Abstract
The Ski-interacting protein SKIP/SNW1 functions as both a splicing factor and a transcriptional coactivator for induced genes. We showed previously that transcription elongation factors such as SKIP are dispensable in cells subjected to DNA damage stress. However, we report here that SKIP is critical for both basal and stress-induced expression of the cell cycle arrest factor p21Cip1. RNAi chromatin immunoprecipitation (RNAi-ChIP) and RNA immunoprecipitation (RNA-IP) experiments indicate that SKIP is not required for transcription elongation of the gene under stress, but instead is critical for splicing and p21Cip1 protein expression. SKIP interacts with the 3′ splice site recognition factor U2AF65 and recruits it to the p21Cip1 gene and mRNA. Remarkably, SKIP is not required for splicing or loading of U2AF65 at other investigated p53-induced targets, including the proapoptotic gene PUMA. Consequently, depletion of SKIP induces a rapid down-regulation of p21Cip1 and predisposes cells to undergo p53-mediated apoptosis, which is greatly enhanced by chemotherapeutic DNA damage agents. ChIP experiments reveal that SKIP is recruited to the p21Cip1, and not PUMA, gene promoters, indicating that p21Cip1 gene-specific splicing is predominantly cotranscriptional. The SKIP-associated factors DHX8 and Prp19 are also selectively required for p21Cip1 expression under stress. Together, these studies define a new step that controls cancer cell apoptosis.
Keywords: SKIP/SNW1, p21Cip1, p53, splicing, cotranscriptional, apoptosis, cancer
Factors that regulate the elongation phase of RNA polymerase II (RNAPII) transcription also play an important role in protecting cells from DNA damage and environmental stress. Global inhibition of transcription elongation activates the p53 tumor suppressor through formation of long single-stranded regions of DNA that recruit RPA and ATR to signal a stress response, even in the absence of DNA damage (Derheimer et al. 2007; Gartel 2008). Transcription elongation is tightly regulated at many induced genes by the positive elongation factor P-TEFb (CycT1:CDK9) (Price 2008; Fuda et al. 2009; Hargreaves et al. 2009). P-TEFb counteracts proteins responsible for RNAPII promoter-proximal pausing (Chiba et al. 2010). As a consequence, p53 is strongly induced in cells treated with P-TEFb/CDK9 inhibitors such as flavopiridol (FP). FP promotes apoptosis through induction of p53 and inhibition of short-lived anti-apoptotic proteins, and is currently in clinical trials as an anti-cancer agent for leukemia and solid tumors (Canduri et al. 2008; Wang et al. 2009). Thus, RNAPII is a genome-wide sensor for DNA damage, through its ability to activate p53 and initiate programmed cell death upon encountering significant blocks to elongation.
The Ski-interacting protein SKIP (Snw1 and NCoA62) is a required transcriptional coactivator for many newly induced genes (Leong et al. 2001, 2004; Zhang et al. 2003; Folk et al. 2004; MacDonald et al. 2004) and counteracts transcriptional repression by retinoblastoma (Prathapam et al. 2002). The SKIP homologs in Saccharomyces cerevisiae (Prp45) and Drosophila (BX42) are essential for cell viability, splicing (Ambrozkova et al. 2001; Makarov et al. 2002; Gahura et al. 2009), and nuclear export of spliced mRNAs (Farny et al. 2008). Although elongation factors can affect splicing indirectly through changes in the rate of elongation, and defects in cotranscriptional splicing can reduce RNAPII elongation rates in vivo (Kornblihtt 2007; Muñoz et al. 2009; Pirngruber et al. 2009), SKIP is recruited to promoters as well as transcribed regions and appears to play a direct role in each process. We reported previously that SKIP associates with P-TEFb and stimulates HIV-1 Tat transcription elongation in vivo and in vitro (Brès et al. 2005). At the HIV-1 promoter, SKIP recruits c-Myc and also interacts with the MLL1:Menin histone methyltransferase to promote H3K4 methylation (Brès et al. 2009). Previous studies found that SKIP also binds U2AF35 (Ambrozkova et al. 2001), the PPIL1 peptidyl-prolyl isomerase (Skruzny et al. 2001; Xu et al. 2006), and the DExH RNA helicase Prp22 (Gahura et al. 2009), which helps release mRNA from the spliceosome (Schwer 2008). SKIP is required for cell survival and stress resistance in plants (Hou et al. 2009), and depletion of human SKIP or hPrp22 results in mitotic spindle defects and accumulation in prometaphase (Kittler et al. 2004, 2005), indicating an important role in cell cycle progression.
We reported previously that neither SKIP nor P-TEFb is needed for stress-induced HIV-1 transcription in vivo (Brès et al. 2009). It is unclear why P-TEFb is dispensable under stress, but it could reflect a loss of RNAPII pause factors or promoter histone modifications, or even locus-wide nucleosome depletion, as observed at heat-shock genes (Petesch and Lis 2008). Similarly, an earlier study found that P-TEFb is not required for p53-induced p21Cip1 (henceforth called p21) gene transcription in cells subjected to DNA damage (Gomes et al. 2006). These studies suggest that a widespread loss of elongation control may accompany environmental or genotoxic stress, such as that leading to G2/M arrest. In contrast, p21 gene transcription is selectively blocked at the level of elongation in cells exposed to the S-phase arrest agent hydroxyurea (Mattia et al. 2007), indicating that different types of stress have distinct effects on elongation in vivo.
Different subsets of p53 target genes specify whether cells will arrest to repair DNA damage, or undergo apoptosis (Vazquez et al. 2008; Vousden and Prives 2009). Key p53 target genes in these opposing pathways are the anti-apoptotic G1 cell cycle arrest factor p21 (Abbas and Dutta 2009) and the proapoptotic BH3-only Bcl-2 protein PUMA. The relative levels of these two proteins help to determine the extent of cell survival in response to DNA damage (Yu and Zhang 2003; Yu et al. 2003; Iyer et al. 2004). Known transcription factors that impact this balance include c-Myc, which represses p21 without affecting PUMA expression (Seoane et al. 2002; Jung and Hermeking 2009), and the bromodomain protein Brd7, which promotes p53 binding to the p21, but not PUMA, gene (Drost et al. 2010). As a consequence, inhibition of p21 or expression of c-Myc predisposes tumor cells to undergo apoptosis in response to DNA damage. Interestingly, the pro- and anti-apoptotic p53 target genes contain different types of core promoters and are therefore regulated by different transcription factors (Gomes and Espinosa 2010). In particular, the p21 genes contain high levels of preloaded (poised) RNAPII at the promoter in the absence of DNA damage, which allows for the rapid induction of these genes following p53 activation (Espinosa et al. 2003; Gomes et al. 2006; Morachis et al. 2010). In contrast, RNAPII elongation complexes must assemble de novo at PUMA and other proapoptotic p53 target genes, which delays their expression. Cell growth arrest arising from rapid p21 induction is an initial protective response to DNA damage or oncogene expression. Although the p21 gene is predominantly regulated at the level of transcription, additional factors control its translation, as well as protein and mRNA stability (Abbas and Dutta 2009).
Here we describe an unusual mechanism for p21 gene expression that involves gene-specific splicing by SKIP and is essential for cancer cell survival under stress. In particular, we found that SKIP is critical for splicing and expression of p21, but not for PUMA or other investigated p53 target genes, in human HCT116 (colon cancer) and U2OS (osteosarcoma) cells. SKIP associates with the 3′ splice site recognition factor U2AF65, but not U2AF35, and recruits it to the p21 gene and mRNA in vivo. In contrast, U2AF65 recruitment and splicing at the PUMA gene is independent of SKIP. As a consequence, siRNA-mediated depletion of SKIP induces p53-dependent apoptosis, which is most pronounced in cells subjected to DNA damage. The regulated binding of 3′ splice site recognition factors we observe here is reminiscent of a central feature of alternative splicing, which controls the expression of different isoforms of cell death pathway proteins (e.g., BCL-X and Caspase-9) with distinct or opposing roles in apoptosis (Schwerk and Schulze-Osthoff 2005). Consequently, alternative splicing factors are well-known regulators of p53-dependent and p53-independent apoptosis (Merdzhanova et al. 2008; Kleinridders et al. 2009; Legerski 2009; Moore et al. 2010). Our results reveal that cancer cell survival upon DNA damage also depends on SKIP and associated factors (DHX8 and Prp19), which function as gene-specific regulators of p21 mRNA splicing.
