Abstract
Macroautophagy (autophagy) is a regulated catabolic pathway to degrade cellular organelles and macromolecules. The role of autophagy in cancer is complex and may differ depending on tumor type or context. Here we show that pancreatic cancers have a distinct dependence on autophagy. Pancreatic cancer primary tumors and cell lines show elevated autophagy under basal conditions. Genetic or pharmacologic inhibition of autophagy leads to increased reactive oxygen species, elevated DNA damage, and a metabolic defect leading to decreased mitochondrial oxidative phosphorylation. Together, these ultimately result in significant growth suppression of pancreatic cancer cells in vitro. Most importantly, inhibition of autophagy by genetic means or chloroquine treatment leads to robust tumor regression and prolonged survival in pancreatic cancer xenografts and genetic mouse models. These results suggest that, unlike in other cancers where autophagy inhibition may synergize with chemotherapy or targeted agents by preventing the up-regulation of autophagy as a reactive survival mechanism, autophagy is actually required for tumorigenic growth of pancreatic cancers de novo, and drugs that inactivate this process may have a unique clinical utility in treating pancreatic cancers and other malignancies with a similar dependence on autophagy. As chloroquine and its derivatives are potent inhibitors of autophagy and have been used safely in human patients for decades for a variety of purposes, these results are immediately translatable to the treatment of pancreatic cancer patients, and provide a much needed, novel vantage point of attack.
Keywords: pancreatic cancer, autophagy, Kras, chloroquine, DNA damage, metabolism
Pancreatic cancer is highly lethal, with >40,000 cases diagnosed each year (Jemal et al. 2010). Unfortunately, the lack of effective treatment options and late diagnosis results in a dismal 5-year survival of ∼5% (Hezel et al. 2006). These tumors show an intense therapeutic resistance to cytotoxic chemotherapies, targeted agents, and radiotherapy (Li et al. 2004; Ben-Josef and Lawrence 2008; Brus and Saif 2010). This extreme resistance to a variety of therapies points to altered cell survival and metabolic pathways in these refractory cancers. Although pancreatic ductal adenocarcinoma (PDAC) has a well-defined spectrum of highly recurrent oncogenic lesions—such as activation of KRAS and loss/silencing/mutation of p53, INK4A/ARF, and SMAD4—this information has yet to be translated into effective therapies for the disease (for review, see Hezel et al. 2006). Activating KRAS mutations are present in the great majority of cases, making this an ideal target for therapeutic intervention. Unfortunately, effective KRAS inhibitors have yet to be developed (Van Cutsem et al. 2004). The inhibition of pathways downstream from KRAS is a potentially viable approach to circumventing the difficulties in KRAS inhibition (Engelman et al. 2008). However, KRAS has a multitude of effectors, many of which are poorly characterized, making it a significant challenge to completely shut off the KRAS pathway.
Macroautophagy (referred to as autophagy) is a regulated catabolic pathway to degrade cellular organelles and macromolecules, allowing the recycling of bioenergetic components (Kundu and Thompson 2008; Levine and Kroemer 2008; Mizushima et al. 2008). Autophagy promotes survival in response to nutrient deprivation, but can also promote cell death (type II programmed cell death), depending on the tissue type and developmental context (Levine and Kroemer 2008). In line with the contextual pro- and anti-survival effects in normal cellular metabolism, the role of autophagy in cancer is also complex, with associations with both tumor suppression and therapeutic resistance in advanced tumors (Liang et al. 1999; White and DiPaola 2009). For example, the key autophagy gene, Beclin1, is a haploinsufficient tumor suppressor gene, as heterozygous mice develop multiple tumor types (Liang et al. 1999). Additionally, loss of autophagy can promote aneuploidy and the development of the transformed phenotype in some cell systems (Mathew et al. 2009). In contrast, inhibition of autophagy can synergize with chemotherapy in a mouse model of lymphoma (Amaravadi et al. 2007). The underlying mechanism for this synergy appears to be that autophagy is low under basal conditions in most tumor types, but is induced upon treatment with chemotherapy as a survival mechanism. However, there are studies in which autophagy activation has an inverse role, contributing to tumor cell killing by a variety of agents (Martin et al. 2009; Hamed et al. 2010). Therefore, it is critical to examine the contributions of autophagy in particular tumor types or genetic contexts.
A number of drugs that effectively inhibit autophagy are available, including chloroquine (CQ) and its derivatives. These compounds block lysosomal acidification and autophagosome degradation (the last step of the autophagy pathway) (Rubinsztein et al. 2007). Therefore, the identification of contexts in which autophagy enhances tumor cell survival would lead to immediately testable strategies for novel therapies.
In this study, we demonstrate that pancreatic cancers have constitutively activated autophagy and a profound requirement for this process, making them uniquely sensitive to autophagy inhibition.
Results
Autophagy is elevated in PDAC cell lines and primary tumors
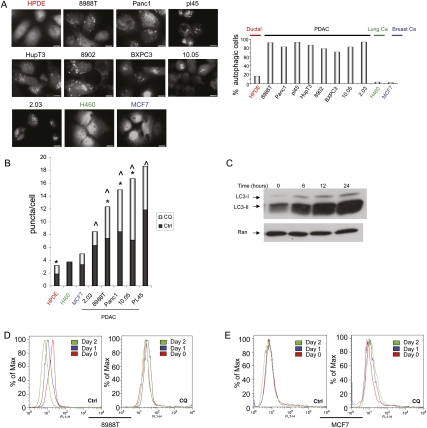
As the role of autophagy in cancer is complex and likely to vary depending on tumor type and other biological contexts, we sought to explore the importance of autophagy in PDAC biology. The microtubule-associated protein 1 light chain 3 (LC3) associates with autophagosome membranes after processing (Ichimura et al. 2000). Total LC3 levels have been reported to be elevated in PDAC and proposed to mark active autophagy in these tumors (Fujii et al. 2008). However, the functional relevance of this observation is not clear, since the relationship between total LC3 levels and autophagy per se is uncertain. To determine more directly the degree of activated autophagy in PDAC cell lines, we first assessed the integration of LC3 into autophagosomes using a GFP-LC3 reporter, a standard assay to measure active autophagy (Klionsky et al. 2008). GFP-LC3 dots were rare in cultures of nontransformed human pancreatic ductal cells (HPDE) and a breast cancer (MCF7) and lung cancer (H460) cell line, with evidence of puncta in 3%–18% of these cells, consistent with the findings of others that high levels of autophagy are typically present in vitro only when cultured cells are deprived of essential nutrients or placed under other stresses (He and Klionsky 2009). On the other hand, in all eight PDAC lines analyzed, GFP-LC3 puncta were detected in 70%–90% of cells (Fig. 1A), indicating a high degree of autophagy activation. PDAC cells also had significantly elevated numbers of puncta per cell as compared with HPDE, H460, and MCF7 cells (Fig. 1B). As further confirmation of this increase in autophagic activity, we demonstrated elevated levels of the lipidated (autophagosome-associated) form of LC3 (LC3-II)—a marker for active autophagy (Klionsky et al. 2008)—in a panel of PDAC lines grown in nutrient-rich conditions (Supplemental Fig. 1A).
Figure 1.
