Abstract
Despite the fact that X-box binding protein-1 (XBP-1) is one of the main regulators of the unfolded protein response (UPR), the modulators of XBP-1 are poorly understood. Here, we show that the regulatory subunits of phosphotidyl inositol 3-kinase (PI3K), p85α (encoded by Pik3r1) and p85β (encoded by Pik3r2) form heterodimers that are disrupted by insulin treatment. This disruption of heterodimerization allows the resulting monomers of p85 to interact with, and increase the nuclear translocation of, the spliced form of XBP-1 (XBP-1s). The interaction between p85 and XBP-1s is lost in ob/ob mice, resulting in a severe defect in XBP-1s translocation to the nucleus and thus in the resolution of endoplasmic reticulum (ER) stress. These defects are ameliorated when p85α and p85β are overexpressed in the liver of ob/ob mice. Our results define a previously unknown insulin receptor signaling pathway and provide new mechanistic insight into the development of ER stress during obesity.
The ER is a large membrane-enclosed cellular organelle in which secretory and membrane-bound proteins are folded into their final three-dimensional structures, lipids and sterols are synthesized, and free calcium is stored1,2. Conditions that interfere with proper functioning of the ER create a state defined as ER stress and lead to activation of the UPR3–5.
The UPR is conveyed to the cell through three main signaling pathways. The first two pathways are initiated by type I transmembrane kinases, PKR-like endoplasmic reticulum kinase (PERK) and inositol requiring enzyme-1 (IRE1), and the third pathway launches with activation of a type II transmembrane protein called activating transcription factor-6 (ATF6)1–5. Activation of PERK during ER stress leads to phosphorylation of eukaryotic translation initiation factor-2α at Ser51 and consequently results in global translational attenuation6,7. IRE1, on the other hand, has both kinase and endoRNase activity8–11. The endoRNase domain of IRE1 splices the mRNA of a transcription factor called X-box–binding protein-1 (XBP-1), removing a 26-bp segment from the full-length XBP-1 messenger RNA that creates a translational frame shift leading to the expression of a higher–molecular-weight protein, XBP-1s12–14. XBP-1s is a highly active transcription factor and is one of the master regulators of ER folding capacity. It upregulates the gene expression of ER chaperones15 and components of the ER-associated degradation pathway15 and is also crucial in ER expansion16,17. Despite its central roles in the UPR response, in cellular machinery and in many disease states, including leptin and insulin resistance18,19, current knowledge regarding the regulation of XBP-1 activity is extremely limited.
PI3K is one of the main nodules in growth factor signaling and consists of a p85 repressive regulatory subunit and a p110 catalytic subunit20–23. This nodule aids in conveying signals from receptor tyrosine kinases to the downstream targets in the cell, leading to activation of divergent signaling pathways that have central roles in various cellular processes, such as glucose homeostasis, protein synthesis, growth and differentiation20–24. The p85 regulatory subunit is crucial in mediating the activation of class IA PI3K by receptor tyrosine kinases22. Binding of p85 to activated receptor tyrosine kinases or adaptor proteins, such as insulin receptor substrate-1 (IRS1) and IRS2, relieves the basal repression of p110, allowing for the activation of the catalytic subunit.
In addition to being a regulatory subunit for p110, it is becoming evident that p85 also interacts with other proteins, such as the small GTPase cdc42 (refs. 25,26), nuclear receptor co-repressor27 and a transmembrane tyrosine phosphatase (CD148)28, and that it is involved in other cellular processes. Here we have identified p85 as an interacting protein with XBP-1s and investigated the consequences of this interaction during ER stress and metabolic disease.
