 Investigation into the hierarchy of the mechanisms of electron capture dissociation mass spectrometry by use of nitrated peptides.
Investigation into the hierarchy of the mechanisms of electron capture dissociation mass spectrometry by use of nitrated peptides.
Abstract
Previously we have shown that the presence of 3-nitrotyrosine within a peptide sequence severely depletes the peptide backbone fragments typically observed following electron capture dissociation (ECD) mass spectrometry. Instead, ECD of nitrated peptides is characterised by abundant losses of small neutrals (hydroxyl radicals, water and ammonia). Here, we investigate the origin of ammonia loss by comparing the ECD behaviour of lysine- and arginine-containing nitrated peptides, and their N-acetylated counterparts, and nitrated peptides containing no basic amino acid residues. The results reveal that ammonia loss derives from the N-terminus of the peptides, however, the key finding of this work is the insight provided into the hierarchy of various proposed ECD mechanisms: the Utah-Washington mechanism, the electron predator mechanism and the Oslo mechanism.
Introduction
Tandem mass spectrometry (MS/MS), in which a precursor ion is characterised according to its fragments, is well-established. The introduction of electron capture dissociation (ECD) in 19981 provided a unique MS/MS technique for biomolecular analysis,2 in particular for peptides and proteins. In peptide/protein ECD, low energy electrons are captured by multiply-charged cations with subsequent cleavage of N–Cα bonds, i.e., cleavage is radically-driven. ECD offers advantages over thermally-driven ‘slow-heating’ MS/MS techniques3 such as collision induced dissociation (CID) and infrared multiphoton dissociation (IRMPD): ECD cleavage is random and therefore sequence coverage tends to be higher,4,5 and labile post-translational modifications (PTMs) are retained on peptide/protein backbone fragments.6 ECD has been successfully applied to the localisation of sites of phosphorylation (serine (S), threonine (T) and tyrosine (Y))7,8 and N- and O-linked glycosylation,9,10 amongst others.
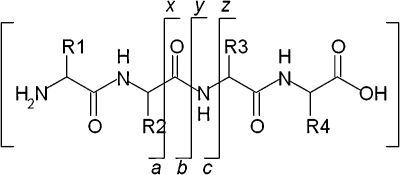
The major product of electron capture by peptide [M + nH]n+ cations is typically the charge-reduced species [M + nH](n–1)+˙, that is, the precursor peptide ion that has captured an electron but has not dissociated. Often, that is accompanied by hydrogen atom loss, i.e., formation of [M + (n – 1)H](n–1)+. The dominant peptide backbone fragmentation pathway proceeds via cleavage of the N–Cα bond to give c and z˙ fragment ions11 (which may be accompanied by hydrogen atom transfer to give c˙ and, more commonly, z fragment ions12). That pathway is in contrast to that observed for CID in which peptide backbone cleavage occurs at N–CO bonds producing b and y fragment ions.13 Scheme 1 shows peptide backbone cleavage sites and fragment ion notation.
Scheme 1. Sites of peptide backbone cleavage and fragment ion notation.