Results
SKIP is essential for p53 stress-induced expression of the p21, but not PUMA, genes
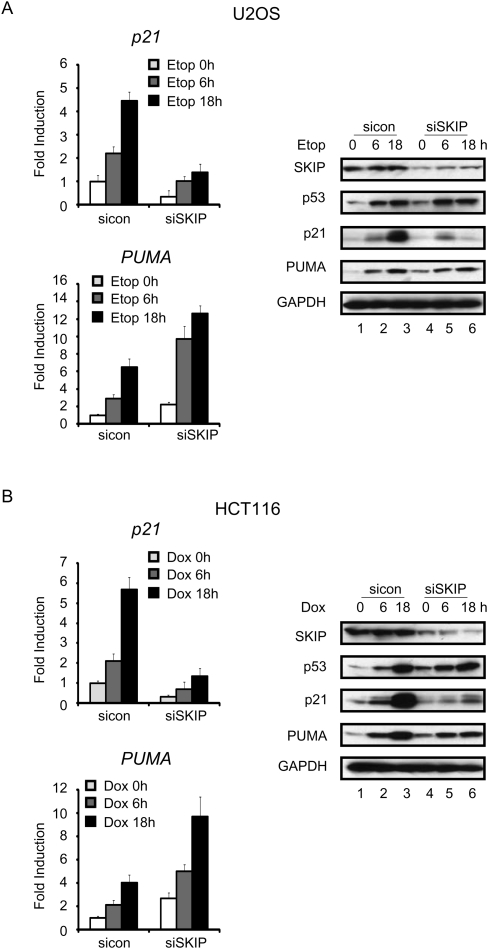
As is observed for many essential proteins, ablation of SKIP by siRNA increases endogenous p53 levels. Immunoblot analysis of extracts from SKIP-depleted U2OS cells revealed a significant increase in the steady-state level of p53, which was phosphorylated at Ser15, a modification that stabilizes the protein (Supplemental Fig. S1A). Levels of the PUMA protein were also elevated, indicating that the induced p53 protein is transcriptionally active. However, we noticed that p21 protein levels were markedly reduced in SKIP knockdown cells compared with cells expressing a control siRNA. To assess whether SKIP plays a role in the normal p53 stress response, endogenous p53 was induced in two human cancer cell lines, U2OS (osteosarcoma) and HCT116 (colon cancer), using the chemotherapeutic DNA damage agents etoposide (U2OS cells) or doxorubicin (HCT116 cells). As expected, DNA damage-induced accumulation of p53 and two of its target genes, p21 and PUMA, was observed in both U2OS (Fig. 1A) and HCT116 (Fig. 1B) cells. Interestingly, p53 levels increased in siRNA-mediated SKIP knockdown cells, and rose further upon exposure of these cells to etoposide or doxorubicin. Consequently, PUMA expression was elevated in SKIP-depleted cells, and increased further with DNA damage (Fig. 1; Supplemental Fig. S1B). In contrast, both basal and stress-induced p21 mRNA levels decreased in SKIP-depleted HCT116 or U2OS cells, compared with cells treated with a control siRNA, accompanied by a strong block to p21 protein expression, as detected by immunoblot (Fig. 1A, 1B, cf. lanes 1–3 and 4–6). Similar results were obtained using two different SKIP siRNAs (Supplemental Fig. S1B). Taken together, these data suggest that SKIP is critical for p53 induction of the anti-apoptotic gene target p21, but not for the proapoptotic PUMA gene.
Figure 1.
SKIP is required for DNA damage-induced p21 gene expression. (A) qRT–PCR analysis of p21 (top panel) and PUMA (botom panel) mRNA levels. U2OS cells were transfected with control or SKIP siRNA for 48 h, and incubated in the presence or absence of etoposide (20 μM) for the indicated times. (Right panel, lanes 1–6) Protein lysates were subjected to immunoblot analysis. (B) qRT–PCR analysis of p21 (top panel) and PUMA (bottom panel) mRNA levels. HCT116 cells were transfected with control or SKIP siRNA for 48 h, and incubated in the presence or absence of doxorubicin (0.5 μM) for the indicated times. (Right panel, lanes 1–6) Protein lysates were subjected to immunoblot analysis. All of the mRNA expression levels were normalized to GAPDH mRNA, and the values represent the fold increase or decrease over untreated cells. Error bars represent the standard deviation obtained from three independent experiments.
SKIP is dispensable for stress-induced transcription of the p21 gene
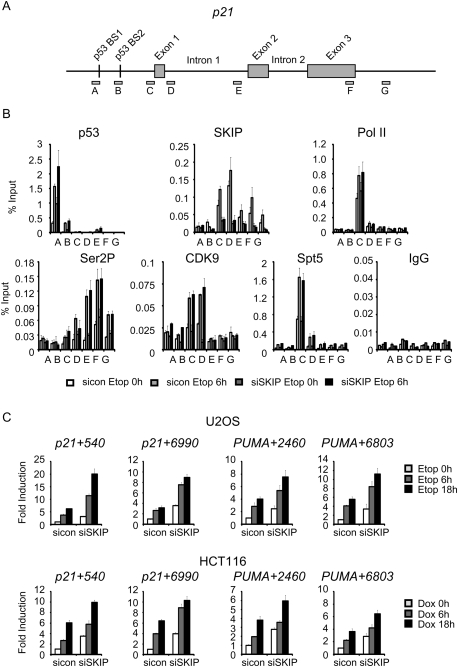
Numerous transcription and chromatin factors, including c-Myc (Seoane et al. 2002) and p300 (Iyer et al. 2004), are known to differentially affect p53 transactivation of the p21 and PUMA genes in vivo. However, it was surprising to find a role for SKIP in the p53 pathway, because other elongation factors, including P-TEFb and FACT, are dispensable for p21 expression under conditions of stress (Gomes et al. 2006; Gomes and Espinosa 2010). Consequently, we used RNAi chromatin immunoprecipitation (RNAi-ChIP) experiments to examine the block to p21 expression in SKIP-depleted U2OS cells before and after exposure to etoposide at the promoter and throughout the coding region (Fig. 2A). The ChIP experiments revealed increased p53 binding to the p21 promoter in SKIP knockdown cells, consistent with the observation that p53 is induced in these cells, and p53 occupancy at the gene increased further following etoposide treatment (Fig. 2B). ChIP analysis of the PUMA gene revealed a similar increase in p53 binding in cells treated with SKIP siRNA, which increased further upon stress induction (Supplemental Fig. S2B). Therefore, the loss of p21 protein expression in SKIP-depleted cells is not due to impaired binding of p53 to its target genes.
Figure 2.
Loss of SKIP does not affect p21 gene transcription. (A) Schematic representation of the p21 gene locus, and the relative locations of the primers used for ChIP. (B) ChIP analysis in U2OS cells transfected with control or SKIP siRNA for 48 h, followed by vehicle or etoposide (20 μM) for a further 6 h. ChIP-enriched DNA was quantified by qPCR with the indicated primers, and values are expressed as percentage of input DNA. Error bars represent the standard deviation obtained from three independent experiments. (C) qRT–PCR analysis of p21 and PUMA primary transcripts. U2OS cells (top panel) or HCT116 cells (bottom panel) were transfected with control or SKIP siRNA, and incubated with etoposide (top panel) or doxorubicin (bottom panel) for the indicated times. Numbers of the primers indicate the position of the first base pair relative to the transcription start site. The mRNA expression levels were normalized to GAPDH. Error bars represent the standard deviation obtained from three independent experiments.
Further ChIP analysis revealed that SKIP is present at the p21 gene in the absence of stress, with the highest levels at the promoter and proximal downstream region, but it is also present at lower levels in the coding region, following a pattern similar to that observed for P-TEFb/CDK9. SKIP binding was slightly enhanced by stress and greatly reduced in SKIP knockdown cells (Fig. 2B), consistent with the overall loss of SKIP protein (Fig. 1A). In contrast, only background levels of SKIP were detected at the PUMA gene, and this signal did not change upon SKIP knockdown (Supplemental Fig. S2B). Thus, SKIP associates specifically with the p21, and not PUMA, gene promoters. As reported previously (Espinosa et al. 2003), we detected high levels of RNAPII at the p21 core promoter in the absence of stress, indicative of a paused RNAPII complex, whereas RNAPII occupancy was low at the PUMA promoter but increased strongly following etoposide treatment (Fig. 2B; Supplemental Fig. S2B). Knockdown of SKIP did not affect recruitment of RNAPII, CDK9, or Spt5 at the stress-induced p21 or PUMA genes. Moreover, Ser2-phosphorylated RNAPII levels were unaffected in SKIP knockdown cells, indicating that SKIP is not required for accumulation of active RNAPII elongation complexes within the transcribed region of the p21 (Fig. 2B) or PUMA genes (Supplemental Fig. S2B). Together, the RNAi-ChIP studies indicate that SKIP is selectively recruited to the basal p21 promoter, but is not required for binding of p53 or transcription elongation at the stress-induced p21 gene in vivo.
To confirm that SKIP is not required for transcription under stress conditions, we asked whether nascent unspliced p21 transcripts accumulate in SKIP knockdown cells. Total RNA was isolated from U2OS cells in the presence or absence of etoposide, and was amplified using intron-specific primers specific for nascent p21 and PUMA transcripts (see the Materials and Methods). Interestingly, primary transcripts derived from the p21 (+540 and +6990 primers) and PUMA (+2460 and +6803 primers) genes increased significantly in SKIP knockdown U2OS cells in the absence of stress, and even more dramatically upon addition of etoposide (Fig. 2C, top row). Virtually identical results were observed in HCT116 cells following doxorubicin treatment (Fig. 2C, bottom row). No significant signals were detected from control PCR reactions programmed with RNA but lacking reverse transcriptase (Supplemental Fig. S2C), indicating that the RNA samples were effectively free of contaminating genomic DNA. We conclude that SKIP is dispensable for stress-induced nascent p21 transcription in vivo.
SKIP is required for pre-mRNA splicing of p21, but not PUMA, transcripts
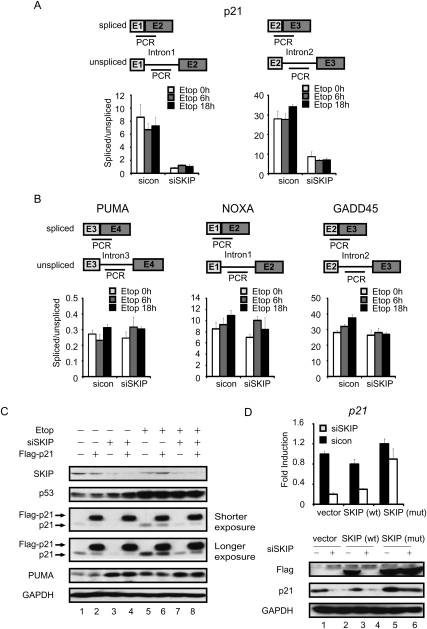
To determine whether SKIP is required for splicing of p21 mRNA, quantitative RT–PCR (qRT–PCR) reactions using intron–exon and exon–exon junction-specific primers were carried out to measure spliced and unspliced mRNA levels, and the ratio of spliced:unspliced transcripts was then used to gage splicing efficiency. As shown in Figure 3A, splicing at either the first or second p21 intron was relatively unchanged upon etoposide treatment in cells treated with a control siRNA, but declined significantly (eightfold and 3.5-fold to fourfold, respectively) in SKIP knockdown cells. The drop in splicing efficiency in SKIP knockdown cells was evident in both the presence and absence of DNA damage. Importantly, loss of SKIP did not affect splicing at the PUMA, NOXA, and GADD45 genes, all of which are direct targets of p53 (Fig. 3B). Virtually identical results were obtained in HCT116 cells exposed to doxorubicin (Supplemental Fig. S3A). We conclude that SKIP is important for efficient splicing of both p21 mRNA introns, but does not affect splicing of PUMA or other tested p53-induced transcripts.