RESULTS
p85α and p85β increase nuclear transport of XBP-1s
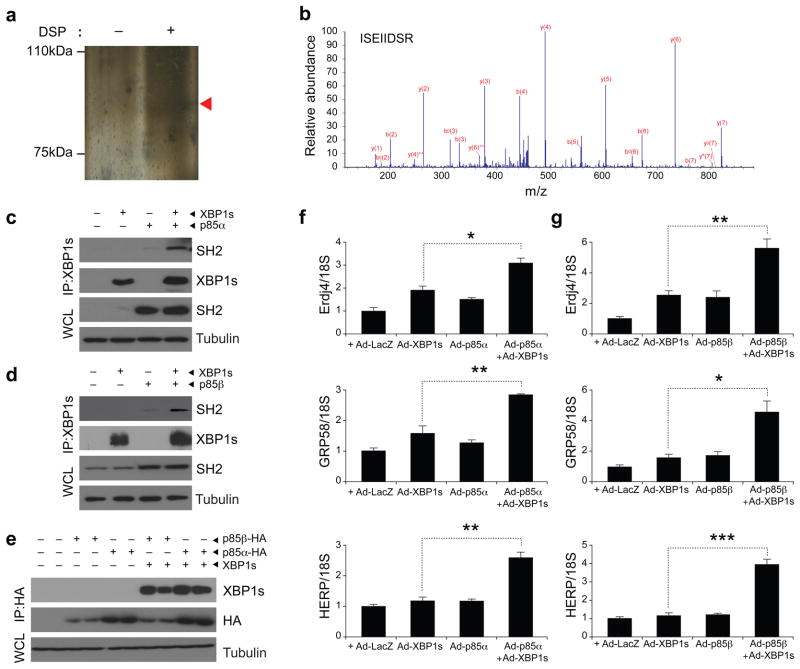
To identify proteins that interact with XBP-1s, we overexpressed XBP-1s in mouse embryo fibroblasts (MEFs) with adenoviruses that encode either a Flag-tagged XBP-1s (Ad-XBP-1s-Flag) or a LacZ (Ad-LacZ) control. After Flag immunoprecipitation, we pooled the eluates and resolved them on an SDS-PAGE gel, and we visualized the bands by silver staining (Fig. 1a). Then we analyzed the bands by tandem mass spectroscopy. An abundant peptide sequence was ISEIIDSR, the amino acid sequence of p85α at residues 535–542 residues (Fig. 1b). In addition, another dominant peptide sequence (VLSEIFSPVLFR) corresponded to residues 263–274 of p85α (data not shown). We next wanted to investigate whether XBP-1s associates directly with p85α or whether the interaction indicated by the tandem mass spectroscopy analysis was an artifact. For this purpose, we expressed XBP-1s and p85α in MEFs by infecting the cells with Ad-XBP-1s and p85α-expressing adenovirus (Ad-p85α) and performed XBP-1 immunoprecipitation. We western blotted the precipitate with an antibody that is specific for the p85 SH2 domain. p85α and XBP-1s co-immunoprecipitated, confirming their interaction (Fig. 1c).
Figure 1.
p85α and p85β interact with XBP-1s. (a) Silver staining of immunoprecipitated proteins with XBP-1 after infecting MEFs with Ad-XBP-1s and cross-linking with DSP (see Online Methods for details). (b) Tandem mass spectroscopy analysis of the band indicated with an arrowhead in a. (c) Immunoblotting for SH2 of p85 and XBP-1s proteins after immunoprecipitation (IP) of XBP-1 from MEFs infected with the indicated adenoviruses. WCL, whole cell lysate. (d) Immunoblots of SH2 and XBP-1 after XBP-1s immunoprecipitation. Total lysates were immunoblotted for SH2 and tubulin. (e) Western blot analysis for XBP-1 and HA after immunoprecipitation with HA-specific antibody crosslinked to beads. (f) Expression of XBP-1 target genes, Dnajb9, Pdia3 and Herpud1, in MEFs infected with Ad-XBP-1s, Ad-p85α, or Ad-XBP-1s and Ad-p85α. (g) mRNA levels of Dnajb9, Pdia3 and Herpud1 in MEFs infected with Ad-XBP-1s, Ad-p85β or Ad-XBP-1s and Ad-p85β. Experiments in c–g were independently repeated three times. Error bars are means ± s.e.m.; P values were determined by Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001).
Considering that p85α and p85β share a marked structural homology22,23, we investigated whether XBP-1s also binds p85β. For this purpose, we infected MEFs with Ad-XBP-1s and p85β-expressing adenovirus (Ad-p85β) and subsequent XBP-1 immunoprecipitation resulted in coimmunoprecipitation of p85β (Fig. 1d). Next, we performed the reverse coimmunoprecipitation experiment by infecting MEFs with either hemagglutinin (HA)-tagged p85α (Ad-p85α-HA) or HA-tagged p85β (Ad-p85β-HA) with or without Ad-XBP-1s and analyzed whether we could detect XBP-1s in these immunoprecipitates. We pulled down the p85α and p85β with an HA-specific antibody that was crosslinked to beads and performed western blotting for XBP-1s. Our results showed that XBP-1s was present in these immunoblots, confirming its interaction with p85α and p85β (Fig. 1e).