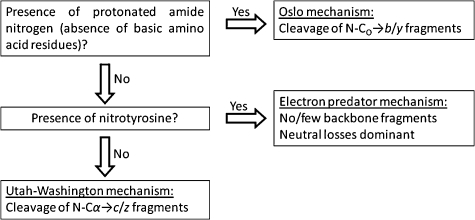
The mechanism by which c and z fragments are generated following electron capture has been the subject of intense debate. Two proposed mechanisms have held precedence: the Cornell mechanism and the Utah-Washington mechanism, both of which are elegantly described in a recent publication by Simons.14 In short, the Cornell mechanism, proposed by McLafferty and co-workers,1 posits initial electron capture to high-n Rydberg states with subsequent localisation to a site of protonation (e.g., lysine or arginine side-chain) forming a hypervalent radical. Hydrogen atom ejection and capture by the amide oxygen occurs followed by cleavage of the adjacent N–Cα bond. A limitation of the Cornell mechanism is the failure to explain the observation of c and z fragments for species in which mobile hydrogen atoms are absent, for example metal-cationised peptides15,16 and peptides carrying fixed charge derivatives.17,18 A further limitation is that hydrogen atom transfer from an arginine radical to an amide carbonyl is an endothermic process.19 The Utah-Washington (UW) mechanism suggests that electron capture to a Coulomb-stabilised amide π* orbital occurs, either directly or via through bond electron transfer from a Rydberg state, rendering the amide bond superbasic, with a proton affinity in the range of 1100–1400 kJ mol–1.20,21 The amide anion radical subsequently abstracts a proton from an accessible site resulting in c and z˙ fragments. Recent theoretical studies14 suggest that initial electron attachment generally occurs to a Rydberg orbital (90–99%) with some (1–10%) direct attachment to amide π* orbitals, and that relaxation to lower energy Rydberg states occurs during which transfer to a (Coulomb-stabilised) amide π* orbital can occur. Experimental evidence for this pathway was previously provided by Beauchamp and co-workers.22 They observed that peptide modifications with an electron affinity (EA) of ≥1.00 eV (specifically benzyl modifications of cysteine) had a deleterious effect on ECD peptide backbone fragmentation and postulated that following initial electron capture to high-n Rydberg states, electron relaxation via through-space or through-bond transfer to the high-EA modification occurs in competition with transfer to the amide π* orbital (UW mechanism). They termed this model the ‘electron predator’ mechanism.
A second minor fragmentation channel often observed in the ECD of peptide ions is the production of a˙ and y fragments. In one of the original ECD papers, it was postulated that a˙/y cleavage occurs via a variation of the Cornell mechanism, i.e., rather than hydrogen atom transfer to an amide oxygen, hydrogen atom transfer to a backbone nitrogen occurs and is followed by heterolytic cleavage of the peptide bond with concomitant loss of CO.23 For certain peptidic structures, e.g., ε-peptides, a˙/y-type cleavage constitutes the major fragmentation pathway.24 Finally, b ions may be observed in the ECD mass spectra of some peptides depending on their structure and the charge-carrier.25 Previous work by us showed that the formation of b ions was not the result of secondary fragmentation of c ions. One possible explanation for the production of b ions is that electron capture by the precursor peptide ion followed by hydrogen atom loss might result in a vibrationally-excited even-electron ion which could dissociate via a mobile proton pathway, as observed in CID. An alternative explanation derives from modelling studies by Uggerud and co-workers.26 They investigated the effect of the initial position of the proton on the ECD fragmentation behaviour and showed that for nitrogen-protonated peptide ions, the expected result was b˙ and y fragments. We term this model the ‘Oslo mechanism’. Experimental studies by Liu and Håkansson on peptides which did not contain basic amino acid residues (BAARs), and therefore must contain protonated amide nitrogens, confirmed the hypothesis: abundant b-type ions were observed.27 It should be noted however that Uggerud's studies suggested radical b ions whereas the b ions observed by us25 and Håkansson27 were even-electron. That observation could be explained by hydrogen atom transfer within a long-lived b˙/y ion complex, similar to that observed for c/z˙ ions.27
We have recently shown that the presence of 3-nitrotyrosine within a peptide sequence has a deleterious effect on ECD backbone cleavage of the c/z type.28 All of the peptides studied contained lysine as the charge-carrier. The effect was particularly severe for doubly-protonated precursor ions and can be explained by the electron predator mechanism proposed by Beauchamp and co-workers.22 In addition, we showed that the ECD mass spectra were dominated by peaks corresponding to the loss of small neutrals (˙OH/H2O/NH3) from the charge-reduced precursor. ECD data from a series of nitrated peptides in which the number of amino acid residues between the nitrotyrosine and the lysine was varied suggested that the proximity of the two was proportional to the abundance of the ammonia loss. Furthermore, 15N-labelling experiments revealed that the nitrotyrosine was not the source of ammonia loss. We postulated that the ammonia losses were the result of non-covalent interactions between the protonated lysine amino acid side-chains and the nitro group. An alternative explanation is that the ammonia loss derives from the protonated N-terminus of the peptides.