PDAC have high levels of basal autophagy. (A) PDAC cells and controls (HPDE [immortalized normal HPDEs], MCF7 [breast cancer], and H460 [lung cancer]) were infected with a retrovirus expressing GFP-LC3, grown in complete media in the presence of serum, and then fixed and analyzed by fluorescence microscopy for the presence of LC3 dots, which represent autophagic vesicles. Numerous discrete autophagic puncta were present in PDAC cells, while each of the control cell lines showed only a diffuse expression of GFP. The percentage of autophagic cells (defined as the presence of more than five foci) is shown in the adjacent histogram. The differences between all eight of the PDAC lines as compared with HPDE cells are statistically significant (P < 0.05 by the Fisher's exact test). Bar, 20 μM. (B) PDAC cells and controls were cultured under normal growth conditions, and the number of LC3 puncta per cell was determined in the absence and presence of the autophagy inhibitor CQ. As in A, note the robust increase in foci of PDAC cells versus HPDE, H460, and MCF7 (dark-gray bars). (^) Statistical significance compared with HPDE cells. Autophagic flux is elevated in PDAC, as evidenced by the increase in puncta per cell when treated with CQ (light-gray bars show increase in foci upon CQ treatment). Asterisk represents a statistically significant increase upon CQ treatment compared with untreated cells. (C) Autophagic flux in 8988T PDAC cells shown by a robust increase in LC3-II expression upon inhibition of lysosomal proteases with E64d + pepstatin A as well as CQ at various time points. (D) Long-term protein degradation was assessed in 8988T cells using a GFP-Neo fusion protein that enters the long-term protein degradation pathway. GFP expression was monitored by FACS. The left panel shows decreased GFP expression on day 1 and day 2 as compared with day 0, indicating activated autophagy under basal conditions. The right panel demonstrates that CQ, an inhibitor of autophagy, blocks the long-term protein degradation in these cells. (E) Long-term protein degradation was assessed in MCF7 cells, as in D. Note the minimal change in GFP expression, indicating low levels of basal autophagy. This is not affected by the addition of CQ.
As autophagy is a dynamic process, it is possible that the accumulation of LC3 puncta and increased LC3-II expression could result from a bona fide increase in autophagy or, alternatively, reflect a block in the later stages of the process, such as impaired autophagosome degradation. To address this issue, we examined autophagic flux in PDAC cell lines. As shown in Figure 1B, inhibiting autophagosome degradation by CQ, which blocks lysosomal acidification and autophagosome degradation (Rubinsztein et al. 2007), further increased the number of GFP-LC3 puncta in PDAC lines. Additionally, treatment of PDAC cells with lysosomal protease inhibitors dramatically increased LC3-II levels under basal conditions (Fig. 1C). Thus, autophagic flux is significantly increased in multiple PDAC lines. Last, as a final measure for highly activated basal autophagy, we assessed long-term protein degradation, a dynamic, functional readout of autophagy. The degradation of normally long-lived proteins can be assayed by monitoring the expression of a GFP-Neo fusion protein (Nimmerjahn et al. 2003; Klionsky et al. 2008). The expression of GFP-Neo was significantly diminished in 8988T PDAC cells over a 2-d period (Fig. 1D). This effect was completely inhibited by CQ, confirming the role of elevated autophagy in this process (Fig. 1D). In contrast, MCF7 cells show only minimal baseline long-term protein degradation, illustrated by stable GFP-Neo expression over the same interval (Fig. 1E). Together, these data demonstrate that autophagy is constitutively activated in PDAC cell lines in a cell-autonomous manner. Consistent with the results of these multiple autophagy assays, we found that key autophagy genes were broadly overexpressed in PDAC cell lines (Supplemental Fig. 1B).
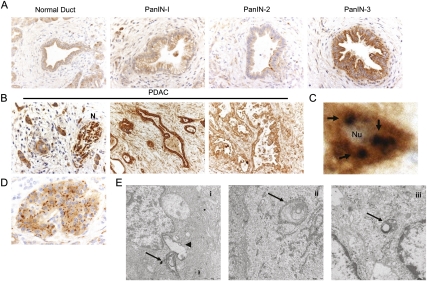
We next sought to monitor autophagy during the progression of human PDAC from premalignant pancreatic intraepithelial neoplasms (PanINs) to advanced tumors. The cleavage of the LC3 protein allows it to be lipidated and targeted to autophagic vesicles and has been used to monitor autophagy (Ketteler and Seed 2008; Sivridis et al. 2010). Using an antibody specific for cleaved LC3, we analyzed a collection of 25 PanINs and 80 primary PDAC by immunohistochemistry (IHC) (Fig. 2A,B). Minimal cytoplasmic staining was observed in the normal pancreatic ductal epithelium or in low-grade PanIN-1 and most PanIN-2 (17 lesions analyzed). In contrast, all high-grade PanIN-3 and PDAC showed elevated cleaved LC3 staining, with moderate to strong staining intensity in 81% (65 of 80 PDAC). The staining was frequently in a microvesicular pattern, and less frequently as large vesicles. This pattern of staining is highly suggestive of autophagosomes (Fig. 2C), a hallmark of autophagy (Mizushima et al. 2010). Comparable high levels of staining were observed in lymph node metastasis (data not shown). We also observed high levels of staining in the nerve fibers infiltrated by tumor (Fig. 2B). Importantly, as an additional control for the specificity of the staining, we observed strong LC3 staining in a microvesicular pattern in pancreatic islets (Fig. 2D), consistent with the known role of constitutive autophagy in β-cell homeostasis (Jung et al. 2008). As further evidence that autophagy was indeed activated in pancreatic primary tumors, electron microscopy showed the presence of autophagosomes fusing to lysosomes, as well as autophagosomes fused to lysosomes (autolysosomes) (Fig. 2E). IHC analysis also revealed that levels of the essential autophagy gene ATG7 (He and Klionsky, 2009) are elevated in the majority of PDAC examined (57 of 80 tumors) (Supplemental Fig. 1C). Thus, the data using human cell lines, as well as primary human tumors, demonstrate that autophagy is up-regulated in the later stages of the progression of PanIN to PDAC.
Figure 2.
(A) Activated autophagy was assessed by IHC for cleaved LC3 at different stages of primary PDAC. LC3 is low or absent in normal exocrine pancreas and in low-grade PanIN-1 and PanIN-2 lesions, whereas staining is up-regulated and exhibits a vesicular staining pattern in all high-grade PanIN-3 (A) and PDAC (B). (N) Nerve bundle with tumor infiltration, demonstrating robust LC3 expression. (C) High-powered magnification of a tumor cell showing vesicular staining (arrows) suggestive of autophagosomes. (Nu) Nucleus. (D) A representative islet showing high levels of vesicular staining and consistent with known constitutive autophagy in β cells. (E) Transmission electron microscopy of a pancreatic tumor showing an autophagosome fusing to a lysosome (panel i; [arrow] autophagosome; [arrowhead] lysosome), and autophagosomes fused to lysosomes (autolysosomes) (panels ii,iii; arrow).
PDAC are sensitive to autophagy inhibition in vitro by CQ
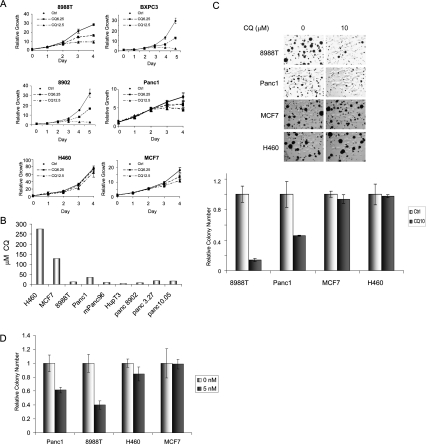
The prominent activation of autophagy in advanced PDAC and in PDAC cell lines grown in nutrient-rich conditions suggests that this process may contribute to PDAC growth. In order to test this hypothesis, we treated a collection of PDAC cell lines with CQ. As a control, CQ was also used to treat cell lines with low basal autophagy—the non-PDAC cell lines H460 and MCF7. We found that CQ markedly decreased the proliferation of all four PDAC cell lines treated, but had minimal effects on H460 and MCF7 cells (Fig. 3A). Correspondingly, the inhibitory concentration 50 (IC50) of CQ was much lower across a larger panel of PDAC cell lines compared with IC50 levels observed in the MCF7 and H460 cells (Fig. 3B). CQ treatment also robustly attenuated anchorage-independent growth of multiple PDAC cell lines while having modest effects on H460 and MCF7 cells (Fig. 3C). Similar results were obtained with a second inhibitor of autophagy, bafilomycin A1, which acts via inhibition of an H+-ATPase responsible for acidification of autophagolysosomes (Fig. 3D; Rubinsztein et al. 2007). Thus, in keeping with their elevated basal autophagy, PDAC cell lines exhibit a marked sensitivity to CQ.