To take the first steps toward understanding the physiological role of the interaction between p85 and XBP-1s, we examined whether p85α or p85β have modulatory effects on XBP-1s activity. We thus expressed XBP-1s with p85α or p85β and analyzed the mRNA levels of various genes that are the targets of XBP-1s, such as Dnajb9 (encoding endoplasmic reticulum-localized DnaJ homologues), Pdia3 (encoding protein disulfide isomerase family A, member 3) and Herpud1 (encoding homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member-1). Quantitative PCR analysis showed that expression of XBP-1s with p85α or with p85β significantly increased its ability to upregulate the transcription of Dnajb9, Pdia3 and Herpud1, suggesting that p85α and p85β both increase XBP-1s activity (Fig. 1f,g). Over-expression of p85α or p85β together with XBP-1s did not increase mRNA levels of XBP-1s (Supplementary Fig. 1).
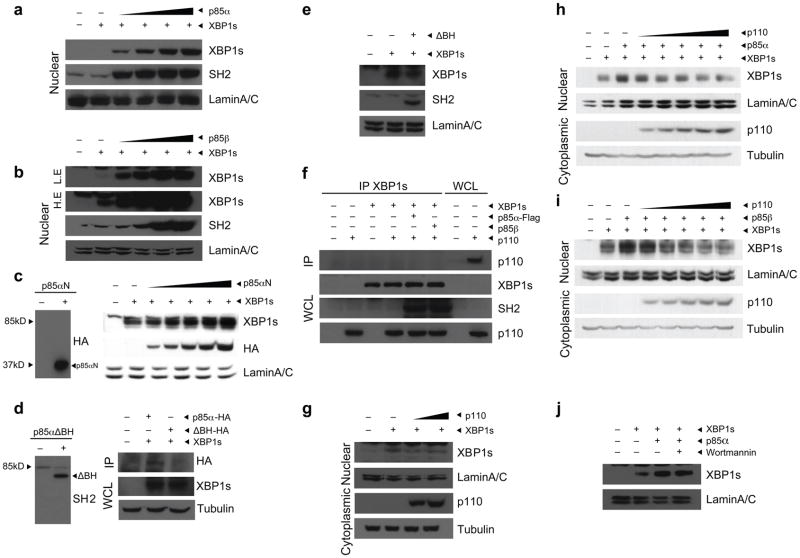
Considering the fact that XBP-1s activity was increased by p85α or p85β and that there are substantial data showing that p85 translocates to the nucleus and can bind nuclear proteins27,29–31, we asked whether p85 regulates the nuclear import of XBP-1s. To investigate this hypothesis, we infected the cells with increasing doses of Ad-p85α or Ad-p85β while keeping the dose of Ad-XBP-1s constant and then analyzed the XBP-1s levels in the nuclear fractions. Increasing the expression of p85α or p85β led to a higher translocation of XBP-1s to the nucleus (Fig. 2a,b). Furthermore, we found that the upregulation of nuclear translocation of XBP-1s in the presence of p85 was not due to an increase in the stability of XBP-1s protein, as p85α did not alter the degradation rate of XBP-1s protein (Supplementary Fig. 2).
Figure 2.
p85α and p85β increase nuclear translocation of XBP-1s. (a,b) Nuclear protein amounts of XBP-1s and p85 in MEFs infected with Ad-XBP-1s and with increasing doses of Ad-p85α (a) or Ad-p85β (b). Lamin A/C was used as a control for nuclear protein level. (c) Left, western blot for HA in MEFs infected with p85αN-HA. Right, nuclear protein amounts of XBP-1s, p85αN-HA and lamin A/C in MEFs expressing XBP-1s together with increasing amounts of p85αN-HA. (d) Left, western blot for SH2 in MEFs infected with p85αΔBH-HA. Right, immunoblotting for HA in XBP-1 immunoprecipitates from MEFs infected with Ad-XBP-1s and with either p85α-HA or p85αΔBH-HA. (e) Immunoblotting of MEFs infected with Ad-XBP-1s or with Ad-XBP-1s and Ad-p85αΔBH-HA together. Nuclear proteins were immunoblotted with the indicated antibodies. (f) Western blot for p110 in XBP-1 immunoprecipitates and for XBP-1s, SH2 and p110 in whole-cell lysates from cells infected with Ad-XBP-1s, Ad-p110, Ad-XBP-1s and Ad-p110, Ad-XBP-1s, Ad-p110 and Ad-p85α or Ad-XBP-1s, Ad-p110 and Ad-p85β. (g) XBP-1s and lamin A/C protein levels after infection of MEFs with a constant dose of Ad-XBP-1s and increasing doses of Ad-p110. (h,i) Nuclear protein amounts of XBP-1s and lamin A/C in cells infected with Ad-XBP-1s and Ad-p85α (h) or Ad-p85β (i) and with increasing doses of Ad-p110. (j) XBP-1s nuclear protein amounts in the presence or absence of wortmannin (100 μM). Experiments were independently repeated three times.