Here, we have investigated the origin of ammonia loss in the ECD of nitrated peptides. We compare the small neutral losses observed for nitrated peptides containing lysine, arginine and no basic amino acid residues. We also investigate the effect of acetylating the peptide N-terminus. Finally, we performed MS3 (ECD of the precursor peptide followed by IRMPD of the neutral loss ECD fragments). The results reveal that ammonia loss derives from the N-terminus. More importantly, the results show how the hierarchy of ECD mechanisms can be deduced in part by considering the fragmentation behaviour of nitrated peptides.
Results and discussion
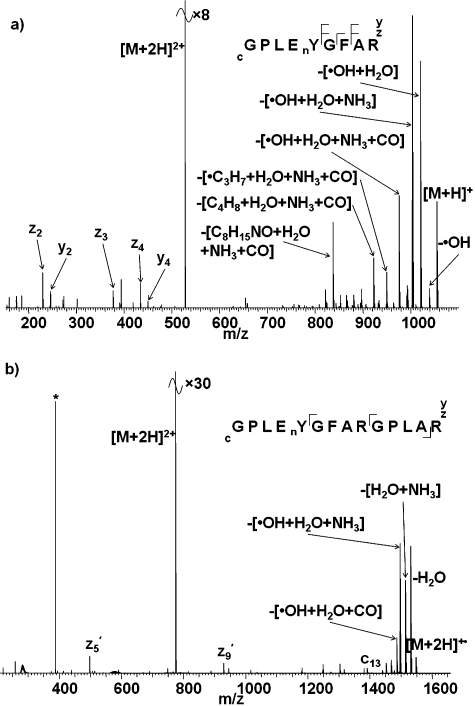
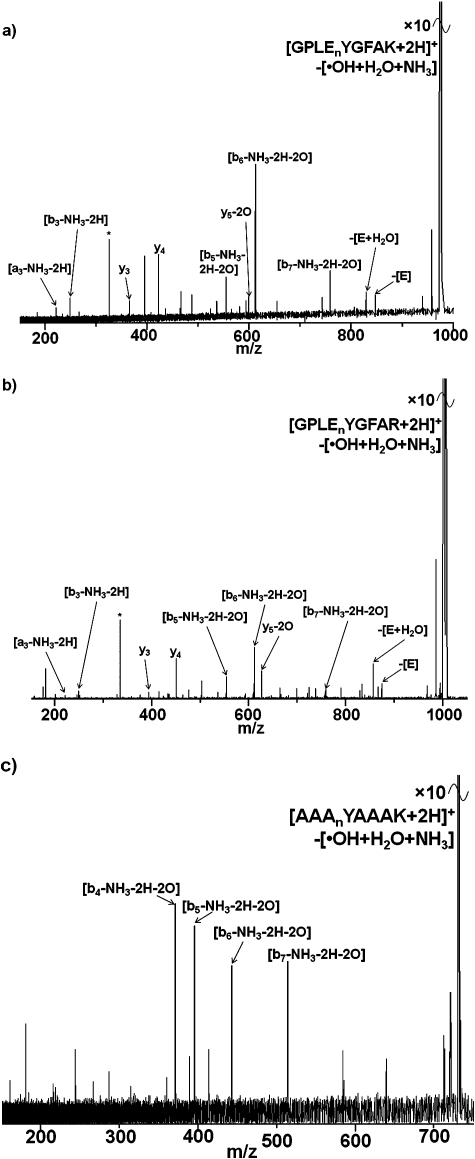
We showed previously that ECD of the doubly-protonated ions of peptides GPLEnYGFAK and GPLEnYGFAKGPLAK (nY is 3-nitrotyrosine), and triply-protonated ions of GPLEnYGFAKGPLAK, resulted in extensive neutral losses from the charge-reduced precursor ions.28 The dominant losses were [˙OH + H2O + NH3] from GPLEnYGFAK and GPLEnYGFAKGPLAK in the 2+ and 3+ charge states, respectively, and [H2O + NH3] from GPLEnYGFAKGPLAK in the 2+ charge state. We postulated that the abundant ammonia loss was the result of interactions between the nitro-group and the lysine side-chain. Fig. 1 shows the ECD mass spectra obtained for the doubly protonated ions of peptides GPLEnYGFAR and GPLEnYGFARGPLAR, i.e., the equivalent peptides in which lysine amino acid residues have been substituted for arginine residues. As seen for the Lys-containing peptides, the presence of nitrotyrosine suppresses ECD peptide backbone cleavage in the Arg-containing peptides, i.e., electron capture proceeds via the electron predator mechanism independent of the nature of the basic amino acid residue. Abundant peaks corresponding to neutral losses from the charge-reduced species were also observed for the Arg-containing nitrated peptides. As for the Lys-containing analogue, the most abundant loss observed on ECD of 2+ ions of GPLEnYGFAR is [˙OH + H2O + NH3], however the neutral loss profiles do vary between the peptides: the relative abundance of the [˙OH + H2O] loss is much greater for the Arg peptide than for the Lys peptide. In addition, neutral losses involving CO were observed for the Arg peptide, as were fragments resulting from cleavages within the Arg side chain. For the longer peptides (GPLEnYGFA(R/K)GPLA(R/K)), variation in the neutral loss profiles is again observed. For the Lys-containing peptides, the most abundant loss was [H2O + NH3] whereas for the Arg-containing peptide the most abundant losses are [˙OH + H2O + NH3] and H2O. Losses involving CO were observed for the Arg-containing peptide.