Figure 3.
SKIP regulates p21 mRNA splicing in vivo. (A, top panel) Schematic diagram of the primer pairs used to detect p21 unspliced and spliced mRNAs. (Bottom panel) qRT–PCR analysis was used to determine the ratio of unspliced to spliced p21 mRNA. U2OS cells were transfected with control or SKIP siRNA, and incubated with etoposide as indicated. (B) The top panel shows a schematic representation of the primer pairs used to detect the PUMA, GADD45, or NOXA unspliced and spliced mRNAs, whereas the bottom panel shows the ratio of unspliced to spliced mRNA for each gene, as determined by qRT–PCR. U2OS cells were treated as in A. (C) U2OS cells were transfected with empty vector or pCMV-Flag-p21, and, 24 h later, were transfected with control or SKIP siRNA for another 48 h. Cells were left untreated or treated with etoposide (20 μM) for a further 18 h prior to Western blot analysis. (D) Rescue of SKIP knockdown with an siRNA-resistant vector. U2OS cells were transfected with control, wild-type, or mutant SKIP vectors, and, 12 h later, were transfected with control or SKIP siRNA prior to mRNA and protein analysis at 48 h.
To examine the effects of SKIP knockdown on the p21 mRNA stability, qRT–PCR was performed to measure p21 mRNA half-life in U2OS cells that were transfected with SKIP or control siRNAs for 48 h, followed by treatment with transcriptional inhibitor actinomycin D for 0, 2, 4, or 6 h (Supplemental Fig. S3B). The results indicate that SKIP has no significant effect on p21 mRNA stability. The SKIP homolog in Drosophila has been shown to promote the export of spliced mRNAs (Farny et al. 2008). To test whether SKIP affects the mRNA export of p21 mRNA in human cells, SKIP or control siRNAs were transfected in U2OS cells for 48 h, followed by the treatment with etoposide for 18 h. Cells were fractionated into nuclear and cytoplasmic fractions, and mRNA levels were monitored. As shown in Supplemental Figure S3C, depletion of SKIP did not significantly affect export of either p21 or GAPDH mRNAs, indicating that the mRNA export pathway used in mammalian cells under DNA damage conditions is not dependent on SKIP.
To determine whether SKIP might also affect p21 protein stability, the rate of p21 protein turnover was measured in SKIP knockdown cells in the absence of stress. Forty-eight hours after transfection with control or SKIP siRNA, U2OS cells were treated with cycloheximide (CHX) to prevent new protein synthesis, and the decay of endogenous p21 protein was measured (Supplemental Fig. S3D). The results indicate that SKIP has no significant effect on p21 stability in the absence of stress. The proteasome inhibitor MG132 elevated p21 protein levels in both control and SKIP siRNA transfected cells (Supplemental Fig. S3E), indicating that it is a short-lived protein and subject to active proteolytic degradation under both conditions. Based on these findings, we reasoned that a cDNA encoding p21 should be expressed independently of SKIP in these cells. To assess this possibility, a Flag-tagged p21 cDNA encoding the full-length p21 protein expressed from a heterologous (CMV) promoter and lacking both introns as well as 5′ untranslated region (UTR) and 3′UTR sequences was transfected into U2OS cells, and, after 24 h, either control or SKIP siRNAs were transfected into the cells for a further 48 h, followed by treatment with or without etoposide for 18 h. As shown in Figure 3C, the basal and stress-induced endogenous p21 protein levels decreased in SKIP-depleted cells, whereas expression of the larger Flag-p21 hybrid protein was unaffected. Similar results were obtained from p53-null H1299 cells in the absence of stress (Supplemental Fig. S3F). Importantly, the decrease of p21 mRNA and protein levels in SKIP knockdown cells was effectively rescued upon expression of a vector encoding an siRNA-resistant form of SKIP, but not the wild-type (siRNA-sensitive) SKIP (Fig. 3D), indicating that these results are not due to off-target effects. Together, these data indicate that SKIP regulates p21 expression through a unique gene-specific splicing mechanism.
SKIP interacts with and recruits U2AF65 to the p21 gene and mRNA
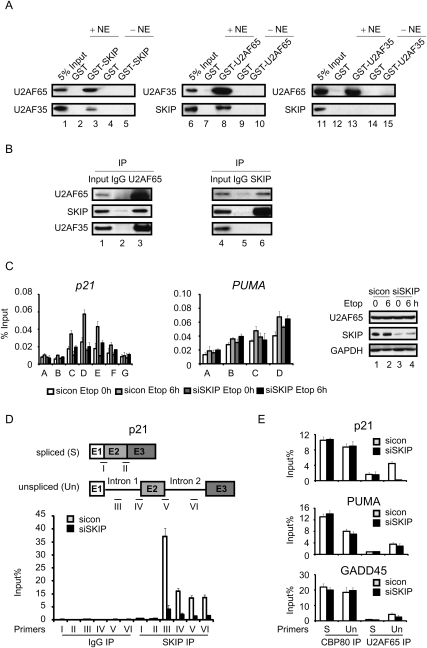
Although SKIP is required for splicing, the steps it regulates are unclear. SKIP is a component of the activated spliceosome complex; however, the fission yeast homolog of SKIP was shown previously to bind U2AF35, the small subunit of the U2AF 3′ splice site recognition complex (Ambrozkova et al. 2001), indicating that it might also function at an early step in splicing. To determine whether human SKIP protein also associates with the U2AF complex, recombinant full-length glutathione-S-transferase (GST)-SKIP was purified and coupled to glutathione-S-sepharose beads for GST pull-down experiments using nuclear extracts from HCT116 cells. Relatively low levels of U2AF35 were recovered in the GST-SKIP pull-down fractions, and this association was disrupted when the beads were treated with RNase A (V Brès and K Jones, unpubl.), indicating that this interaction may be indirect. Interestingly, we observed much stronger binding of the endogenous U2AF65 protein to the GST-SKIP beads (Fig. 4A, left panel), and this association was unaffected by RNase A (V Brès and K Jones, unpubl.). No U2AF65 was recovered in the control GST-bead fraction, indicating that the interaction is specific for SKIP. In reciprocal pull-down experiments, GST-U2AF65 bound avidly to nuclear U2AF35 and SKIP, whereas none of these factors bound to GST alone (Fig. 4A, middle panel, cf. lanes 7 and 8). Interestingly, SKIP was not detected in GST-U2AF35 pull-down fractions (Fig. 4A, right panel), which otherwise contained high levels of nuclear U2AF65. To examine this association further, reciprocal coimmunoprecipitation experiments were performed with U2OS whole-cell lysates. As shown in Figure 4B, both SKIP and U2AF35 coimmunoprecipitated with U2AF65 (left panel), whereas U2AF65, but not U2AF35, was recovered in the SKIP immunoprecipitate (right panel). These results indicate that SKIP interacts with U2AF65 independently of U2AF35.
Figure 4.
SKIP associates with unspliced p21 mRNA and recruits U2AF65. (A) Immunoblot analysis of the interaction between SKIP, U2AF35, and U2AF65 in GST pull-down experiments from HCT116 cell nuclear extract. (B) Total proteins were extracted from U2OS cells for coimmunoprecipitation. Immunoprecipitates were examined by Western blot using antibodies against SKIP, U2AF35, or U2AF65. (C) ChIP analysis of U2AF65 binding on the p21 and PUMA genes. U2OS cells were transfected with control or SKIP siRNA for 48 h, followed by treatment with vehicle or etoposide (20 μM) for 6 h. Protein extracts were immunoprecipitated with antibodies against U2AF65. ChIP-enriched DNA was quantified by qPCR with the indicated primers in Figure 2A and Supplemental Figure S2A. (Right panel) Immunoblot analysis. Error bars represent the standard deviation obtained from three independent experiments. (D, top panel) Schematic representation of the primer pairs used to detect p21 unspliced and spliced mRNAs. RNA-IP analysis of binding of the SKIP protein to p21 unspliced or spliced mRNA. U2OS cells were transfected with control or SKIP siRNA for 48 h. RNA samples were purified from nonprecipitated cellular lysates (input), or extracts precipitated with control IgG or SKIP antibody. Immunoprecipitated p21 mRNA was detected using qRT–PCR with the indicated primers. Values were expressed as percentage of input RNA. Error bars represent the standard deviation obtained from three independent experiments. (E) RNA-IP analysis of binding of CBP80 or U2AF65 to p21, PUMA, or GADD45 unspliced or spliced mRNA. Experiments were performed as in D. The primers used for detecting p21 transcripts were primer IV (unspliced) and primer I (spliced) as in D. The primers used for detecting PUMA or GADD45 transcripts were the same as in Figure 3B.