To investigate whether p110 is also involved in p85-mediated XBP-1s nuclear translocation, we first infected the cells with Ad-XBP-1s along with increasing doses of adenovirus that encodes the amino terminus of p85α (Ad-p85N-HA), which lacks the p110-binding region. In spite of its inability to bind p110, this version of p85α was still capable of increasing the nuclear translocation of XBP-1s (Fig. 2c).
The B cell receptor homology (BH) domain within the amino terminus of p85 was previously shown to mediate the interactions of p85 with other proteins such as cdc42 (refs. 25,26). Taking this information into account, we investigated whether p85α lacking the BH domain (p85αΔBH-HA) could still interact with XBP-1s. We infected MEFs with either Ad-XBP-1s and Ad-p85α-HA or Ad-XBP-1s and Ad-p85αΔBH-HA and performed XBP-1s immunoprecipitation. p85αΔBH-HA did not immunoprecipitate with XBP-1s (Fig. 2d), indicating that p85 interacts with XBP-1s through its BH domain. Indeed, p85αΔBH-HA failed to increase XBP-1s nuclear translocation (Fig. 2e). We next investigated whether p110 exists in the p85–XBP-1s complexes. For this purpose, we infected MEFs with Ad-p110, Ad-XBP-1s, Ad-XBP-1s and Ad-p110, Ad-XBP-1s, Ad-p85α and Ad-p110 or Ad-XBP-1s, Ad-p85β and Ad-p110 and immunoprecipitated the XBP-1s. We found no evidence of the existence of p110 in the p85–XBP-1 complex (Fig. 2f). Next, we infected MEFs with a constant dose of Ad-XBP-1s while treating with increasing doses of Ad-p110. We found that p110 did not increase nuclear translocation of XBP-1s and even led to a slight decrease in the nuclear abundance of XBP-1s (Fig. 2g).
As our data to this point indicated that the p85–XBP-1s and p85-p110 complexes are independent of each other, we were interested in determining whether additional expression of p110 would create a squelching effect on p85 by decreasing the availability of p85 to interact with XBP-1s. Our results indeed showed that increasing the expression of p110, while keeping the abundance of p85α or p85β and of XBP-1s constant, reduced the p85-mediated upregulation of nuclear translocation of XBP-1s (Fig. 2h,i). In addition, treatment of cells with wortmannin, inhibiting p110 catalytic activity, did not affect the p85α-mediated nuclear translocation of XBP-1s (Fig. 2j).
Next, to test whether overexpression of p85α or p85β increases the ability of the cells to recover from the ER stress, we infected MEFs with Ad-LacZ, Ad-p85α or Ad-p85β and stimulated them with 100 mM dithiothreonine (DTT). We analyzed the recovery from ER stress by determining the degree of PERK phosphorylation at residue Thr980. Stimulation of the LacZ-infected cells led to robust activation of ER stress (Supplementary Fig. 3a,b). The degree of PERK Thr980 phosphorylation in the LacZ-infected group declined over time but still remained high even 4 h after the removal of DTT from the cells (Supplementary Fig. 3a,b). In contrast, overexpression of p85α or p85β markedly shortened the recovery time from the ER stress (Supplementary Fig. 3a,b). Two hours after DTT removal, PERK Thr980 phosphorylation was barely visible, and it was completely diminished at the 4-h time point in the Ad-p85α– or Ad-p85β– infected cells (Supplementary Fig. 3a,b). These data indicate that p85α and p85β increase the capacity of cells to cope with ER stress. Furthermore, overexpression of neither p85αΔBH nor p85βΔBH improved the ability of cells to recover from ER stress after DTT stimulation (Supplementary Fig. 3c,d).
p85α and p85β migrated to the nucleus when we infected MEFs with Ad-p85α or Ad-p85β (Fig. 2a,b). To analyze whether the translocation of p85α or p85β was affected by XBP-1s, we compared the nuclear abundance of p85α or p85β in the absence or presence of XBP-1s. We found that XBP-1s did not alter the p85α and p85β migration pattern to the nucleus even when very highly expressed, which suggests that XBP-1s cannot promote the nuclear transport of the p85 isoforms (Supplementary Fig. 4a,b).