Fig. 1. ECD mass spectra of (a) [GPLEnYGFAR + 2H]2+ ions; (b) ECD of [GPLEnYGFARGPLAR + 2H]2+ ions. nY denotes 3-nitrotyrosine. Inset: summary of observed fragments. * corresponds to the 2nd harmonic peak.
The results obtained for the Arg-containing peptides appear to corroborate the hypothesis that ammonia loss derives from the basic amino acid side-chain. To determine unambiguously whether that was the case we performed two experiments. Firstly, we examined the ECD behaviour of the nitrated peptide GPLEnYGFAL in which Lys/Arg has been substituted with Leu, i.e., contains no basic amino acid residue (BAAR). Secondly, we investigated the ECD behaviour of the Arg- and Lys-containing peptides which had been N-acetylated thus preventing any ammonia loss from the N-terminus. (We have shown previously, via 15N isotope labelling experiments, that ammonia loss does not derive from the nitro group28).
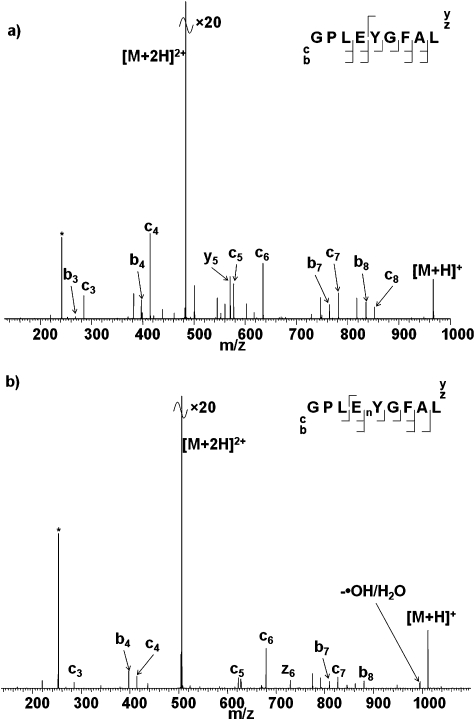
Fig. 2 shows the ECD mass spectra of doubly-charged ions of the non-BAAR-containing unmodified and nitrated peptides. There are two main points of interest: the abundant loss of small neutrals is not observed for the nitrated peptide and the presence of the nitrotyrosine does not affect the backbone cleavage observed. Moreover, for both the nitrated and unmodified peptides significant b-type fragmentation is observed. At first glance the substantial decrease in neutral losses (no losses involving ammonia and low level loss of ˙OH and water) appears to confirm that the losses derive from basic amino acid side-chains. However, the similarity between the ECD mass spectra from the two peptides suggests that caution should be exercised before drawing such a conclusion. The electron predator mechanism does not appear to apply to nitrated GPLEnYGFAL because ECD backbone fragments are observed. The peptide does not contain a basic amino acid and presumably is therefore protonated on a backbone amide nitrogen. As described above, Liu and Håkansson27 have previously shown that such a situation leads to b-type fragmentation via the Oslo mechanism. This result suggests that the Oslo mechanism takes precedence over the electron predator mechanism.