Figure 3.
Inhibition of autophagy in PDAC cell lines attenuates growth and tumorigenicity in vitro. (A) Growth curves of four different PDAC cell lines (8988T, BXPC3, 8902, and Panc1) and control cell lines H460 and MCF7 treated with CQ (6.25 μM and 12.5 μM) to inhibit autophagy or with PBS as a control. The PDAC cells showed a dose-dependent robust suppression of growth, whereas H460 and MCF7 cells with low basal autophagy were only minimally affected. (B) IC50 of CQ in micromolar in a panel of PDAC lines and H460 and MCF7 cells. Note the low IC50 values for the PDAC lines as compared with MCF7 and H460. (C) Soft agar assays were performed to assess for the ability of CQ (10 μM) to inhibit anchorage-independent growth. Colony formation was suppressed in 8988T and Panc1 cells, but not MCF7 and H460 cells. The histogram below shows quantitation of these assays relative to untreated cells. Error bars represent triplicates. (D) Bafilomycin A1, an inhibitor of lysosomal acidication and autophagy, attenuates PDAC anchorage-independent growth. Cells were seeded in soft agar and treated with 5 nM bafilomycin A1 (charcoal bar) or vehicle (light-gray bar). Data are expressed relative to control, with error bars representing standard deviations of triplicates. Note the robust reduction in Panc1 and 8988T PDAC cells but not in H460 and MCF7 cells.
Autophagy inhibition by RNAi attenuates PDAC growth
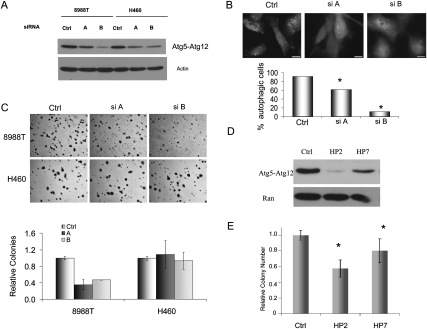
As the chemical inhibitors of autophagy used above affect lysosomal function, they may impact other cellular processes in addition to autophagy. To further validate the importance of autophagy in PDAC tumorigenicity, we genetically inactivated this process using two different siRNAs to ATG5, a ubiquitin-like protein essential for autophagosome expansion and completion (Levine and Kroemer 2008). These siRNAs suppressed expression of the ATG5 protein and inhibited autophagy (Fig. 4A,B). Additionally, ATG5 siRNAs inhibited soft agar growth of 8988T cells by >50%, while not significantly affecting H460 cells (Fig. 4C). Similar results were obtained with suppression of ATG5 expression by retroviral shRNAs to ATG5 (Fig. 4D,E). RNAi to ATG5 also suppressed growth in multiple additional PDAC cell lines, as did siRNAs to Lamp2, a dominant-negative mutant of Rab7, and shRNAs to ATG3—proteins critical for autophagy (Supplemental Fig. 2; Tanaka et al. 2000; Gutierrez et al. 2004; Radoshevich et al. 2010). Taken together, these data illustrate that PDAC lines depend on autophagy for continued growth and tumorigenesis.
Figure 4.
(A) Cells were transfected with two different siRNAs against the essential autophagy gene ATG5 (siA and B) or with a control, scrambled siRNA. Shown is a representative Western blot comparing ATG5 expression with a control siRNA. The bottom panel is an actin loading control. (B) ATG5 siRNA transfected 8988T cells were assessed for basal autophagy using the GFP-LC3 assay. The histogram below shows the percentage of autophagic cells (more than five puncta), and reveals statistically significant reductions in the siATG5-expressing cells (asterisks) compared with the control (P < 0.05 Fisher's exact test). (C) Cells were transfected with control or siRNAs to ATG5 and soft agar were assays performed. Both siRNAs significantly inhibited anchorage-independent growth of 8988T PDAC cells but not H460 cells. The histogram shows quantitation expressed relative to control. Error bars represent triplicates. (D) Two shRNAs to ATG5 (HP2 and HP7) suppress expression of ATG5. (E) ATG5 shRNAs decrease soft agar growth of 8988T PDAC cells, corresponding to the efficiency of knockdown. Results shown are from three independent experiments performed in triplicate. Asterisks show a statistically significant decrease as compared with control (P < 0.05 by t-test).
Autophagy inhibition in PDAC results in increased reactive oxygen species (ROS), DNA damage, and altered cell metabolism
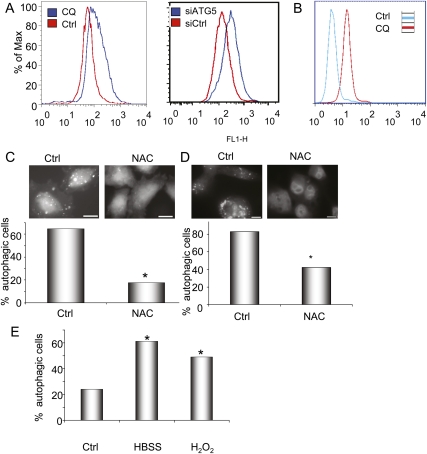
Autophagy has been shown to be regulated by ROS (Chen et al. 2007; Scherz-Shouval et al. 2007; Dewaele et al. 2010). In turn, the loss of autophagy can induce ROS, leading to DNA damage (Mathew et al. 2009). As inflammation and ROS are associated with both Ras transformation and the initiation of PDAC (Irani et al. 1997; Chu et al. 2007; Guerra et al. 2007), it is possible that the elevated basal autophagy in PDAC may also serve as an adaptation to prevent the accumulation of cytotoxic levels of ROS, thereby allowing sustained tumor growth. Correspondingly, inhibition of autophagy in 8988T cells by CQ treatment and siRNA to ATG5 resulted in an increase in total ROS levels (Fig. 5A) as well as mitochondrial ROS (Fig. 5B). Conversely, inhibition of ROS with the antioxidant N-Acetyl cysteine (NAC) significantly attenuated the levels of basal autophagy in 8988T and Panc1 cells (Fig. 5C,D). Consistent with a cross-regulation of ROS and autophagy in PDAC, treatment of HPDE cells with hydrogen peroxide induced autophagy in a robust manner (Fig. 5E).
Figure 5.
Autophagy is regulated by ROS in PDAC. (A) The left panel shows 8988T cells treated with CQ (25 μM) to inhibit autophagy and stained with DCF-DA to determine ROS levels by measuring fluorescence by FACS. Note the greater ROS levels, indicated by the increased fluorescence upon CQ treatment (blue curve). The right panel shows a similar increase in ROS when autophagy is inhibited by siRNA to ATG5. (B) Inhibition of autophagy by treatment with CQ increases mitochondrial ROS, as evidenced by increased MitoSOX staining. (C) Treatment of 8988T PDAC cells with 3 mM NAC diminishes basal autophagy, shown by the disappearance of GFP-LC3-labeled autophagosomes and the presence of only diffuse signal in the top panels. Below, a histogram shows quantitation of these results, expressed as percentage of autophagic cells. Asterisks indicate that the difference is statistically significant (P < 0.05 by Fisher's exact test). (D) Treatment of Panc1 cells with NAC attenuates basal autophagy similar to 8988T cells. The histogram below shows quantitation of the assay, with the asterisk representing a statistically significant decrease compared with control by Fisher's exact test (P < 0.05). (E) Treatment of HPDE immortalized human ductal cells with 0.5 mM hydrogen peroxide (H2O2) leads to a significant increase in autophagy, shown by a significant increase in GFP-LC3 autophagosomes (asterisks show a significant difference as compared with control by Fisher's exact test; P < 0.05). (HBSS) Hank's buffered salt solution (serum and amino acid starvation) is included as a control for autophagy induction.