Based on these findings, we next used RNAi-ChIP experiments to analyze whether SKIP is responsible for cotranscriptional recruitment of mRNA splicing factors at the p21 gene. Interestingly, U2AF65 occupancy within the coding region of the p21 gene decreased significantly in SKIP knockdown cells (Fig. 4C, left panel). In contrast, loss of SKIP had no effect on binding of U2AF65 to the PUMA gene (Fig. 4C, center panel). Steady-state U2AF65 protein levels were unaffected in SKIP-depleted cells, as measured by immunoblot (Fig. 4C, right panel). Unfortunately, we were unable to monitor U2AF35 occupancy at the p21 gene due to lack of a suitable antibody. We conclude that SKIP is required for stable binding of U2AF65 at the p21, but not PUMA, genes in vivo.
These data strongly suggest that SKIP regulates cotranscriptional loading of U2AF65 and splicing at both introns of the p21 gene, and that spliceosomal complexes formed in the absence of SKIP may be unable to splice p21 mRNAs whether on or off of the gene. To examine U2AF65 binding to p21 mRNA directly, RNA immunoprecipitation (RNA-IP) experiments were carried out in U2OS cell extracts. As shown in Figure 4D, high levels of the p21 transcript were recovered in SKIP antibody, and not control immunoglobin G (IgG), immunoprecipitates. Importantly, the SKIP immunoprecipitation (SKIP-IP) fractions contained significantly higher levels of unspliced (detected with primers III–VI) than spliced (detected with primers I–II) transcripts. Furthermore, the level of unspliced transcript bound to SKIP declined greatly in SKIP knockdown cells, whereas the low background level of spliced mRNA in the SKIP-IP fraction was unaffected, indicating that this latter signal is nonspecific. Because the mRNA in these experiments was not sonicated, it was not possible to localize the position of SKIP binding in these experiments. Thus, the higher signal detected with primer III likely reflects an increased efficiency in binding to p21 mRNA. Interestingly, SKIP also bound to PUMA mRNA introns. Thus, SKIP binds preferentially to introns, presumably as part of the spliceosome complex, but does not discriminate between the p21 and PUMA mRNA. Therefore, SKIP selectivity in splicing is likely conferred by its ability to bind to the core promoter and recruit U2AF65 cotranscriptionally to the p21 gene and mRNA.
Promoter-proximal intron splicing is strongly influenced by 5′-mRNA capping (Lewis et al. 1996), and, consequently, we asked whether SKIP affects loading of the mRNA cap-binding protein CBP80. As shown in Figure 4E, p21, PUMA, and GADD45 mRNAs were efficiently recovered in CBP80 immunoprecipitates, and ablation of SKIP had no affect on CBP80 binding to either the spliced or unspliced mRNAs. In contrast, U2AF65 bound preferentially to the unspliced mRNAs. Most interestingly, the binding of U2AF65 to unspliced p21 mRNA was largely abolished in si-SKIP-treated cells (Fig. 4E), consistent with the ChIP results, whereas U2AF65 binding to the PUMA or GADD45 mRNAs was only modestly affected in SKIP knockdown cells. To assess whether U2AF65 is required for expression of p21 and PUMA genes, mRNA and protein levels for these genes were analyzed in cells transfected with si-U2AF65. Knockdown of U2AF65 significantly reduced pre-mRNA splicing (Supplemental Fig. S4A) and protein expression (Supplemental Fig. S4B) of both p21 and PUMA mRNAs in control and DNA-damaged cells, confirming its role as a general splicing factor. These data indicate that SKIP binds to introns at both target and nontarget mRNAs, and is required for binding of U2AF65 to p21 mRNA. Although U2AF65 is also required for splicing of PUMA mRNA, it is recruited to the gene and mRNA independently of SKIP.
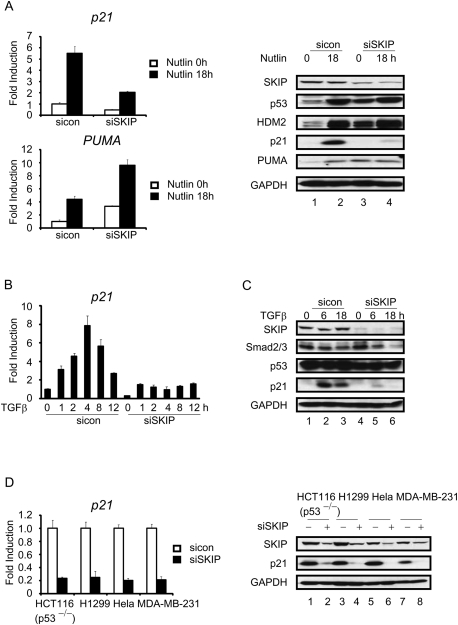
SKIP is also required for p21 induction by Nutlin3a or TGF-β signaling
To assess whether SKIP-regulated p21 gene expression is restricted to conditions of stress, we used the nongenotoxic drug Nutlin3 to activate p53 and induce p21 gene expression in U2OS cells. Nutlin3 disrupts binding of p53 to the HDM2 ubiquitin ligase, and therefore can stabilize p53 in the absence of stress. As shown in Figure 5A, Nutlin3 induced p53 activation of several downstream target genes, including p21, PUMA, and HDM2. Nutlin3-induced expression of PUMA and HDM2 was further increased in si-SKIP cells, while the induction of p21 was strongly suppressed. Thus, SKIP is required for p53-induced p21 expression, irrespective of DNA damage. To address whether SKIP regulation depends on the activator, p21 induction was studied in the human breast cancer cell line MDA-MB-231, which expresses a mutant p53 protein, treated with anti-mitogenic cytokine transforming growth factor-β (TGF-β). In these cells, p21 mRNA was induced rapidly in response to TGF-β signaling, and mRNA levels peaked 4 h after induction (Fig. 5B). Addition of TGF-β did not affect the mutant p53 protein levels (Fig. 5C). Strikingly, this increase of p21 mRNA and protein was completely abolished in SKIP knockdown cells (Fig. 5B,C). These findings in H1299 (p53-null) cells were compared with two cell lines that are deficient for p53 signaling: HeLa (p53 inactivated by the E6 protein of HPV-18) and HCT116 p53−/− (p53 gene deleted by homologous recombination). In all of these cells, loss of SKIP gave rise to a strong inhibition of endogenous p21 mRNA and protein expression (Fig. 5D). In the absence of stress, SKIP likely affects both p21 transcription elongation and splicing. Together, these findings highlight the general role for SKIP as a critical regulator of p21 expression.
Figure 5.
SKIP is required for Nutlin and TGF-β-induced p21 gene expression. (A, left panel) qRT–PCR analysis of p21 and PUMA mRNA levels. U2OS cells were transfected with control or SKIP siRNA for 48 h, and incubated in the presence or absence of Nutlin (10 μM) for 18 h. (Right panel, lanes 1–4) Protein lysates were subjected to immunoblot analysis. (B) qRT–PCR analysis of p21 mRNA levels. MDA-MB-231 cells were transfected with control or SKIP siRNA for 48 h, followed by incubation in the presence or absence of TGF-β (5 ng/mL) for the indicated times. (C, lanes 1–6) Immunoblot analysis of SKIP, Smad2/3, p53, p21, or GAPDH in cells transfected with control or SKIP siRNA for 48 h, followed by treatment with TGF-β (5 ng/mL) for the indicated times. (D, left) qRT–PCR analysis of p21 mRNA levels in HCT116 p53−/− cells, H1299 cells, HeLa cells and MDA-MB-231 cells transfected with control or SKIP siRNA for 48 h. (Right, lanes 1–8) Protein lysates were subjected to immunoblot analysis. All of the mRNA expression levels were normalized to GAPDH mRNA, and are represented as fold increase or decrease over untreated cells. Error bars represent the standard deviation obtained from three independent experiments.
SKIP is an essential cancer cell survival factor that counteracts DNA damage-induced apoptosis
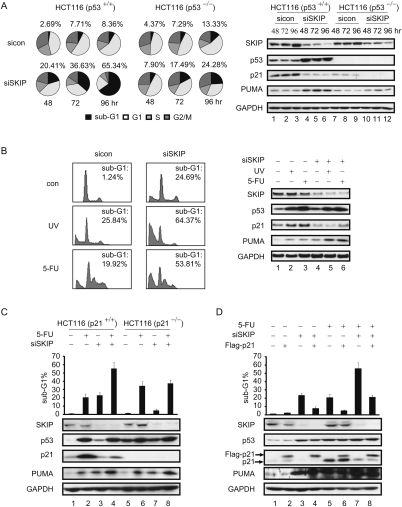
The observation that SKIP is critical for p21, but not PUMA, gene expression indicates that loss of SKIP should predispose cells to undergo p53-dependent apoptosis. To test this directly, HCT116 cells were transfected with SKIP siRNA or control siRNA for 48, 72, and 96 h. The cells were collected and the percentage of cells in each phase of the cell cycle was quantified by flow cytometric analyses. As shown in Figure 6A, knockdown of SKIP did not lead to cell cycle arrest at the G1, S, or G2/M phase of the cell cycle. Rather, the SKIP-depleted cells were subjected to massive DNA fragmentation and cell apoptosis, as measured by the sub-G1 DNA content, with >70% cell death at 96 h following transfection of SKIP siRNA. Next, we asked whether SKIP depletion can induce apoptosis in the isogenic HCT116 p53−/− cell line. As observed in the HCT116 parental cells, the cell cycle progression of the SKIP-depleted cells is similar to that of the cells transfected with control siRNA. However, cell death triggered by knockdown of SKIP is largely attenuated, but not absent, in the HCT116 p53−/− cells, with the percentage of cells in the sub-G1 fraction reduced to 25% after 96 h of treatment with SKIP siRNA (Fig. 6A, bottom panel). The expression of endogenous SKIP was identical in these two cell lines, whereas both p21 and PUMA protein levels were higher in the HCT116 parental cells compared with the p53-null cells (Fig. 6A, right panel). Detailed quantification of the effect of si-SKIP on the cell cycle is presented in Supplemental Table 1. We conclude that SKIP is required for cancer cell survival through its role in p21 expression, which counteracts p53-mediated apoptosis.