Fig. 2. ECD mass spectra of (a) [GPLEYGFAL + 2H]2+ ions; (b) [GPLEnYGFAL + 2H]2+ ions. * corresponds to the 2nd harmonic peak.
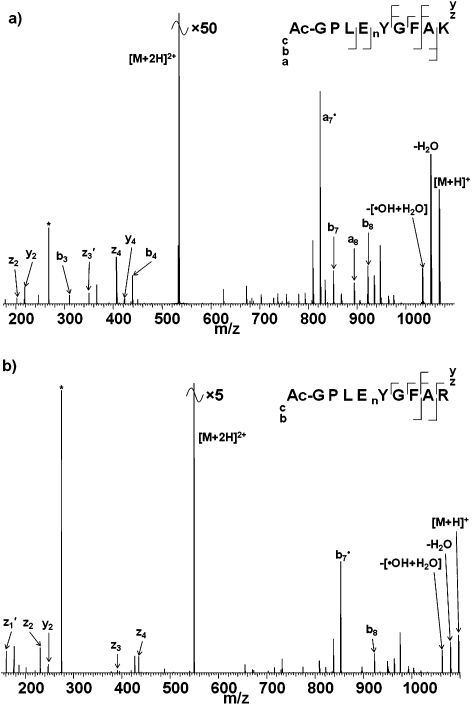
Fig. 3 shows ECD mass spectra of the doubly-charged ions of the N-acetylated nitrated peptides Ac-GPLEnYGFAK/R. (Acetylation solely of the N-terminus was confirmed by CID, see ESI†, Fig. S1.) Three salient features of the ECD mass spectra can be identified. Firstly, neutral losses involving NH3 were not observed in the ECD of N-acetylated nitrated peptides. Secondly, abundant losses of ˙OH radicals and water are observed. Finally, the backbone cleavage observed differs from that observed in the absence of N-acetylation. The observation that neutral losses involving ammonia were not observed suggests that NH3 loss derives from the N-terminus, in contrast to the results for the non-BAAR-containing peptide. The continued losses of ˙OH radicals and water suggest that these derive from interactions between the nitro group and the basic amino acid chain.22,29 For the Lys-containing nitrated peptide, N-acetylation results in the appearance of additional a- and b-type fragments, b 3, b 4, b 7, b 8, a 7˙ and a 8 (cf. b 7 for GPLEnYGFAK). The a 7˙ fragment ion is the most abundant peak in the mass spectrum. Similar observations were made for the Arg-containing peptide: N-acetylation results in the appearance of b 7˙ and b 8 fragments in the ECD mass spectrum, with b 7˙ being the most abundant product ion. These observations can be explained in terms of the protonation state of the precursor ions. For the non-acetylated nitrated peptides, the sites of protonation are presumed to be the basic amino acid side-chain (Lys/Arg) and the N-terminus for doubly-charged ions. Acetylation precludes protonation at the N-terminus and therefore doubly-charged acetylated nitrated peptide ions must contain one proton on the BAAR side-chain and one on a backbone amide nitrogen. As shown previously, the presence of a protonated nitrogen promotes the Oslo mechanism of ECD and that is confirmed here by the presence of b-type ions. Again this result suggests that the Oslo mechanism takes precedence over the electron predator and UW (or Cornell) mechanism.
Fig. 3. ECD mass spectra of doubly-charged N-terminal acetylated peptide ions: (a) [Ac-GPLEnYGFAK + 2H]2+; (b) [Ac-GPLEnYGFAR + 2H]2+. * corresponds to the 2nd harmonic peak.