p85α and p85β form an insulin-sensitive heterodimer
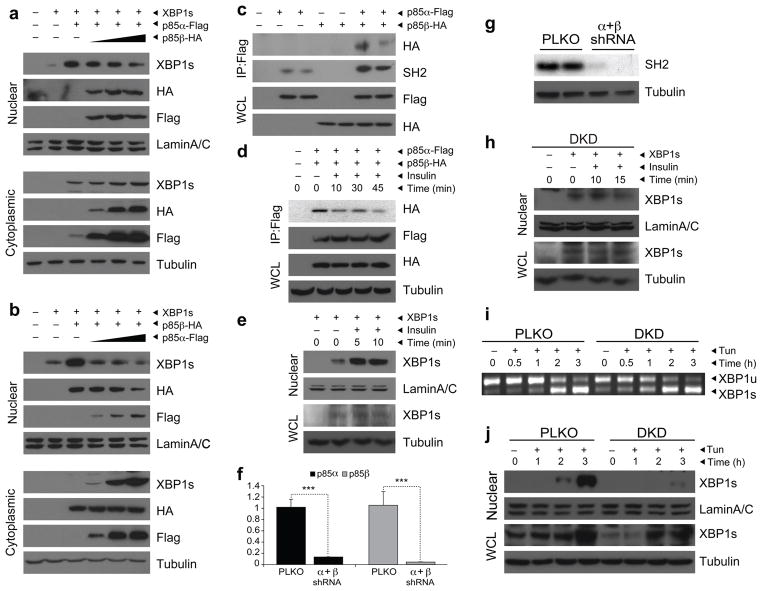
Next, we examined whether expression of p85α and p85β together with XBP-1s would further increase the nuclear translocation of XBP-1s. We found that when we increased the p85β input while keeping p85α and XBP-1s expression constant, p85α-mediated upregulation of XBP-1s in the nucleus was reduced (Fig. 3a). We obtained similar results when we gradually increased the dose of Ad-p85α while keeping the expression of p85β and XBP-1s constant (Fig. 3b). These results suggested a competitive interaction among p85α, p85β and XBP-1. Considering that both isoforms of p85 neutralized the effect of the other on XBP-1s, we hypothesized that p85α and p85β might bind each other and that this interaction is stronger than their interaction with XBP-1s. To test this postulate, we expressed a Flag-tagged p85α (p85α-Flag) with an HA-tagged p85β (p85β-HA) in MEF cells by adenovirus-mediated infection and performed Flag immuno-precipitation. This procedure did not lead to precipitation of p85β-HA when it was expressed alone (Fig. 3c). However, immunoprecipitation of the Flag tag successfully brought down the p85β-HA when it was expressed with p85α-Flag, suggesting that p85α and p85β interact with each other. Indeed, when we reblotted the same membrane with SH2-specific antibody which recognizes both p85α and p85β, the amount of SH2 signal in the p85α and p85β coexpression condition was markedly more than that in the p85α-Flag alone condition (Fig. 3c). This result provided additional support that immunoprecipitation of p85α also pulls down another isoform of p85, p85β, in our experimental conditions (Fig. 3c).
Figure 3.
Insulin increases nuclear transport of XBP-1s. (a) Nuclear and cytoplasmic protein amounts of XBP-1s, p85α and p85β in MEFs infected with a constant dose of XBP-1s and p85α-Flag and increasing doses of p85β-HA. (b) Nuclear and cytoplasmic protein amounts of XBP-1s, p85α and p85β in MEFs infected with a constant dose of XBP-1s and p85β-HA and increasing doses of p85α-Flag. (c) HA and SH2 blotting in Flag immunoprecipitates of cells infected with Ad-p85α-Flag and Ad-p85β-HA. (d) HA and Flag immunoblotting in p85α-Flag immunoprecipitates after insulin (500 nM) stimulation. (e) Nuclear protein amounts of XBP-1s in insulin (500 nM)-stimulated XBP-1s–infected MEFs. (f) mRNA levels of p85α and p85β in the p85α and p85β DKD cells. (g) Protein amount of p85 in the DKD cells. (h) Nuclear and total protein levels of XBP-1s in DKD cells that were infected with Ad-XBP-1s and stimulated with insulin for 10 and 15 min. (i) XBP-1 splicing assay in PLKO and DKD cells that were stimulated with tunicamycin (0.75 μg ml–1) for the indicated time periods. (j) Nuclear and total XBP-1s protein amount in PLKO and DKD cells after tunicamycin (2 μg ml–1) stimulation. Each experiment was independently repeated three times.