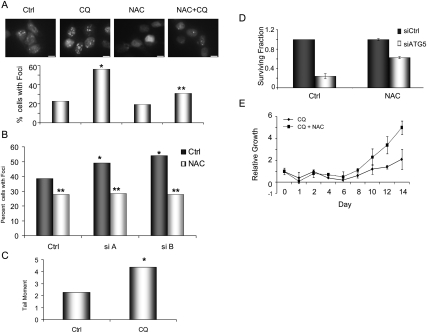
To determine if the accumulation of ROS in these cells results in DNA damage, we assessed for markers of DNA double-strand breaks (DSBs). Strikingly, the inhibition of autophagy resulted in an increase in DNA damage, as reflected by the greater number of 53BP1 foci in CQ or ATG5 siRNA-treated PDAC cells (Fig. 6A,B). Neutral comet assays revealed increased tail moments in CQ-treated cells (Fig. 6C), confirming the presence of increased DSBs. These effects were mitigated by concurrent treatment with NAC, demonstrating that they are mediated by ROS (Fig. 6A,B). Importantly, NAC also partially rescued cell proliferation and clonogenic growth of the CQ or ATG5 siRNA-treated cells (Fig. 6D,E). Together, these data demonstrate that autophagy serves to control ROS levels and, ultimately, DNA damage in PDAC cells, allowing for continued tumor growth.
Figure 6.
(A) Inhibition of autophagy in 8988T PDAC cells with CQ leads to an increase in DNA damage. The top panels show the results from cells expressing a GFP-53BP1 fusion construct. Note the increase in foci upon CQ treatment, indicating an increase in DNA DSBs. This was mitigated by concurrent treatment with NAC. The histogram below shows quantitation from a representative assay, with a single asterisk representing statistical significance compared with control and a double asterisk representing statistical significance compared with CQ treatment (P < 0.05 Fisher's exact test). (B) Inhibition of autophagy using two siRNAs to ATG5 (siA and B) increases DNA DSBs, measured by 53BP1 foci (charcoal bars) and expressed as percentage of cells with >10 foci. A single asterisk indicates that the increase in 53BP1 foci is statistically significant compared with control (P < 0.05 Fisher's exact test). The light-gray bars show the effect of NAC on 53BP1 foci in a particular experiment. The double asterisk indicates a statistically significant decrease in foci as compared with the corresponding untreated cells (charcoal bar), demonstrating that NAC inhibits the DNA damage caused by autophagy inhibition. (C) 8988T PDAC cells were treated with CQ and subjected to a neutral comet assay to measure the amount of DSBs. Data are expressed as average tail moment (tail moment is the tail length multiplied by the percentage of DNA in the tail). Note the increase upon treatment with CQ as compared with control (asterisk indicates statistical significance; P < 0.05 by t-test). (D) Clonogenic survival assays were performed on 8988T cells transfected with either a control siRNA (charcoal bar) or an siRNA to ATG5 (light-gray bar), and results are expressed as surviving fraction relative to control. Note the significant reduction in surviving fraction with inhibition of autophagy by RNAi. Identical assays were performed in the presence of 1 mM NAC to inhibit ROS. This led to an increase in surviving fraction in ATG5-suppressed cells, indicating a partial rescue by NAC. (E) 8988T cells were treated with CQ with (broken line) or without (solid line) NAC, and proliferation was measured. There was a robust increase in growth with concomitant NAC treatment.
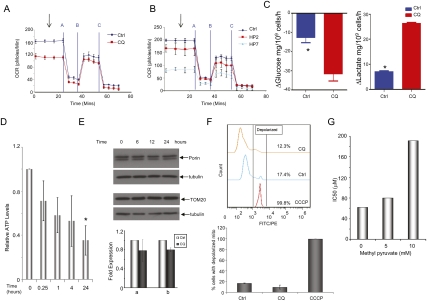
We speculated that the increased mitochondrial ROS seen with autophagy inhibition by CQ and RNAi may arise from impairment in mitochondrial function. As autophagy has a known role in mitochondrial quality control by degrading damaged mitochondria (Priault et al. 2005), we assessed whether inhibition of autophagy leads to a decrease in mitochondrial function. To this end, we measured the effect of either CQ treatment or shRNAs to ATG5 on oxidative phosphorylation in 8988T PDAC cells. As shown in Figure 7A, oxygen consumption was severely decreased upon CQ treatment. Consistent with these data, inhibition of autophagy using shRNAs to ATG5 also decreased oxygen consumption (Fig. 7B). Therefore, autophagy inhibition in PDAC cells leads to a decrease in oxidative phosphorylation. Consistent with the decrease in oxidative phosphorylation was that CQ treatment of these cells caused a significantly elevated uptake of glucose and production of lactate (Fig. 7C), indicating that there was a compensatory switch to glycolysis in response to impaired oxidative phosphorylation. Importantly, these cells show a significant decrease in intracellular ATP (Fig. 7D) upon autophagy inhibition, likely reflecting the inability of these cells to maintain normal energy production.
Figure 7.
PDAC metabolism is altered by autophagy inhibition. (A) Oxidative phosphorylation, as measured by oxygen consumption in 8988T PDAC cells. (Blue) Control; (red) CQ. CQ treatment robustly decreases the basal oxygen consumption ratio (OCR), normalized to cell number or protein concentration. Data represent the mean of four independent experiments, with error bars representing standard deviations. The graph shows basal mitochondrial respiration (3 mM glucose) (arrow) and leak (respiration nonlinked to mitochondrial ATP synthesis, 2 μM oligomycin) (line A), nonmitochondrial OCR (2 μM anti-mycin A) (line C) and respiration after FCCP (5 μM) (line B). Identical experiments as in A were performed using two shRNAs to ATG5 (HP2 and HP7). Both shRNAs decrease basal oxygen consumption similar to CQ. Data represent the mean of three independent experiments. (C) 8988T cells were treated with CQ, and glucose uptake was measured and compared with control cells (left panel) and lactate secretion (right panel). Note the significant increase in glucose uptake as well as increased lactate secretion in cells in which autophagy is inhibited by CQ, indicating an increase in glycolysis. (Asterisk shows a statistically significant change by t-test.) (D) Inhibition of autophagy results in a decrease in intracellular ATP. Results are expressed as a fold of control and are normalized to protein concentration. Data are from three independent experiments, with error bars representing standard deviations. The asterisk shows a statistically significant decrease compared with control by a t-test. (E) Autophagy inhibition does not result in an increase in mitochondrial mass. Western blotting for TOM20 or Porin (mitochondrial proteins) does not show increases upon inhibition of lysosomal proteases plus CQ for the indicated time periods. Mitochondrial mass was also determined by quantitative real-time PCR using two different primers for mitochondrial DNA (a and b). Data shown are from two independent experiments performed in triplicate. Expression was normalized using primers for nuclear-derived DNA and expressed as fold of control. There was no significant increase in mitochondrial mass in response to CQ treatment. (F) Mitochondrial membrane potential was measured using JC1. The uncoupler CCCP was included as a positive control for depolarized mitochondria. The top panel shows a representative experiment showing no increase in mitochondrial membrane depolarization upon CQ treatment. The graph below shows data from two independent experiments. (G) The addition of methyl pyruvate (MP) protects PDAC cells from autophagy inhibition by CQ. 8988T cells were treated with the indicated concentration of MP and subjected to IC50 assays with increasing doses of CQ. As depicted in the bar graph, the IC50s markedly increase with increasing concentrations of MP, indicating that it is protecting cells from the inhibitory effects of CQ.