ECD experiments on the non-BAAR-containing nitrated peptide and N-acetylated nitrated peptides appeared to give conflicting results with regard to the origin of ammonia loss (although as discussed the differences can be explained in terms of competing mechanisms). Nevertheless, in order to unambiguously determine the origin of ammonia loss, we performed MS3 experiments.
Fig. 4 shows the MS3 (IRMPD of ECD fragments) spectra of three nitrated doubly-charged peptides; in all cases ECD resulted in the [[M + 2H]–[˙OH + H2O + NH3]]+ fragment, which was subsequently selected for fragmentation by IRMPD.30 Consider first the MS3 spectrum for GPLEnYGFAK: two peaks corresponding to singly-charged y 3 and y 4 fragments of the precursor peptide are observed suggesting that none of the neutral losses originate C-terminal of the nitrotyrosine. A peak corresponding to the y 5 fragment of the precursor peptide less the mass of two oxygen atoms is observed suggesting that the water and ˙OH losses derive from the nitrotyrosine residue. The peak corresponding to the b 3 fragment of the precursor less ammonia and two hydrogen atoms suggests that ammonia loss occurs N-terminal of the leucine residue. Also noteworthy is the presence of a peak corresponding to the loss of a glutamic acid residue. That suggests that the loss of ammonia may involve reaction of the N-terminus with nitrotyrosine and concomitant cleavage of the glutamic acid–tyrosine, or leucine–glutamic acid, peptide bond. Similar results were observed for the MS3 of GPLEnYGFAR: the data suggest that hydroxyl radical and water losses originate from nitrotyrosine whereas ammonia loss originates at the N-terminus. The MS3 results for AAAnYAAAK are less conclusive. Nevertheless they show that ˙OH, water and ammonia losses all arise N-terminal of Ala4.
Fig. 4. MS3 (IRMPD of ECD fragments) mass spectra of doubly-charged peptide ions: (a) [[GPLEnYGFAK + 2H]+–[˙OH + H2O + NH3]] ions; (b) [[GPLEnYGFAR + 2H]+–[˙OH + H2O + NH3]] ions; (c) [[AAAnYAAAK + 2H]+–[˙OH + H2O + NH3]] ions. * denotes noise peak.
Experimental
Preparation of synthetic peptides
The nitrated peptides, GPLEnYGFAK, GPLEnYGFAKGPLAK, GPLEnYGFAR, GPLEnYGFARGPLAR, and GPLEnYGFAL (where nY indicates 3-nitrotyrosine), their unmodified counterparts and AAAnYAAAK were synthesised by Alta Bioscience (Birmingham, UK). All synthetic peptides were used without further purification. Selective N-terminus acetylation was completed by incubating 1 ∶ 1 acetic anhydride (Sigma-Aldrich, UK): peptide in 100 mM ammonium bicarbonate overnight at 37 °C. The peptides were diluted to 2 pmol μL–1 in 49.5 ∶ 49.5% methanol (Fisher Scientific, Leicestershire, UK): water (J.T. Baker, Deventer, The Netherlands), and 1% formic acid (Fisher Scientific).
Mass spectrometry
ECD
Analyses were performed on a Thermo Finnigan LTQ FT Ultra mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Samples were injected by use of an Advion Biosciences Triversa electrospray source (Advion Biosciences, Ithaca, NY, USA) at a flow rate of ∼200 nL min–1. MS/MS spectra were acquired in the ICR cell with a resolution of 100 000 at m/z 400. Precursor ions were isolated in the linear ion trap and transferred to the ICR cell for ECD. Automatic gain control (AGC) target was 2 × 105 with maximum fill time 1 s. Isolation width was m/z 5. Electrons for ECD were produced by an indirectly heated barium–tungsten cylindrical dispenser cathode (5.1 mm diameter, 154 mm from the cell, 1 mm off axis) (Heat-Wave Labs, Watsonville, CA, USA). The current across the electrode was ∼1.1 A. Ions were irradiated with electrons for 70 ms at 5% energy (corresponding to a cathode potential of –2.775 V). Each ECD scan comprises 4 co-added microscans. Mass spectra shown comprise 30 averaged scans.