Figure 6.
SKIP is required for cell survival and modulates DNA damage-induced cell apoptosis. (A, left panel) FACS analysis of the cell cycle profile. HCT116 cells or HCT116 p53−/− cells were harvested at indicated times after transfection with control or SKIP siRNA, and the DNA content was determined by FACS. Pie charts display the percentage of cells in each stage of the cell cycle. The percentage of sub-G1 cells is indicated above each chart; Supplemental Table S1 lists the data for other cell cycle phases. (Right panel) Cell extracts were subjected to immunoblot analysis. (B, left) FACS analysis of cell apoptosis. Twenty-four hours after control or SKIP siRNA transfection, HCT116 cells were left untreated or treated with UV (60 J/m2) or 5-FU (50 μM) for 24 h. The percentage of cells in the sub-G1 phase was quantified for the plots. (Right panel, lanes 1–6) Cell extracts were subjected to immunoblot analysis. Role of p21 in SKIP-regulated cell apoptosis. (C, top panel) FACS analysis of cell apoptosis. HCT116 cells or HCT116 p21−/− cells were transfected with control or SKIP siRNA for 24 h, and left untreated or treated with 5-FU (50 μM) for 24 h. The percentage of cells in the sub-G1 phase was quantified by FACS and is represented in the graph. (Bottom panel, lanes 1–8) Cell extracts were subjected to immunoblot analysis. (D, top panel) FACS analysis of cell apoptosis. HCT116 cells were first transfected with empty vector or pCMV-Flag-p21, and, 24 h later, were transfected with control or SKIP siRNA for another 24 h, and then the cells were left untreated or treated with 5-FU (50 μM) for a further 24 h. The percentage of cells in the sub-G1 phase was quantified and is represented in the graph. (Bottom panel, lanes 1–8) Cell extracts were subjected to immunoblot analysis. Error bars represent the standard deviation obtained from three independent experiments.
The observation that SKIP remains essential for p21 protein expression even under conditions of stress led us to ask whether loss of SKIP sensitizes cells to apoptosis induced by chemotherapeutic DNA damage agents. Therefore, HCT116 cells were treated either with si-control or si-SKIP RNA, and, 48 h after transfection, the cells were treated with UVC or 5-FU for a further 24 h. FACS analysis of these cells revealed that apoptosis induced by UVC or 5-FU treatment was much higher in cells containing reduced levels of SKIP (Fig. 6B, left panel). Immunoblots were also used to monitor the protein levels of SKIP, p53, PUMA, and p21 in these experiments (Fig. 6B, right panel), and confirmed that p21 expression remains SKIP-dependent under UVC and 5-FU stress conditions. These findings indicate that SKIP loss strongly augments chemotherapy-induced cell killing.
Conversely, we also asked whether ectopic expression of SKIP would render cells resistant to p53-mediated apoptosis. To address this question, HCT116 cells were engineered to stably express a V5-tagged SKIP protein (HCT116-SKIP). HCT116 and HCT116-SKIP cells were treated with either UVC or 5-FU for 48 h, and apoptosis was monitored by FACS sorting. Strikingly, HCT116-SKIP cells were much more resistant to DNA damage-induced cell death (Supplemental Fig. S5A). However, immunoblot analysis of protein expression in these cells indicates that activation of p53 is significantly impaired in these cells, and, consequently, the mechanism is distinct from that observed in SKIP knockdown cells. Similar results were observed in HCT116 cells that overexpress SKIP through transient expression (Supplemental Fig. S5B). Thus, excessively high levels of SKIP may inactivate factors that are normally required for p53 activation. Taken together, these results suggest that SKIP is critical for cell viability, and that changes in SKIP expression can strongly modulate the cell response to DNA damage.
The anti-apoptotic function of SKIP is primarily due to its ability to regulate p21 expression
Together, these findings suggest that SKIP depletion sensitizes cells to undergo apoptosis through its ability to prevent p21 expression. To test this model, we asked whether knockdown of SKIP affects apoptosis in HCT116 p21−/− cells, which lack the p21 protein and are more prone to undergo apoptosis in response to DNA damage. Although p53 was induced more strongly in 5-FU-treated HCT116 cells, levels of the anti-apoptotic p21 protein were also much higher in these cells than in the SKIP knockdown cells (Fig. 6C, cf. lanes 2 and 3), and consequently, the overall extent of apoptosis was comparable in 5-FU and SKIP-depleted cells. Knockdown of SKIP in the 5-FU-treated cells resulted in high levels of p53 and low levels of p21, further enhancing apoptosis (Fig. 6C, lane 4). In contrast, in the HCT116 p21−/− cells, basal p53 levels are higher (Fig. 6C, lane 5), and increase further upon exposure to 5-FU (Fig. 6C, lane 6), but only modestly, if at all, in the si-SKIP-treated cells (Fig. 6C, lane 7). Consequently, 5-FU treatment increases apoptosis more readily in HCT116 p21−/− cells (Fig. 6C, lane 6), whereas apoptosis is only modestly increased upon SKIP depletion (Fig. 6C, lane 7), consistent with the fact p53 levels are only marginally higher in these cells. Moreover, 5-FU-mediated apoptosis was not enhanced further by SKIP knockdown in the HCT p21−/− cells (Fig. 6C, lane 8). Thus, the enhanced apoptosis seen in SKIP-depleted cells is predominantly linked to down-regulation of p21 expression, which appears to be a major target for SKIP in HCT116 cells, whereas 5-FU-induced cell death is linked to the strong induction of p53.
In addition, we asked whether overexpression of Flag-p21 could block the apoptotic effect of SKIP knockdown in HCT116 cells. As shown in Figure 6D, expression of the Flag-p21 protein significantly reduced cell death induced by depletion of SKIP (cf. lanes 3 and 4) or treatment with 5-FU (cf. lanes 5 and 6), as well as the enhanced level of apoptosis observed in cells exposed to both 5-FU and SKIP-siRNA (cf. lanes 7 and 8). The expression of SKIP, p53, and PUMA under these different experimental conditions was monitored by immunoblot (Fig. 6D, bottom panel), and confirmed that ectopic p21 blocks apoptosis without influencing expression of any of these factors, presumably through induction of cell cycle arrest. Together, these findings indicate that the primary mechanism by which SKIP controls p53 apoptosis is through its ability to regulate p21 expression.
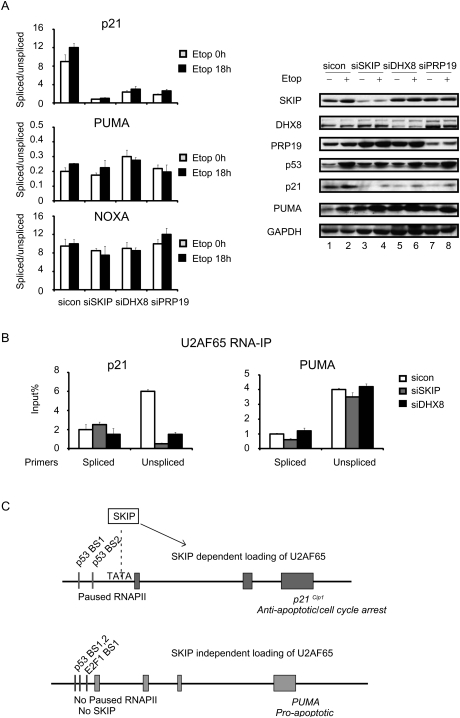
The SKIP-associated factors DHX8 and Prp19 are also selectively required for p21 splicing
Although SKIP has been shown to regulate the catalytic step in splicing as a component of the activated spliceosome, our findings indicate that it also functions at an earlier step to regulate loading of U2AF65 at the p21 gene. Consequently, we wondered whether other SKIP-interacting splicing factors also control p21 gene-specific splicing. Previous studies have shown that SKIP interacts with DHX8 (hPrp22), the human homolog of a yeast RNA helicase implicated in branch point recognition and removal of the spliceosome from the transcript (Gahura et al. 2009), and both proteins were detected in a genome-wide RNAi screen for factors required for mitotic progression through prometaphase (Kittler et al. 2004). Within the spliceosome, SKIP also associates with Prp19 complex proteins (Wahl et al. 2009). Interestingly, these experiments revealed that siRNA-mediated knockdown of human DNX8 or Prp19 leads to a selective down-regulation of splicing of p21 transcripts, without affecting splicing of PUMA or NOXA mRNAs (Fig. 7A, left panel); a corresponding decline in p21 protein expression was also evident by immunoblot (Fig. 7A, right panel). Moreover, RNA-IP analysis established that U2AF65 loading on p21 mRNA is strongly reduced in the DHX8 knockdown cells (Fig. 7B). These findings indicate that other spliceosome components also function selectively in p21 expression, and contrast with siRNA knockdown of U2AF65, which disrupts splicing of both p21 and PUMA mRNAs. We conclude that a subset of SKIP-associated spliceosomal proteins is not universally required for splicing under stress, but rather functions in a gene-specific manner to regulate cotranscriptional p21 mRNA splicing.
Figure 7.