The reduction in oxidative phosphorylation could reflect an accumulation of damaged mitochondria due to suppression of mitophagy (the autophagic degradation of mitochondria). However, we were unable to demonstrate significant basal mitophagy in PDAC cells even in the presence of CQ (Supplemental Fig. 3A; data not shown). Consistent with these findings, there did not appear to be a significant increase in mitochondrial mass upon autophagy inhibition by CQ treatment (Fig. 7E). Additionally, as damaged (depolarized) mitochondria are typically targeted for mitophagy (Twig et al. 2008), we next assessed mitochondrial membrane potential in PDAC cells upon autophagy inhibition. As shown in Figure 7F, there was no significant increase in membrane depolarization upon autophagy inhibition. Together, these data suggest that the mitochondria themselves are functional, and that the decrease in oxidative phosphorylation may reflect a decrease in carbon substrates that fuel the TCA cycle. To determine if this was indeed the case, we attempted to rescue the effect of inhibiting autophagy in PDAC cells with CQ by supplementing the cells with excess pyruvate, which would provide substrates for the TCA cycle. Indeed, pyruvate was able to partially rescue CQ sensitivity, as illustrated by markedly increased IC50 values in pyruvate-treated cells (Fig. 7G). The addition of pyruvate also mitigated the decrease in ATP by CQ treatment (Supplemental Fig. 3C). Therefore, the data indicate that PDAC rely on autophagy for proper energy homeostasis by supplying bioenergetic intermediates to fuel oxidative phosphorylation.
In vivo inhibition of autophagy has a potent effect on PDAC growth
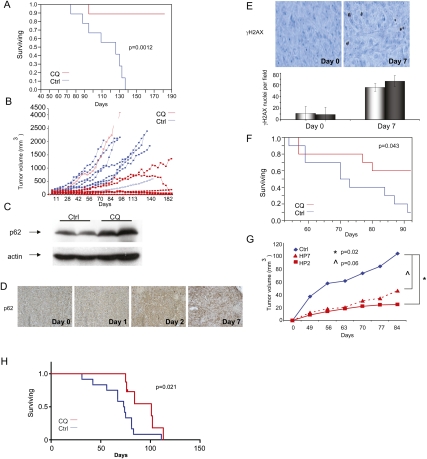
We next sought to test whether inhibition of autophagy is a potential therapeutic approach against PDAC in vivo. 8988T PDAC cells and H460 lung cancer cells were used to represent “high” and “low” basal autophagy lines, respectively. These were grown as xenografts until tumors were ∼0.5–1 cm and then separated into two cohorts, treated by intraperitoneal injection of PBS or CQ. Mirroring the in vitro studies, CQ had minimal effect both on tumor-specific survival and tumor volumes in H460 xenografts (Supplemental Fig. 4A,B). In contrast, 8988T PDAC xenografts had a robust response to CQ treatment (Fig. 8A,B). All eight control mice died from PDAC by 140 d (median 117 d), whereas only one of eight CQ-treated animals died of PDAC by the time the experiment was terminated at ∼180 d (Fig. 8A). Remarkably, there was a sustained complete tumor regression in four of eight mice (Fig. 8B). CQ did not cause any evident side effects in the treated animals. Thus, CQ can both prolong survival and eradicate tumors in mice at dosages that are well tolerated. We also observed a robust response to CQ in Panc1 xenografts (Fig. 8F) and in an orthotopic PDAC model with 8988T cells grown in the pancreata of nude mice (Supplemental Fig. 4C,D).
Figure 8.
Inhibition of autophagy using CQ robustly suppresses PDAC tumor growth in vivo. (A) 8988T PDAC cells were grown as xenograft tumors in the flanks of nude mice. When tumors reached 0.5–1 cm, mice were divided into two treatment groups of 10 mice per group (PBS or 60 mg/kg per day CQ intraperitoneally). Pancreatic cancer survival analysis is depicted in graphic form. (Blue) Control; (red) CQ. Note the increased survival of the CQ-treated cohort (P = 0.0012 by the log-rank test). Only one mouse died of pancreatic cancer in the CQ cohort over a period of >6 mo. (B) Tumor volumes were measured twice per week and plotted as a function of time. Each line represents the growth kinetics of an individual tumor. (Blue) Control; (red) CQ. Note the CQ-treated tumors segregate together at the bottom of the graph, indicating slower growth kinetics. Multiple tumors have completely regressed in the CQ cohort. (C) Western blot on two untreated control xenografts and two from mice treated with CQ. Notice the increase in p62 expression, indicating autophagy was successfully inhibited in the tumor. Actin is shown below as a loading control. (D) IHC analysis of xenografts harvested at different time points after CQ treatment (days 0, 1, 2, and 7). Similar to the Western blot analysis, p62 expression is increased over time, as indicated by the stronger brown staining, as compared with untreated (day 0). (E) IHC was performed on xenografts to assess γH2AX expression, a marker for DNA damage. This expression, depicted by brown nuclear staining, is highly up-regulated in the CQ-treated xenografts, indicating an increase in DNA damage. Shown here is a representative image from a xenograft harvested from a mouse treated with CQ for 7 d or an untreated control. The histogram below shows quantitation of these results from two control tumors and two treated tumors, shown as the average number of positive staining cells per 20× field (at least 10 fields were counted for each sample). Error bars represent standard deviations. (F) CQ treatment prolongs PDAC-specific survival in mice carrying Panc1 xenografts. Experiment was performed as in A, with 10 mice per group. The difference in survival was significant by log-rank test. Sixty percent of the treated mice were alive at 90 d compared with 10% of the controls. (G) Suppression of ATG5 expression by shRNAs decreases 8988T xenograft growth. Cells were infected with two different shRNAs to ATG5 (HP2 and HP7) or control retroviruses and injected into the flanks of nude mice (n = 12 for each group). Tumor volume was measured weekly. Note the robust decrease in volume as compared with control. This was statistically significant by a t-test for HP2 versus Ctrl ([*] P = 0.02) and a strong statistical trend for HP7 versus Ctrl ([^] P = 0.06). (H) Survival is prolonged in the Kras-driven genetic mouse model of PDAC by CQ treatment. Mice began treatment at 8 wk of age (n = 8). Survival is compared with an IP PBS-treated control cohort of identical genotype (n = 12). The difference in survival is significant by log-rank test.
To confirm that we were inhibiting autophagy in tumors, we measured p62 expression, a scaffold protein known to be degraded by autophagy (Bjorkoy et al. 2009). Western blot and IHC showed that the levels of p62 were increased in treated 8988T xenografts compared with untreated controls (Fig. 8C,D). Additionally, there was an increase in DNA damage in CQ-treated tumors, as assessed by IHC for γH2AX expression (Fig. 8E). Therefore, as in our in vitro studies, the anti-proliferative effect of CQ in PDAC xenografts is associated with attenuated autophagy and elevated DNA damage.
As further confirmation of the importance of autophagy in PDAC growth in vivo, we inhibited autophagy in 8988T cells using two shRNAs to ATG5 and assessed their ability to grow as xenografts. As shown in Figure 8G, both shRNAs robustly diminished tumor growth, as evidenced by decreased tumor volumes as compared with controls.