MS3 (IRMPD of ECD fragments)
Analyses were performed on a Bruker 12 T Apex Qe Ultra (Bruker, Billerica, MA, USA). Samples were injected by use of an Advion Biosciences Triversa electrospray source (Advion Biosciences). Precursor ions were isolated by use of the mass resolving quadrupole. For ECD, 1.8 A was applied to the dispenser cathode filament (Heatwave Technologies), 20 V to the lens, 0.8 V to the bias, and a pulse of 70 ms was applied. For MS3, ions were isolated in the ICR cell via correlated sweep excitation (COSE) and exposed to a 25 W CO2 laser (Synrad Inc., Mukilteo, WA, USA) at 70% power for 100 ms. Mass spectra shown are the sum of 300 acquisitions.
ECD MS/MS data were analysed using Xcalibur 2.10 software (Thermo Fisher Scientific), MS3 data were analysed using DataAnalysis 4.0 software (Bruker Daltonics). All mass spectra were manually searched for a, b, c˙/c, y, z/z′ fragment ions using Protein Prospector ver. 5.5.0 software (UCSF, San Francisco, CA, USA).
Conclusion
Our results show that 3-nitrotyrosine-containing peptides provide insight into the hierarchy of the mechanisms of electron capture dissociation. The findings are summarised in Scheme 2. Regardless of whether a peptide contains nitrotyrosine or not, if the peptide does not contain a basic amino acid residue, ECD of doubly-protonated ions will proceed via the Oslo mechanism to produce b- and y-type fragments. If a peptide contains a basic amino acid residue and nitrotyrosine (or other modification with positive electron affinity), the electron predator mechanism will take precedence, resulting in no, or few, backbone fragments, and abundant losses of small neutral species. Losses involving ammonia are only observed if the N-terminus of the peptide is able to interact with the nitrotyrosine group. Only in the absence of nitrotyrosine (or other modification with positive electron affinity) and presence of a basic amino acid residue will ECD proceed via the UW (or the Cornell) mechanism.
Scheme 2. Hierarchy of ECD mechanisms.
Acknowledgments
This work has been funded by EPSRC (A.W.J. and H.J.C.) and the Wellcome Trust (074131, H.J.C.). The Thermo Fisher Scientific LTQ FT Ultra mass spectrometer used in this research was obtained through the Birmingham Science City Translational Medicine: Experimental Medicine Network of Excellence project, with support from Advantage West Midlands (AWM). The authors thank Dr Logan Mackay, Dr Dave Clarke and Dr Pat Langridge-Smith for access to the FT-ICR facility at SIRCAMS, School of Chemistry, University of Edinburgh, and for their technical assistance.
Footnotes
†Electronic supplementary information (ESI) available: CID MS/MS spectra of N-acetylated peptides; CID experimental data. See DOI: 10.1039/c0cp00623h
References
- Zubarev R. A., Kelleher N. L., McLafferty F. W. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- Cooper H. J., Hakansson K., Marshall A. G. Mass Spectrom. Rev. 2005;24:201–222. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]
- McLuckey S. A., Goeringer D. E. J. Mass Spectrom. 1997;32:461–474. doi: 10.1002/(SICI)1096-9888(199610)31:10<1093::AID-JMS393>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Axelsson J., Palmblad M., Hakansson K., Hakansson P. Rapid Commun. Mass Spectrom. 1999;13:474–477. doi: 10.1002/(SICI)1097-0231(19990330)13:6<474::AID-RCM505>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Zubarev R. A., Horn D. M., Fridriksson E. K., Kelleher N. L., Kruger N. A., Lewis M. A., Carpenter B. K., McLafferty F. W. Anal. Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- Kelleher N. L., Zubarev R. A., Bush K., Furie B., Furie B. C., McLafferty F. W., Walsh C. T. Anal. Chem. 1999;71:4250–4253. doi: 10.1021/ac990684x. [DOI] [PubMed] [Google Scholar]
