Bishop AJ, Schiestl RH 2007. Involvement of homologous recombination in carcinogenesis. Adv Genet 58: 67–87 [DOI] [PubMed] [Google Scholar]
- Richardson C, Moynahan ME, Jasin M 1998. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev 12: 3831–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue A, Lafrance M, Gauthier MC, McDonald D, Hendzel M, West SC, Jasin M, Masson JY 2006. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J 25: 222–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 146: 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Livingston DM 2000. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408: 429–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM 1997a. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90: 425–435 [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM 1997b. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88: 265–275 [DOI] [PubMed] [Google Scholar]
- Scully R, Puget N, Vlasakova K 2000. DNA polymerase stalling, sister chromatid recombination and the BRCA genes. Oncogene 19: 6176–6183 [DOI] [PubMed] [Google Scholar]
- Shao G, Patterson-Fortin J, Messick TE, Feng D, Shanbhag N, Wang Y, Greenberg RA 2009. MERIT40 controls BRCA1–Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev 23: 740–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P 2001. Mediator function of the human Rad51B–Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev 15: 3308–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP, Dimitrov SD, Feunteun J, Gelman R, Drapkin R, Lu SD, Shestakova E, Velmurugan S, Denunzio N, Dragomir S, et al. 2007. Further evidence for BRCA1 communication with the inactive X chromosome. Cell 128: 991–1002 [DOI] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316: 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol 19: 5166–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M 2004. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol 24: 9305–9316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421: 961–966 [DOI] [PubMed] [Google Scholar]
- Stucki M, Jackson SP 2006. γH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 5: 534–543 [DOI] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell 12: 209–219 [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC 1997. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem 272: 7940–7945 [DOI] [PubMed] [Google Scholar]
- Sung P, Klein H 2006. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol 7: 739–750 [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL 1995. DNA strand exchange mediated by a RAD51–ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82: 453–461 [DOI] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Chen J 2009. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci 106: 7155–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Nakamura K, Taniguchi Y, Paull TT 2007. Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell 28: 351–352 [DOI] [PubMed] [Google Scholar]
- Thompson LH, Schild D 2001. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res 477: 131–153 [DOI] [PubMed] [Google Scholar]
- Thompson LH, Schild D 2002. Recombinational DNA repair and human disease. Mutat Res 509: 49–78 [DOI] [PubMed] [Google Scholar]
- Vilenchik MM, Knudson AG 2003. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci 100: 12871–12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, King MC 2007. Ten genes for inherited breast cancer. Cancer Cell 11: 103–105 [DOI] [PubMed] [Google Scholar]
- Wang B, Elledge SJ 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci 104: 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316: 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Hurov K, Hofmann K, Elledge SJ 2009. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev 23: 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Richardson CA, Elliott B, Jasin M 2006. Modeling oncogenic translocations: distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amst) 5: 1065–1074 [DOI] [PubMed] [Google Scholar]
- Wong AK, Ormonde PA, Pero R, Chen Y, Lian L, Salada G, Berry S, Lawrence Q, Dayananth P, Ha P, et al. 1998. Characterization of a carboxy-terminal BRCA1 interacting protein. Oncogene 17: 2279–2285 [DOI] [PubMed] [Google Scholar]
- Wu L 2008. Wrestling off RAD51: a novel role for RecQ helicases. Bioessays 30: 291–295 [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
- Wu L, Davies SL, Levitt NC, Hickson ID 2001. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem 276: 19375–19381 [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22: 719–729 [DOI] [PubMed] [Google Scholar]
- Xie A, Puget N, Shim I, Odate S, Jarzyna I, Bassing CH, Alt FW, Scully R 2004. Control of sister chromatid recombination by histone H2AX. Mol Cell 16: 1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, et al. 2007. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell 28: 1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Litman R, Wang S, Peng M, Guillemette S, Rooney T, Cantor SB 2010. Targeting the FANCJ–BRCA1 interaction promotes a switch from recombination to poleta-dependent bypass. Oncogene 29: 2499–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, Jetten AM 2007. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res 67: 6647–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonetani Y, Hochegger H, Sonoda E, Shinya S, Yoshikawa H, Takeda S, Yamazoe M 2005. Differential and collaborative actions of Rad51 paralog proteins in cellular response to DNA damage. Nucleic Acids Res 33: 4544–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Shi LZ, Zhu Q, Wu P, Zhang YW, Basilio A, Tonnu N, Verma IM, Berns MW, Hunter T 2009. CtIP links DNA double-strand break sensing to resection. Mol Cell 36: 954–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Baer R 2000. Nuclear localization and cell cycle-specific expression of CtIP, a protein that associates with the BRCA1 tumor suppressor. J Biol Chem 275: 18541–18549 [DOI] [PubMed] [Google Scholar]
- Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R 1998. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem 273: 25388–25392 [DOI] [PubMed] [Google Scholar]
- Yun MH, Hiom K 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459: 460–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jasin M 2011. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol 18: 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ma Z, Treszezamsky A, Powell SN 2005. MDC1 interacts with Rad51 and facilitates homologous recombination. Nat Struct Mol Biol 12: 902–909 [DOI] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X 2009. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol 19: 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Chen CF, Li S, Chen Y, Wang CC, Xiao J, Chen PL, Sharp ZD, Lee WH 1999. Association of BRCA1 with the hRad50–hMre11–p95 complex and the DNA damage response. Science 285: 747–750 [DOI] [PubMed] [Google Scholar]