We next sought to determine whether autophagy inhibition was effective in blocking tumor growth in an autochthonous PDAC model, which may provide a more faithful system for preclinical studies (Hingorani et al. 2003; Bardeesy et al. 2006). These models recapitulate the extreme therapeutic resistance seen in these tumors, with minimal prolongation of survival with the standard therapy, gemcitabine (Olive et al. 2009). Other targeted therapies have had negligible or little effect on median survival in these models: cyclopamine (6-d increase) (Feldmann et al. 2008), and dasatinib (no increase) (Morton et al. 2010). To date, the most effective strategy of combining inhibition of the Hedgehog pathway with gemcitabine increased median survival by 14 d (Olive et al. 2009) and is the basis for several human clinical trials currently under development. Our studies employed a similar Kras-driven genetically engineered mouse model (GEMM) of PDAC (Hingorani et al. 2003; Bardeesy et al. 2006). Mice were treated with CQ following the establishment of advanced PanINs or focal PDAC and monitored for PDAC progression. As observed in the xenograft and orthotopic models, CQ treatment led to a significant increase in survival in the Kras-driven PDAC GEMM, leading to a robust 27-d increase in median survival as a monotherapy (Fig. 8H).
Discussion
In summary, we showed that autophagy has a critical role in PDAC pathogenesis. Autophagy is highly activated in the later stages of PDAC transformation in a cell-autonomous fashion, and is required for continued malignant growth in vitro and in vivo. Whereas the prevailing view is that autophagy is induced by environmental stress stimuli, including nutrient deprivation and chemotherapeutic agents (Kundu and Thompson 2008; Mizushima et al. 2008), PDAC cells exhibit constitutive autophagy under basal conditions. Our results suggest that this is an acquired alteration that enables PDAC growth by preventing the accumulation of genotoxic levels of ROS as well as sustaining oxidative phosphorylation by providing bioenergetic intermediates. The dependence of PDAC on this pathway is exploitable for therapeutic benefit.
Autophagy may act to promote tumorigenesis in other types of cancer, but it may not be as prevalent or as pronounced as in PDAC, in which the overwhelming majority of tumors are dependent on this process. In this regard, our data are consistent with recent work from White and colleagues (Guo et al. 2011) showing that transformation by oncogenic RAS may cause an addiction to autophagy to maintain energy balance.
However, further work must be performed to determine the specific roles of autophagy in other tumor types.
The positive role for autophagy in the maintenance of advanced PDAC stands in contrast to a number of other malignancies, in which genetic evidence from human specimens and mouse models shows that inactivation of autophagy can promote tumorigenesis (Liang et al. 1999; Mathew et al. 2009). However, our results do not necessarily preclude such a tumor suppression function in PDAC. Since ROS is required to promote cellular transformation, yet at very high levels is cytotoxic, it is possible that the role of autophagy is biphasic. A loss of autophagy during early stages of cancer may promote the protumorigenic genomic instability seen in PDAC (Kimmelman et al. 2008), whereas in highly metabolically active PDAC cells, autophagy may restrain oxidative damage from reaching cytotoxic levels as well as maintain energy homeostasis.
Previous studies have suggested that autophagy can contribute to chemotherapeutic resistance rather than tumor maintenance per se in certain contexts. For example, Thompson and colleagues (Amaravadi et al. 2007) have elegantly demonstrated in a lymphoma model that treatment of mice with CQ resulted in a growth impairment of tumors, but no tumor regression unless combined with restoration of p53 expression or akylating chemotherapy. Cooperation of autophagy inhibition with chemotherapy has also been observed in leukemia (Carew et al. 2007; Bellodi et al. 2009), and may reflect the up-regulation of autophagy as a possible survival mechanism in response to chemotherapeutic agents (Hippert et al. 2006). PDAC, in contrast, is characterized by a critical role of autophagy for tumorigenicity under basal conditions. We believe that this represents a unique aspect of the biology of PDAC and possibly other Ras-driven tumors. As cytotoxic agents (e.g., H2O2) increase autophagy in pancreatic cells, this process could have an additional function, potentially contributing to the pronounced therapeutic resistance that characterizes PDAC.
The dependence of PDAC on autophagy may provide a much needed target for therapeutic intervention in a disease with limited effective therapies. In fact, CQ and its derivatives—which effectively inhibit autophagy and, in our studies, result in PDAC regression—have been safely used in patients for many years as anti-malarial therapies or for rheumatologic conditions. In fact, the doses in the range of those used in our studies are safely achievable in human patients (Tett et al. 1989; Munster et al. 2002). Given our findings, there is compelling rationale to begin trials in PDAC using these drugs targeting autophagy.
Materials and methods
Cell culture and reagents
Tumor cell lines were obtained from the American Type Culture Collection or the German Collection of Microorganisms and Cell Cultures. The establishment and characterization of the near-normal immortalized HPDE cells were reported previously (Furukawa et al. 1996; Liu et al. 1998). Typically, Lipofectamine 2000 (Invitrogen, 11668) was used to transfect cells with plasmids, and RNAiMax (Invitrogen, 13778) was used for siRNAs. All siRNAs were used at a final concentration of 20 nM.
Plasmids and RNAi
pBabe-LC3-GFP was constructed by subcloning EGFP-LC3 from EGFP-LC3 (Addgene, 11546) into pBabe-puro. 53BP1-GFP fusion plasmid was a generous gift from Randall King (Harvard School of Public Health). The GFP-Neo construct has been described previously (Nimmerjahn et al. 2003). The Rab7 dominant-negative (T22N) was a generous gift from Dr. Andrew Thorburn (University of Colorado Cancer Center). ATG5 and Lamp2 siRNAs were synthesized by Invitrogen: ATG5: siA, CAAUCCCAUCCAGAGUUGCUUGUGA; siB, AGUGAACAUCUGAGCUACCCGGAUA. Lamp2: siA, UCAGGAUAAGGUUGCUUCAGUUAUU; siB, GCAGCACCAUUAAGUAUCUAGACUU. ATG5 shRNAs were a generous gift from Xiao-Feng Zhu (Zhou et al. 2009).
Autophagy assays
Long-term protein degradation monitoring using Neo-GFP has been described previously (Nimmerjahn et al. 2003). In brief, cells were transfected with Neo-GFP using lipid-based transfection. Fresh medium containing the indicated drugs was added daily. At the specified time point, cells were trypsinized and fixed in 70% ethanol and stored at 4°C until the day of analysis. Fixed cells were washed twice with cold PBS twice and analyzed by FACS for GFP expression. Ten-thousand cells were analyzed in each experiment. Virus encoding EGFP-LC3 was packaged in 293T cells and stable cell lines were created by standard infection protocols in the presence of polybrene and selected with puromycin for at least 2 d. To assess for autophagy, cells were plated on multitest slides, treated as indicated, and fixed with 4% paraformaldehyde. Typically, at least 200 cells were counted, and cells with more than five puncta were considered autophagy-positive. To determine the number of foci per cell, deconvolution images were taken using a Zeiss Axio Imager Z1 and a three-dimensional reconstruction was created. The total number of foci was counted in a minimum of 60 cells. To detect mitophagy, two different assays were used. First, cells were transfected with an mCherry-Parkin fusion protein and Parkin foci were scored. Alternatively, 8988T cells stably expressing LC3-GFP were treated with or without CQ (25 μM) for 4 h. Cells were fixed with 4% paraformaldehyde, followed by immunofluorescence staining with antibody against TOM20 and Cy3-conjugated secondary antibody. Deconvolution images were taken using a Zeiss Axio Imager Z1 to detect colocalization of TOM20-labeled mitochondria and GFP-LC3-labeled autophagosomes.
Soft agar assay
Five-thousand cells per well were seeded in medium containing 0.5% agarose on top of bottom agar containing 1% agar (Nobel Agar, BD 214230). After 10–14 d, colonies were stained with p-iodonitrotetrazolium violet (Sigma, 18377) and five fields were counted under low power, with positive colonies scored based on control colony size. If indicated, inhibitors were added at the time agar plates were made, with 200 μL of medium containing inhibitors added every 3 d. For RNAi experiments, cells were seeded day after transfection. Experiments were performed in triplicate.