- Shi S. D. H., Hemling M. E., Carr S. A., Horn D. M., Lindh I., McLafferty F. W. Anal. Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- Stensballe A., Jensen O. N., Olsen J. V., Haselmann K. F., Zubarev R. A. Rapid Commun. Mass Spectrom. 2000;14:1793–1800. doi: 10.1002/1097-0231(20001015)14:19<1793::AID-RCM95>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Hakansson K., Cooper H. J., Emmett M. R., Costello C. E., Marshall A. G., Nilsson C. L. Anal. Chem. 2001;73:4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- Mirgorodskaya E., Roepstorff P., Zubarev R. A. Anal. Chem. 1999;71:4431–4436. doi: 10.1021/ac990578v. [DOI] [PubMed] [Google Scholar]
- Zubarev R. A., Haselmann K. F., Budnik B., Kjeldsen F., Jensen F. Eur. J. Mass Spectrom. 2002;8:337–349. [Google Scholar]
- Savitski M. M., Kjeldsen F., Nielsen M. L., Zubarev R. A. J. Am. Soc. Mass Spectrom. 2007;18:113–120. doi: 10.1016/j.jasms.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Roepstorff P., Fohlman J. Biol. Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- Simons J. Chem. Phys. Lett. 2010;484:81–95. [Google Scholar]
- Iavarone A. T., Paech K., Williams E. R. Anal. Chem. 2004;76:2231–2238. doi: 10.1021/ac035431p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudgins R. R., Hakansson K., Quinn J. P., Hendrickson C. L. and Marshall A. G., 50th ASMS Conference on Mass Spectrometry and Allied Topics, Orlando, FL, 2002.
- Chamot-Rooke J., Malosse C., Frison G., Turecek F. J. Am. Soc. Mass Spectrom. 2007;18:2146–2161. doi: 10.1016/j.jasms.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Li X., Cournoyer J. J., Lin C., O'Connor P. B. J. Am. Soc. Mass Spectrom. 2008;19:1514–1526. doi: 10.1016/j.jasms.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. H., Turecek F. J. Am. Chem. Soc. 2006;128:12520–12530. doi: 10.1021/ja063676o. [DOI] [PubMed] [Google Scholar]
- Syrstad E. A., Turecek F. J. Am. Soc. Mass Spectrom. 2005;16:208–224. doi: 10.1016/j.jasms.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Sobczyk M., Anusiewicz W., Berdys-Kochanska J., Sawicka A., Skurski P., Simons J. J. Phys. Chem. A. 2005;109:250–258. doi: 10.1021/jp0463114. [DOI] [PubMed] [Google Scholar]
- Sohn C. H., Chung C. K., Yin S., Ramachandran P., Loo J. A., Beauchamp J. L. J. Am. Chem. Soc. 2009;131:5444–5459. doi: 10.1021/ja806534r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubarev R. A., Kruger N. A., Fridriksson E. K., Lewis M. A., Horn D. M., Carpenter B. K., McLafferty F. W. J. Am. Chem. Soc. 1999;121:2857–2862. [Google Scholar]
- Cooper H. J., Hudgins R. R., Marshall A. G. Int. J. Mass Spectrom. 2004;234:23–35. [Google Scholar]
- Cooper H. J. J. Am. Soc. Mass Spectrom. 2005;16:1932–1940. doi: 10.1016/j.jasms.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Bakken V., Helgaker T., Uggerud E. Eur. J. Mass Spectrom. 2004;10:625–638. doi: 10.1255/ejms.665. [DOI] [PubMed] [Google Scholar]
- Liu H. C., Håkansson K. J. Am. Soc. Mass Spectrom. 2007;18:2007–2013. doi: 10.1016/j.jasms.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Jones A. W., Mikhailov V. A., Iniesta J., Cooper H. J. J. Am. Soc. Mass Spectrom. 2010;21:268–277. doi: 10.1016/j.jasms.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Polasek M., Turecek F. J. Am. Chem. Soc. 2000;122:9511–9524. [Google Scholar]
- Woodlin R. L., Bomse D. S., Beauchamp J. L. J. Am. Chem. Soc. 1978;100:3248–3250. [Google Scholar]