DHX8 and Prp19 selectively regulate p21 mRNA splicing, and DHX8, like SKIP, is required for binding of U2AF65 to p21 unspliced mRNA. (A, left) qRT–PCR was used to monitor the ratio of unspliced to spliced p21, PUMA, or NOXA mRNAs. U2OS cells were transfected with control, SKIP, DHX8, or Prp19 siRNA for 48 h, and incubated in the presence or absence of etoposide (20 μM) for the indicated times. (Right panel, lanes 1–6) Protein lysates were subjected to immunoblot analysis. (B) RNA-IP analysis of binding of U2AF65 to p21 or PUMA unspliced or spliced mRNA. U2OS cells were transfected with control, SKIP, or DHX8 siRNA for 48 h. RNA samples were purified from nonprecipitated cellular lysates (input) or extracts precipitated with U2AF65 antibody. Immunoprecipitated p21 transcript was detected using qRT-PCR with the primers used in Figure 4E. Values are expressed as percentage of input RNA. Error bars represent the standard deviation obtained from three independent experiments. (C) Model for the role of SKIP in the regulation of p21 gene-specific splicing.
Discussion
The CDK inhibitor p21 is a potent cell cycle arrest factor that counteracts p53-dependent apoptosis and predisposes cells to undergo differentiation or cellular senescence. Transcriptional induction of the p21 gene plays a central role in TGF-β/SMAD-mediated G1 cell cycle arrest, as well as DNA damage/p53-induced inhibition of cell division. Conversely, the p21 gene is transcriptionally repressed by c-Myc to override the cell cycle checkpoint and promote proliferation. Here, we show that basal and stress-induced p21 expression requires the SKIP/SNW1 transcription elongation and splicing factor. These results were unexpected, given that p53 induction of p21 does not require the P-TEFb elongation factor (Gomes et al. 2006), and that neither P-TEFb nor SKIP is required for stress-induced HIV-1 transcription (Brès et al. 2009). RNAi-ChIP experiments revealed that SKIP is not required for p53 binding or accumulation of Ser2P-RNAPII in the body of the p21 gene, and qRT–PCR analysis with intron-specific primers confirmed that it is not needed for nascent p21 transcription. Thus, SKIP, like P-TEFb, is dispensable for transcription at the p21 gene in cells exposed to DNA damage, supporting the idea that elongation control is lost in cells subjected to DNA damage. At heat-shock genes, P-TEFb predominantly affects 3′-mRNA end processing, rather than promoter-proximal elongation (Ni et al. 2004). At the stress-induced HIV-1 promoter, transcription is accompanied by loss of typical histone modifications, including trimethylation of H3K4 (H3K4me3) and H2BUb, and activation of heat-shock genes is preceded by widespread nucleosome depletion (Petesch and Lis 2008). Thus, profound changes in chromatin structure may alleviate the need for elongation factors under DNA damage.
SKIP selectively regulates p21 pre-mRNA splicing under stress
To define the block to p21 expression, qRT–PCR experiments were carried out using intron–exon and exon–exon junction primers to monitor the level of spliced mRNA, which revealed a strong block to splicing at both p21 introns in SKIP knockdown cells. Following an earlier report that the fission yeast SKIP homolog associates with the U2AF recognition factor (Ambrozkova et al. 2001), we discovered that human SKIP interacts with U2AF65, the polypyrimidine tract-binding factor required for 3′ splice site recognition. Interestingly, SKIP appears to recognize U2AF65 independently of the small U2AF subunit U2AF35. ChIP studies revealed that SKIP recruits U2AF65 to the p21 gene, and RNA-IP experiments indicated that it is also required for U2AF65 binding to the mRNA. However, SKIP is not required for splicing or binding of U2AF65 to the PUMA gene and mRNA. The RNA-IP experiments further revealed that SKIP preferentially associates with introns rather than exons, presumably as part of the spliceosome; however, it is present at both target (p21) and nontarget (PUMA) mRNAs. In contrast, SKIP is present at the p21, but not PUMA, genes both before and after DNA damage, indicating that the specificity is determined by the core promoter.
Previous studies have shown that the p21 promoter, like the HIV-1 promoter, contains high levels of paused RNAPII prior to induction, whereas the PUMA promoter assembles the RNAPII transcription complex de novo upon gene activation (Gomes and Espinosa 2010). Many transcription factors discriminate between these two promoter types, including p300 and c-Myc, which activate and repress p21 expression, respectively, without affecting PUMA gene transcription (Seoane et al. 2002; Iyer et al. 2004). ChIP experiments show that SKIP binds to the p21 gene with a pattern similar to that observed for P-TEFb, peaking at the core promoter and proximal region. The absence of SKIP at the PUMA gene establishes that it is not recruited through p53. It is unclear how SKIP is recruited to the p21 gene; however, we showed previously that it is recruited to the basal HIV-1 promoter via the H2B ubiquitin ligase hRNF20 (Shema et al. 2008). It will be interesting to learn whether any other transcription or chromatin regulators at the p21 promoter also affect splicing and cotranscriptional loading of U2AF65. We did not observe any effect of SKIP on p21 mRNA or protein stability or mRNA export; however, it remains possible that it could affect p21 translation, which depends on cotranscriptional loading of the CUGBP1 5′UTR factor (Iakova et al. 2004). Translation could also be affected by mRNA-capping defects, although we did not detect any defect in binding of the cap-binding protein CBP80 to either p21 or PUMA mRNA.
Evidence that p21 splicing is cotranscriptional
Although it is widely recognized that elongation factors can indirectly affect mRNA splicing patterns through changes in the rate of nascent transcription, SKIP appears to directly affect each process. SKIP is an essential factor in many organisms (Folk et al. 2004), and studies of the S. cerevisiae (Prp45) or Drosophila (BX42) homologs have focused mainly on its roles in mRNA splicing, spliceosome assembly, and export of spliced mRNAs (Farny et al. 2008). Consequently, it was not surprising to find a role for SKIP in splicing of the p21 gene. What is remarkable is the gene-specific activity of SKIP under stress, where it is dispensable for splicing of many p53 target genes, including PUMA, GADD45, and NOXA. Moreover, SKIP differentially affects p21 and not PUMA expression even in the absence of stress, and therefore appears not to be universally required for splicing in human cells. Our data indicate that SKIP is required to load U2AF65 onto the p21 gene and mRNA, and we found no evidence for selective binding of SKIP to the p21 mRNA, indicating that splicing is predominantly cotranscriptional in this case. This conclusion is consistent with recent studies showing that RNAPII undergoes pausing and release—accompanied by changes in RNAPII phosphorylation—at 3′ splice sites, and that cotranscriptional splicing may be widespread in yeast (Alexander et al. 2010, Oesterreich et al. 2010), and is also consistent with studies showing that the SC35 splicing factor can affect RNAPII elongation and pausing (Lin et al. 2008; Xiao et al. 2008). These studies also raise the question of whether the 3′ splice site recognition factors might also play a role in promoter-proximal pausing at some genes. In addition, although the p21 intron sequences appear to conform to the consensus, it is possible that the intron also contributes to SKIP-dependent binding of U2AF65. Unfortunately, the p21 reporter genes that we tested are not responsive to stress, and therefore it is unclear whether the p21 promoter is sufficient to confer SKIP-dependent splicing to a heterologous intron. We also show that two SKIP-associated splicing factors, DHX8 (hPrp22) and Prp19, also selectively regulate p21 splicing under stress conditions. The yeast homolog of DHX8, Prp22, promotes the second catalytic step of splicing at nonconsensus splice sites (Gahura et al. 2009), and is also involved in mRNA release from the spliceosome (Schwer 2008). Thus, a subset of splicing factors may function with SKIP to control cotranscriptional loading of U2AF65 at target genes.
Interestingly, we found that SKIP selectively associates with U2AF65, and not with its heterodimeric partner, U2AF35. In this respect, SKIP resembles certain other regulatory factors, including the Wilms' tumor protein, which binds selectively to U2AF65 and not U2AF35 (Davies et al. 1998). In contrast, the histone H3.3 chaperone and oncogene DEK (Sawatsubashi et al. 2010) regulates the 3′ splice site checkpoint through selective binding to U2AF35 (Soares et al. 2006) and not U2AF65. Binding of DEK to U2AF35 confers its specificity for the 3′-AG dinucleotide, and is required for U2AF35 binding at selected introns (Soares et al. 2006). In addition, the transcription coregulator SNIP1—which controls CycD1 expression and cell cycle progression (Bracken et al. 2008), as well as c-Myc stability and transactivation (Fujii et al. 2006)—functions to recruit U2AF65 and other RNA processing factors to the 3′ end of the CycD1 gene and mRNA to control mRNA stability. Interestingly, substoichiometric amounts of SKIP were detected in the SNIP1 RNA processing complex (Bracken et al. 2008). SNIP1 is also required for p53 expression and ATR substrate phosphorylation (Roche et al. 2007). Because SNIP1 inhibits TGF-β signaling (Kim et al. 2000), opposite to the role of SKIP, it will be interesting to examine whether competition for U2AF65 might influence RNAPII pausing and elongation. SKIP also associates with the MLL1:Menin histone methyltransferase and is required for H3K4 methylation at the HIV-1 promoter (Brès et al. 2009), indicating that it may, like DEK, provide a link between splicing and chromatin. In this respect, it is interesting that chromatin modifications can directly impact splicing specificity (Luco et al. 2011). Cotranscriptional loading of the U2snRNP complex has been shown to depend on H3K4me3 and the Chd1 chromatin remodeling factor (Sims et al. 2007), as well as SAGA/Gcn5 acetylation of histone H3 (Gunderson and Johnson 2009), and it will be interesting to learn whether SKIP might also regulate splicing through changes in chromatin structure.