Cell proliferation assay
Cells were plated in 24-well plates at 5000 cells per well. The day after plating, cells were treated with CQ at the indicated concentrations. Cells were fixed in 10% formalin and stained with 0.1% crystal violet. Dye was extracted with 10% acetic acid and the relative proliferation was determined by OD at 595 nm.
IC50 assay
Cells were plated in 96-well plates and treated by serial dilution of CQ the day after plating for 48–72 h. Cell viability was measured using the Cell-Titer-Glow assay (Promega, G7570) according to the manufacturer's instructions. The IC50 was calculated using a sigmoidal model using BioDataFit 1.02.
Clonogenic survival assay
Cells were plated in 6-cm dishes at 200 cells per dish in growth medium with 15% FBS and treated with CQ the day after seeding. Cells were pretreated with NAC the day before seeding. After 7 d, cells were fixed in 80% methanol and stained with 0.2% crystal violet and colonies were counted. The surviving fraction was calculated using the plating efficiency.
Expression analysis
For quantitative RT–PCR, mRNA was isolated by using Trizol per the manufacturer's instructions and was DNase-treated. cDNA was reverse-transcribed using the Thermoscript RT–PCR system (Invitrogen, 11146). Real-time PCR were performed using SYBR Green in a Bio-Rad Chromo4 Thermocycler. Primer sequences are available on request. Western blot analysis was performed according to standard protocols. IHC was performed via standard protocols. The following antibodies were used for Western or IHC: LC3-II (Novus Biologicals, NB600-1384), cleaved LC3 (Abgent, Ap1805a), p62 (Abnova, H00008878-M01), actin (Sigma, A2066), ATG5 (Cell Signaling, 2630), γH2AX (Millipore, 05-636), ATG7 (Epitomics, 2504-1), and Lamp2 (Abcam, ab25631).
Electron microscopy
Fresh tissue from the primary lesion was fixed immediately in 2.5% glutaraldehyde for 3 h at room temperature. After post-fixation in 1% osmium tetroxide for 1 h and dehydration, the samples were embedded in a mixture of epon–araldite. Conventional thin sections from four blocks were collected on uncoated grids, stained with uranil and lead citrate, and examined in a Leo912 electron microscope.
DNA damage assays
8988T cells were transduced with retrovirus encoding a p53BP1-GFP fusion protein containing a portion of the 53BP1 protein (∼1700 base pairs, including a tudor domain, which we and others have shown to form foci in response to DNA damage). Cells were treated as indicated, plated on multitest slides, and fixed with 4% paraformaldehyde. Cells with >10 GFP foci were scored as positive. Neutral comet assays were performed according to the manufacturer's instructions (Trevigen). Briefly, cells were treated as indicated for 24 h, combined with low-melting agarose (catalog no. 4250-050-02), and then mounted on CometSlide (catalog no. 4250-200-03). Following cell lysis (catalog no. 4250-050-01) and unwinding of DNA, the cells were electrophoresed for 10 min. Slides were fixed with ethanol and stained by DAPI. One-hundred-fifty randomly selected cells from each sample were analyzed using CometScore software (http://autocomet.com). DNA damage was determined by tail moment (tail length multiplied by the percentage of DNA in the tail).
Mouse treatment studies
For xenografts, 2 million cells suspended in 100 μL of Hanks buffered saline solution were injected subcutaneously into the lower flank of NCr nude mice (Taconic). Tumors were grown until they reached 0.5–1 cm in greatest dimension, and mice were separated into two groups matched for tumor volume. Daily intraperitoneal injection was performed with CQ at 60 mg/kg in 100 μL of PBS or 100 μL of PBS only daily. Tumors were measured twice weekly after the start of treatment using calipers, and volume was calculated by the formula (length × width2)/2. All xenograft and orthotopic animal experiments were approved by the Institutional Animal Care and Use Committee under protocol numbers 04-114 and 04-605. Tumor-specific death was determined by a tumor reaching >2 cm in maximal dimension, a tumor causing the mouse to be moribund, skin ulceration, and death caused by the tumor. For shRNA studies, 8988T cells were infected with the two retroviral shRNAs or control and subjected to a short puromycin selection, and then 2 million cells were injected into the flanks of nude mice. Measurements were taken as above. For orthotopic assays, 500,000 8988T cells expressing luciferase were implanted into the tail of the pancreas in Matrigel. Mice were randomized to receive CQ or PBS daily (as above), beginning 10 d after injection, and luciferase imaging was performed weekly at the Longwood Small Animal Imaging Facility. Cohorts of genetically engineered mice were generated from a single colony (LSL-KrasG12D; p53 L/+) (Bardeesy et al. 2006) under protocol 2005N00148. At 8 wk of age, treatment was initiated (IP PBS for one cohort and IP CQ for the other). Pancreatic cancer survival was compared using Kaplan-Meier analysis.
Mitochondrial/metabolism studies
Oxygen consumption measurements: Cells were seeded in a 24-well Seahorse plate, and oxygen consumption rates (OCRs) were measured using the Seahorse XF24 instrument (Seahorse Biosciences). Basal mitochondrial respiration (3 mM glucose) and leak (respiration nonlinked to mitochondrial ATP synthesis, 2 μM oligomycin) were measured. Nonmitochondrial OCRs were obtained by adding 2 μM antimycin A and uncoupled respiration was obtained by FCCP (5 μM). To determine the membrane potential of the mitochondria, JC1 dye was used (Invitrogen) according to the manufacturer's instructions. In brief, after the indicated treatments, cells were stained with 2 μM JC1 for 15 min. The uncoupler CCCP (50 μM) was included as a positive control. Cells were resuspended in PBS and FACS was performed using a BD FACSCanto II HTS. Data were analyzed by FlowJO. To determine mitochondrial biomass changes upon CQ treatment, cells were treated for the indicated periods of time and then subjected to Western blot analysis with antibodies to mitochondrial proteins, TOM20 (Santa Cruz Biotechnology), and Porin (MitoSciences). Alternatively, quantitative PCR was performed on genomic DNA (as above) using primers for mitochondrial-derived DNA and was normalized using sequences from nuclear-derived DNA. Primer sequences are available on request. For the metabolite flux studies, ∼2 × 105 to 3 × 105 cells were plated in a six-well plate and treated with CQ as indicated for 3 d. Medium was collected, and glucose and lactate levels in the medium were analyzed using a metabolite analyzer (Nova Flex BioProfiles). The rates shown were calculated from the difference in metabolite concentration to the medium from a blank well and normalized to cell number for each individual well. ATP was measured using the ATP Bioluminescence Assay kit CLS II (Roche, 11699695001) per the manufacturer's instructions. In brief, cells were boiled in boiling buffer at 100°C and supernatants were collected after a quick spin. ATP levels were analyzed by measuring luminescence after addition of luciferase reagent to supernatant and were normalized by protein concentration.
Acknowledgments
We thank Drs. Alan Wang and David Sabatini for critical reading of the manuscript. We thank Drs. Florian Muller, Dipanjan Chowdhury, Kwok Wong, Brian Alexander, and Ron DePinho for insightful discussion. We thank Dr. Xiao-Feng Zhu for shRNAs to ATG5 and Dr. Randall King for the GFP-53BP1 fusion protein. We thank Dr. Ming Tsao for the HPDE cells. We thank Dr. Andrew Thorburn for the Rab7 dominant-negative. A.C.K., S.Y., and Y.L. are supported by the Dana-Farber Cancer Institute. Funding was also provided by the National Cancer Institute Grants (5P50CA127003-03; developmental project grant to A.C.K.), Kimmel Scholar Award (to A.C.K.), and AACR-PanCAN Career Development Award (to A.C.K.). G.C. is supported by an American-Italian Cancer Foundation post-doctoral fellowship and a grant from Fondazione Umberto Veronesi. M.L. is a recipient of a post-doctoral fellowship from Fundación Ramón Areces.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.2016111.