SKIP is an essential cancer cell survival factor
We show here that ablating SKIP expression results in p53-mediated apoptosis of HCT116 colon cancer or U2OS osteosarcoma cells. In contrast, SKIP knockdown in HeLa cells, which lack a functioning p53 pathway, results in G2/M arrest in prometaphase (Kittler et al. 2004, 2005). Thus, the p53 pathway appears to be a prime target for SKIP in colon cancer cells. Although SKIP may regulate splicing at many genes, the observation that HCT116 p21−/− cells are largely insensitive to apoptosis by si-SKIP, and that Flag-p21 overexpression is sufficient to block apoptosis in wild-type HCT116 cells, strongly indicates that p21 is a major target for SKIP in these cells.
Numerous studies have also identified potent anti-apoptotic roles for various splicing factors in the control of alternative splicing that commonly regulate splice site choice through effects on binding of the U2AF65:35 complex (Chen and Manley 2009). Alternative splicing of Bcl2 mRNAs regulates the balance of expression of pro- and anti-apoptotic family members, and also mediates the differential expression of various death receptors, death ligands, and caspases. Splicing factor activity is subject to inhibition by stress, with different effects on the constitutive and alternative splicing pathways (for review, see Giul and Cáceres 2007; Biamonti and Cáceres 2008). We observed previously that ectopic expression of the SKIP SNW domain strongly favors the use of the HIV-1 A3 splice acceptor site (Brès et al. 2005), indicating that it might also play a role in alternative splicing. Consequently, it will be important to determine whether SKIP regulates the alternative splicing pattern of genes involved in apoptosis, and, similarly, whether alternative splicing factors, including the SR proteins, regulate cotranscriptional p21 gene-specific splicing under stress conditions.
Taken together, our findings indicate that inhibitors of SKIP could be of therapeutic benefit by augmenting DNA damage chemotherapy-induced apoptosis. Like certain other short-lived anti-apoptotic factors, we found that SKIP levels decline in cells treated with the CDK inhibitor FP (V Brès and K Jones, unpubl.), which has shown clinical benefit in leukemia and as a combination chemotherapy for colon cancer. Importantly, we show here that apoptosis associated with SKIP ablation is greatly enhanced when combined with DNA damage agents that further induce p53 levels, such as 5-FU and UV (Fig. 7). Thus, SKIP and associated enzymes that control p21 splicing, such as DHX8 and Prp19, may be useful anti-cancer targets, as would be small molecule inhibitors that selectively block the protein–protein interactions needed to recruit U2AF65 to the p21 gene. Further studies on the mechanism of SKIP-regulated p21 mRNA splicing, and identification of other factors that control this step, may suggest new approaches to enhance chemotherapy-induced cell killing.
Materials and methods
Plasmids, siRNAs, drugs, and antibodies
Mammalian expression constructs of human pV5-SKIP and pFlag-SKIP were generated by subcloning SKIP cDNA into pcDNA6 (Invitrogen) and pCMV-Tag2 (Stratagene) vectors, respectively. Human Flag-p21 was obtained from Addgene (plasmid no. 16240). The bacterial expression construct encoding full-length SKIP was described previously (Brès et al. 2009). For rescue experiments, siRNA-resistant vector was prepared by site-directed mutagenesis using the primer 5′-AATCTGGACAAGGACATGTATGGCGACGATCTCGAAGCCAGAATAAAGACCAACAG-3′ with substituted nucleotides (underlined). The resultant cDNA fragment replaces the original nucleotide sequence targeted by SKIP siRNA without changing the amino acid sequence, and was subcloned into the pCMV-Tag2 vector. The mutations were confirmed by sequence analysis. Synthetic dsRNA oligonucleotides targeting SKIP, U2AF65, and CDK9 were purchased from Ambion and are listed in Supplemental Table S2. Etopside, doxorubicin, Nutlin3, 5-FU, CHX, actinomycin D, and MG132 were purchased from Sigma, and TGF-β was obtained from R&D Systems. The antibodies for Western blots, ChIP, and RNA-IP are listed in Supplemental Table S3.
Cell lines and cell culture
U2OS, HCT116 (wild type, p21−/−, and p53−/−) (Polyak et al. 1996), H1299, HeLa, and MDA-MB-231 cells were maintained in DMEM supplemented with 10% FBS. The HCT116-SKIP stable cells were generated by transfecting the expression construct pV5-SKIP into the parental HCT116 cell line. Stable clones were selected in medium containing 10 μg/mL blasticidin (Invitrogen) for 3 wk.
Cell cycle and apoptosis analysis
Cells were plated in 100-mm dishes and treated with the siRNAs, UV, or 5-FU. At the indicated time points, cells were trypsinized, washed with phosphate-buffered saline (PBS), and fixed in 70% ethanol overnight at 4°C. After being washed with PBS, cells were incubated with propidium iodide (PI)/RNase-staining buffer (BD Bioscience) for 15 min at room temperature. Cell distribution across the cell cycle was analyzed with FACScan (Becton Dickinson) and CellQuest software.
GST pull-down experiments
GST fusion constructs were expressed in BL21 Escherichia coli cells, and crude bacterial lysates were prepared by sonication in GST lysis buffer (25 mM Tris at pH 7.5, 150 mM NaCl, 1 mM EDTA, protease inhibitor). Approximately 10 μg of the appropriate GST fusion proteins was incubated with precleared HCT116 nuclear extract for 2 h at 4°C. The binding reaction was then added with 30 μL of glutathione-Sepharose beads and mixed for another 1 h at 4°C. The beads were washed four times with the above GST lysis buffer, separated on a 10% SDS-PAGE, and analyzed by Western blotting.
Subcellular fractionation, qRT–PCR, and ChIP
Cell fractionation was performed using the PARIS kit (Ambion) according to the manufacturer's instructions. Total RNAs were isolated using Trizol and were subjected to DNaseI treatment prior to reverse transcription using random hexamers and SuperScript III reverse transcriptase (Invitrogen). The resulting cDNAs were subjected to qPCR with the indicated primer sets (Supplemental Table S4). Values were normalized to those of GAPDH. ChIP assays were performed essentially the same as described previously (Brès et al. 2009). Briefly, cells were fixed with 1% formaldehyde, and then whole-cell lysates were prepared. Protein lysate was subjected to ChIP with the indicated antibodies (Supplemental Table S3), followed by DNA purification. ChIP-enriched DNA was analyzed with qPCR with the indicated primer sets (Supplemental Table S5).
Coimmunoprecipitation and RNA-IP
Cells were lysed in cold lysis buffer (50 mM Tris-Cl at pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% sodium deoxycholate, protease inhibitor mixture). Cell extracts (500 μg) were incubated with the first antibodies or control normal IgG on a rotator overnight at 4°C, followed by addition of protein A/G Sepharose CL-4B beads for 2 h at 4°C. Beads were then washed four times using the lysis buffer. The immune complexes were subjected to SDS-PAGE followed by immunoblotting with the secondary antibody. For RNA-IP experiments, cells were lysed in ice-cold NET-2 buffer (50 mM Tris-HCL at pH 7.4, 300 mM NaCl, 0.5% [vol/vol] Nonidet P-40, 1× complete protease inhibitors [Roche], 100 U/mL RNase OUT [Invitrogen]). The lysate was incubated with the indicated antibodies (Supplemental Table S3) or control normal rabbit/mouse IgG on a rotator overnight at 4°C, followed by addition of protein A/G agarose (Invitrogen) for 2 h at 4°C. Beads were then washed four times using the NET-2 buffer. Immunoprecipitated RNA was then extracted using Trizol and reverse-transcribed with random hexamers. The resulting cDNA was analyzed with the indicated primer sets (Supplemental Table S6).
Acknowledgments
We thank Dr. Bert Vogelstein (Johns Hopkins University) for the HCT116 wild-type, p53−/−, and p21−/− cell lines; Dr. Iain Mattaj (EMBL-Heidelberg) for antisera to CBP80; and Dr. Don Rio (University of California at Berkeley) for discussions on splicing mechanisms. This study was funded by a grant from the NIH (CA125535).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2002611.
Supplemental material is available for this article.