Supplemental material is available for this article.
References
- Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB 2007. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest 117: 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, et al. 2006. Both p16(Ink4a) and the p19(Arf)–p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci 103: 5947–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, Ronchetti M, Galavotti S, Young KW, Selmi T, Yacobi R, et al. 2009. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest 119: 1109–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Josef E, Lawrence TS 2008. Chemoradiotherapy for unresectable pancreatic cancer. Int J Clin Oncol 13: 121–126 [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Pankiv S, Øvervatn A, Brech A, Johansen T 2009. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 452: 181–197 [DOI] [PubMed] [Google Scholar]
- Brus C, Saif MW 2010. Second line therapy for advanced pancreatic adenocarcinoma: where are we and where are we going? Highlights from the ‘2010 ASCO Annual Meeting.’ Chicago, IL, USA. June 4–8, 2010. JOP 11: 321–323 [PubMed] [Google Scholar]
- Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, Houghton JA, Huang P, Giles FJ, Cleveland JL 2007. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood 110: 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB 2007. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci 120: 4155–4166 [DOI] [PubMed] [Google Scholar]
- Chu GC, Kimmelman AC, Hezel AF, DePinho RA 2007. Stromal biology of pancreatic cancer. J Cell Biochem 101: 887–907 [DOI] [PubMed] [Google Scholar]
- Dewaele M, Maes H, Agostinis P 2010. ROS-mediated mechanisms of autophagy stimulation and their relevance in cancer therapy. Autophagy 19: 1–17 [DOI] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. 2008. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 14: 1351–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann G, Habbe N, Dhara S, Bisht S, Alvarez H, Fendrich V, Beaty R, Mullendore M, Karikari C, Bardeesy N, et al. 2008. Hedgehog inhibition prolongs survival in a genetically engineered mouse model of pancreatic cancer. Gut 57: 1420–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Mitsunaga S, Yamazaki M, Hasebe T, Ishii G, Kojima M, Kinoshita T, Ueno T, Esumi H, Ochiai A 2008. Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcomes. Cancer Sci 99: 1813–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, Tsao MS 1996. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol 148: 1763–1770 [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Schuhmacher AJ, Cañamero M, Grippo PJ, Verdaguer L, Pérez-Gallego L, Dubus P, Sandgren EP, Barbacid M 2007. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11: 291–302 [DOI] [PubMed] [Google Scholar]
- Guo JY, Chen H-Y, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JMS, Karantza V, et al. 2011. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 25: 460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Munafó DB, Berón W, Colombo MI 2004. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci 117: 2687–2697 [DOI] [PubMed] [Google Scholar]
- Hamed HA, Yacoub A, Park MA, Eulitt P, Sarkar D, Dimitrie IP, Chen CS, Grant S, Curiel DT, Fisher PB, et al. 2010. OSU-03012 enhances Ad.7-induced GBM cell killing via ER stress and autophagy and by decreasing expression of mitochondrial protective proteins. Cancer Biol Ther 9: 526–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ 2009. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA 2006. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 20: 1218–1249 [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. 2003. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4: 437–450 [DOI] [PubMed] [Google Scholar]
- Hippert MM, O'Toole PS, Thorburn A 2006. Autophagy in cancer: good, bad, or both? Cancer Res 66: 9349–9351 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Irani K, Xia Y, Zweier JL, Sollott SJ, Der CJ, Fearon ER, Sundaresan M, Finkel T, Goldschmidt-Clermont PJ 1997. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science 275: 1567–1568 [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegal R, Xu J, Ward E 2010. Cancer statistics, 2010. CA Cancer J Clin 60: 277–300 [DOI] [PubMed] [Google Scholar]
- Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, Nguyen YH, Kang TM, Yoon KH, Kim JW, et al. 2008. Loss of autophagy diminishes pancreatic β cell mass and function with resultant hyperglycemia. Cell Metab 8: 318–324 [DOI] [PubMed] [Google Scholar]
- Ketteler R, Seed B 2008. Quantitation of autophagy by luciferase release assay. Autophagy 4: 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman AC, Hezel AF, Aguirre AJ, Zheng H, Paik JH, Ying H, Chu GC, Zhang JX, Sahin E, Yeo G, et al. 2008. Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc Natl Acad Sci 105: 19372–19377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4: 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Thompson CB 2008. Autophagy: basic principles and relevance to disease. Annu Rev Pathol 3: 427–455 [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G 2008. Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Xie K, Wolff R, Abbruzzese JL 2004. Pancreatic cancer. Lancet 363: 1049–1057 [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402: 672–676 [DOI] [PubMed] [Google Scholar]
- Liu N, Furukawa T, Kobari M, Tsao MS 1998. Comparative phenotypic studies of duct epithelial cell lines derived from normal human pancreas and pancreatic carcinoma. Am J Pathol 153: 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AP, Mitchell C, Rahmani M, Nephew KP, Grant S, Dent P 2009. Inhibition of MCL-1 enhances lapatinib toxicity and overcomes lapatinib resistance via BAK-dependent autophagy. Cancer Biol Ther 8: 2084–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ 2008. Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B 2010. Methods in mammalian autophagy research. Cell 140: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton JP, Karim SA, Graham K, Timpson P, Jamieson N, Athineos D, Doyle B, McKay C, Heung MY, Oien KA, et al. 2010. Dasatinib inhibits the development of metastases in a mouse model of pancreatic ductal adenocarcinoma. Gastroenterology 139: 292–303 [DOI] [PubMed] [Google Scholar]
- Munster T, Gibbs JP, Shen D, Baethge BA, Botstein GR, Caldwell J, Dietz F, Ettlinger R, Golden HE, Lindsley H, et al. 2002. Hydroxychloroquine concentration-response relationships in patients with rheumatoid arthritis. Arthritis Rheum 46: 1460–1469 [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, Mautner J 2003. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol 33: 1250–1259 [DOI] [PubMed] [Google Scholar]
- Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, et al. 2009. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 324: 1457–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC 2005. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ 12: 1613–1621 [DOI] [PubMed] [Google Scholar]
- Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, Debnath J 2010. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 142: 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ 2007. Potential therapeutic applications of autophagy. Nat Rev Drug Discov 6: 304–312 [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z 2007. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26: 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivridis E, Koukourakis MI, Zois CE, Ledaki I, Ferguson DJ, Harris AL, Gatter KC, Giatromanolaki A 2010. LC3A-positive light microscopy detected patterns of autophagy and prognosis in operable breast carcinomas. Am J Pathol 176: 2477–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lüllmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P 2000. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406: 902–906 [DOI] [PubMed] [Google Scholar]
- Tett SE, Cutler DJ, Day RO, Brown KF 1989. Bioavailability of hydroxychloroquine tablets in healthy volunteers. Br J Clin Pharmacol 27: 771–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E, van de Velde H, Karasek P, Oettle H, Vervenne WL, Szawlowski A, Schoffski P, Post S, Verslype C, Neumann H, et al. 2004. Phase III trial of gemcitabine plus tipifarnib compared with gemcitabine plus placebo in advanced pancreatic cancer. J Clin Oncol 22: 1430–1438 [DOI] [PubMed] [Google Scholar]
- White E, DiPaola RS 2009. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res 15: 5308–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou WJ, Deng R, Zhang XY, Feng GK, Gu LQ, Zhu XF 2009. G-quadruplex ligand SYUIQ-5 induces autophagy by telomere damage and TRF2 delocalization in cancer cells. Mol Cancer Ther 8: 3203–3213 [DOI] [PubMed] [Google Scholar]