References
- Abbas T, Dutta A 2009. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 9: 400–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RD, Innocente SA, Barrass JD, Beggs JD 2010. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell 40: 582–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrozkova M, Puta F, Fukova I, Skruzny M, Brabek J, Folk P 2001. The fission yeast ortholog of the coregulator SKIP interacts with the small subunit of U2AF. Biochem Biophys Res Commun 284: 1148–1154 [DOI] [PubMed] [Google Scholar]
- Biamonti G, Cáceres JF 2008. Cellular stress and RNA splicing. Trends Biochem Sci 34: 146–153 [DOI] [PubMed] [Google Scholar]
- Bracken CP, Wall SJ, Barre B, Panov KI, Ajuh PM, Perkins ND 2008. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res 68: 7621–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brès V, Gomes N, Pickle L, Jones KA 2005. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev 19: 1211–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brès V, Yoshida T, Pickle L, Jones KA 2009. SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation. Mol Cell 36: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canduri F, Perez PC, Caceres RA, de Azevedo WF Jr 2008. CDK9 a potential target for drug development. Med Chem 4: 210–218 [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba K, Yamamoto J, Yamaguchi Y, Handa H 2010. Promoter-proximal pausing and its release: molecular mechanisms and physiological functions. Exp Cell Res 316: 2723–2730 [DOI] [PubMed] [Google Scholar]
- Davies RC, Calvio C, Bratt E, Larrson SH, Lamond AI, Hastie ND 1998. WT1 interacts with the splicing factor U2AF65 in an isoform-dependent manner and can be incorporated into the spliceosome. Genes Dev 12: 3217–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derheimer FA, O'Hagan HM, Kreuger HM, Hanasoge S, Paulsen MT, Ljungman M 2007. RPA and ATR link transcriptional stress to p53. Proc Natl Acad Sci 104: 12778–12783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, Mantovani F, Tocco F, Elkon R, Comel A, Holstege H, Kerkhoven R, Jonkers J, Veerhoeve PM, Agami R, et al. 2010. BRD7 is a candidate tumor suppressor required for p53 function. Nat Cell Biol 12: 380–391 [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Verdun RE, Emerson BM 2003. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell 12: 1015–1027 [DOI] [PubMed] [Google Scholar]
- Farny NG, Hurt JA, Silver PA 2008. Definition of global and transcript-specific mRNA export pathways in metazoans. Genes Dev 22: 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folk P, Puta F, Skruzny M 2004. Transcriptional coregulator SNW/SKIP: the concealed tie of dissimilar pathways. Cell Mol Life Sci 61: 629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461: 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Lyakh LA, Bracken CP, Fukuoda J, Hayakawa M, Tsukiyama T, Soll SJ, Harris M, Rocha S, Roche KC, et al. 2006. SNIP1 is a candidate modifier gene of the transcriptional activity of c-Myc on E box-dependent target genes. Mol Cell 24: 771–783 [DOI] [PubMed] [Google Scholar]
- Gahura O, Abrhamova K, Skruzny M, Valentova A, Munzarova V, Folk P, Puta F 2009. Prp45 affects Prp22 partition in spliceosomal complexes and splicing efficiency of non-consensus substrates. J Cell Biochem 106: 139–151 [DOI] [PubMed] [Google Scholar]
- Gartel AL 2008. Transcriptional inhibitors, p53 and apoptosis. Biochim Biophys Acta 1786: 83–86 [DOI] [PubMed] [Google Scholar]
- Giul S, Cáceres JF 2007. Stressful splicing. Mol Cell 28: 180–181 [DOI] [PubMed] [Google Scholar]
- Gomes NP, Espinosa JM 2010. Differential regulation of p53 target genes: it's (core promoter) elementary. Genes Dev 24: 111–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev 20: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson FQ, Johnson TL 2009. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet 5: e1000682 doi: 10.1371/journal.pgen.1000682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves D, Horng T, Medzhitov R 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138: 129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Xie K, Yao J, Qi Z, Xiong L 2009. A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance. Proc Natl Acad Sci 106: 6410–6415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA 2004. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J 23: 406–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NG, Chin SF, Ozdag H, Daigo Y, Hu DE, Cariati M, Brindle K, Aparicio S, Caldas C 2004. p300 regulates p53-dependent apoptosis after DNA damage in colorectal cancer cells by modulation of PUMA/p21 levels. Proc Natl Acad Sci 101: 7386–7391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung P, Hermeking H 2009. The c-Myc-AP4-p21 cascade. Cell Cycle 8: 982–989 [DOI] [PubMed] [Google Scholar]
- Kim RH, Wang D, Tsang M, Martin J, Huff C, de Caestecker MP, Parks WT, Meng X, Lechleider RJ, Wang T, et al. 2000. A novel smad nuclear interacting protein, SNIP1, suppresses p300-dependent TGF-β signal transduction. Genes Dev 14: 1605–1616 [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, Fischer S, Konstantinova I, Habermann B, Grabner H, et al. 2004. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 432: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Ma C, Poser I, Fischer S, Hyman AA, Buchholz F 2005. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc Natl Acad Sci 102: 2396–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinridders A, Pogoda HM, Irlenbusch S, Smyth N, Koncz C, Hammerschmidt M, Brüning JC 2009. PLRG1 is an essential regulator of cell proliferation and apoptosis during vertebrate development and tissue homeostasis. Mol Cell Biol 29: 3173–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR 2007. Coupling transcription and alternative splicing. Adv Exp Med Biol 623: 175–189 [DOI] [PubMed] [Google Scholar]
- Legerski RJ 2009. The Pso4 complex splices into the DNA damage response. Cell Cycle 8: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Leong GM, Subramaniam N, Figueroa J, Flanagan JL, Hayman MJ, Eisman JA, Kouzmenko AP 2001. Ski-interacting protein interacts with Smad proteins to augment transforming growth factor-β-dependent transcription. J Biol Chem 276: 18243–18248 [DOI] [PubMed] [Google Scholar]
- Leong GM, Subramaniam N, Issa LL, Barry JB, Kino T, Driggers PH, Hayman MJ, Eisman JA, Gardiner EM 2004. Ski-interacting protein, a bifunctional nuclear receptor coregulator that interacts with N-CoR/SMRT and p300. Biochem Biophys Res Commun 315: 1070–1076 [DOI] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′-splice site. Genes Dev 10: 1683–1698 [DOI] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu X-D 2008. The splicing factor SC35 has an active role in transcription elongation. Nat Struct Mol Biol 15: 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T 2011. Epigenetics in alternative pre-mRNA splicing. Cell 144: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald PN, Dowd DR, Zhang C, Gu C 2004. Emerging insights into the coactivator role of NCoA62/SKIP in Vitamin D-mediated transcription. J Steroid Biochem Mol Biol 89-90: 179–186 [DOI] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208 [DOI] [PubMed] [Google Scholar]
- Mattia M, Gottifredi V, McKinney K, Prives C 2007. p53-dependent p21 mRNA elongation is impaired when DNA replication is stalled. Mol Cell Biol 27: 1309–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdzhanova G, Edmond V, De Seranno S, Van den Broeck A, Corcos L, Brambilla C, Brambilla E, Gazzeri E, Eymin B 2008. E2F1 controls alternative splicing pattern of genes involved in apoptosis through upregulation of the splicing factor SC35. Cell Death Differ 15: 1815–1823 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Wang Q, Kennedy CJ, Silver PA 2010. An alternative splicing network links cell cycle control to apoptosis. Cell 142: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morachis JM, Murawsky CM, Emerson BM 2010. Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev 24: 135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz MJ, Santangelo MSP, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ, et al. 2009. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 137: 708–720 [DOI] [PubMed] [Google Scholar]
- Ni Z, Schwartz BE, Werner J, Suarez J-R, Lis JT 2004. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell 13: 55–65 [DOI] [PubMed] [Google Scholar]
- Oesterreich FC, Preibisch S, Neugebauer KM 2010. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell 40: 571–581 [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT 2008. Rapid transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirngruber J, Shchebet A, Johnsen SA 2009. Insights into the function of the human P-TEFb component CDK9 in the regulation of chromatin modifications and co-transcriptional mRNA processing. Cell Cycle 8: 3636–3642 [DOI] [PubMed] [Google Scholar]
- Polyak K, Waldman T, He T-C, Kinzler KW, Vogelstein B 1996. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev 10: 1945–1952 [DOI] [PubMed] [Google Scholar]
- Prathapam T, Kühne C, Banks L 2002. Skip interacts with the retinoblastoma tumor suppressor and inhibits its transcriptional repression activity. Nucleic Acids Res 30: 5261–5268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DH 2008. Poised polymerases: on your mark…get set…go! Mol Cell 30: 7–10 [DOI] [PubMed] [Google Scholar]
- Roche KC, Rocha S, Bracken CP, Perkins ND 2007. Regulation of ATR-dependent pathways by the FHA domain containing protein SNIP1. Oncogene 26: 4523–4530 [DOI] [PubMed] [Google Scholar]
- Sawatsubashi S, Murata T, Lim J, Fujiki R, Ito S, Suzuki E, Tanabe M, Zhao Y, Kimura S, Fujiyama S, et al. 2010. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev 24: 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B 2008. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell 30: 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk C, Schulze-Osthoff K 2005. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell 19: 1–13 [DOI] [PubMed] [Google Scholar]
- Seoane J, Le H-V, Massagué J 2002. Myc suppression of the p21Cip1 Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419: 729–734 [DOI] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et al. 2008. The histone H2B-specific ubiquitin ligase hRNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 22: 2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ III, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skruzny M, Ambrozkova M, Fukova I, Martinkova K, Blahuskova A, Hamplova L, Puta F, Folk P 2001. Cyclophilins of a novel subfamily interact with SNW/SKIP coregulator in Dictyostelium discoideum and Schizosaccharomyces pombe. Biochim Biophys Acta 1521: 146–151 [DOI] [PubMed] [Google Scholar]
- Soares LMM, Zanier K, Mackereth C, Sattler M, Valcarcel J 2006. Intron removal requires proofreading of U2AF/3′-splice site recognition by DEK. Science 312: 1961–1964 [DOI] [PubMed] [Google Scholar]
- Vazquez A, Bond EE, Levine AJ, Bond GL 2008. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov 7: 979–987 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C 2009. Blinded by the light: the growing complexity of p53. Cell 137: 413–431 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu XY, De Clercq E 2009. Role of the HIV-1 positive elongation factor P-TEFb and inhibitors thereof. Mini Rev Med Chem 9: 379–385 [DOI] [PubMed] [Google Scholar]
- Xiao R, Sun Y, Ding J-H, Lin S, Rose DW, Rosenfeld MG, Fu X-D, Li X 2008. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol Cell Biol 27: 5393–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Zhang J, Huang X, Sun J, Xu Y, Tang Y, Wu J, Shi Y, Huang Q, Zhang Q 2006. Solution structure of human peptidyl prolyl isomerase-like protein 1 and insights into its interaction with SKIP. J Biol Chem 281: 15900–15908 [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang L 2003. No PUMA, no death: implications for p53-dependent apoptosis. Cancer Cell 4: 248–249 [DOI] [PubMed] [Google Scholar]
- Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L 2003. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci 100: 1931–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Dowd DR, Staal A, Gu C, Lian JB, van Wijnen AJ, Stein GS, MacDonald PN 2003. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J Biol Chem 278: 35325–35336 [DOI] [PubMed] [Google Scholar]