We next wanted to determine whether insulin, one of the central regulators of PI3K activity, modulates the interaction between p85α and p85β. To address this question, we infected MEFs with low doses of Ad-p85α-Flag and Ad-p85β-HA, starved them overnight and then stimulated them with insulin for various lengths of time. Subsequently, we pulled down p85α by immunoprecipitation of Flag and immunoblotted for HA to analyze whether the association between p85α and p85β was affected by insulin treatment. Exposure of cells to insulin dissociated these two proteins in a time-dependent manner (Fig. 3d). Thus, our results have identified a previously unknown role for insulin signaling in modulating the interaction between the two isoforms of p85. Furthermore, chemical inhibition of p38 mitogen-activated protein kinase, c-Jun amino terminal kinase or PI3K did not block insulin-induced dissociation of p85α and p85β (Supplementary Fig. 5).
Insulin increases nuclear translocation of XBP-1s
Considering that p85α and p85β each interact with XBP-1s and that insulin stimulation dissociates p85α from p85β, we then asked whether insulin stimulation could increase the binding of p85α and p85β to XBP-1s and consequently increase the latter’s nuclear translocation. Thus, we infected MEF cells with Ad-XBP-1s and exposed them to insulin for 5 or 10 min after 14 h of serum deprivation. Insulin stimulation markedly increased the translocation of XBP-1s to the nucleus without altering its total protein levels (Fig. 3e). These results define a previously undescribed insulin receptor signaling pathway.
We next wanted to determine whether reductions in p85α and p85β inhibits insulin-mediated nuclear translocation of XBP-1s and whether deficiency of p85α and p85β increases the vulnerability of cells to ER stress. We knocked down p85α and p85β in MEFs with an shRNA lentivirus system specific for p85α and p85β mRNA (Supplementary Fig. 6) to create double-knockdown cells (DKD) and used an empty virus (pLKO) to create a control cell line, PLKO. We analyzed gene expression and protein abundance of p85α and p85β in the DKD and PLKO cell lines. mRNA and protein levels for p85 were markedly reduced in the DKD cells, relative to those in the PLKO controls (Fig. 3f,g). After establishing the stable cell lines, we infected the DKD cells with Ad-XBP-1s and examined whether insulin was still capable of increasing the nuclear transport of XBP-1s. Insulin stimulation did not increase nuclear translocation of XBP-1s in -double-knockdown cells (Fig. 3h), indicating that p85α and p85β have a key role in insulin-stimulated nuclear translocation of XBP-1s. We also investigated the responses of DKD and PLKO cells to ER stress. After overnight starvation, we subjected cells to tunicamycin (0.75 μg ml–1) treatment and analyzed the splicing of XBP-1. XBP-1s mRNA began to form after 2 h of tunicamycin stimulation and reached maximal levels at 3 h in the PLKO cells (Fig. 3i). However, the splicing of XBP-1 was initiated in the DKD cells much more rapidly, starting at 30 min after exposure to the same dose of tunicamycin (Fig. 3i).
To investigate whether these results were indeed due to a defect in XBP-1s translocation to the nucleus, we stimulated the PLKO and DKD cells with 2 μg ml–1 of tunicamycin for 1, 2 and 3 h and isolated nuclear fractions to analyze the XBP-1s levels. Stimulation of PLKO cells with tunicamycin led to a substantial increase in migration of XBP-1s to the nucleus at the 2-h and 3-h time points (Fig. 3j). However, tunicamycin-stimulated XBP-1s translocation to the nucleus was severely retarded in the DKD cells, indicating a crucial role of p85α and p85β in this process (Fig. 3j). There was also a reduction in XBP-1s protein amounts in whole-cell lysates of the DKD cells; however, this reduction was not comparable to the reduction in nuclear translocation of XBP-1s in DKD cells (Fig. 3j).
Defective XBP-1s action and chaperone response in obesity
Under normal circumstances, the spliced form of XBP-1 is present at extremely low levels in tissues, including in the liver. Physiologic ER stress can be created in the liver by feeding the mice after a fasting period32; this process not only creates ER stress but also activates insulin receptor signaling, a condition that is ideal for testing whether XBP-1s and p85 associate in vivo and whether this interaction is defective in a genetic model of obesity.
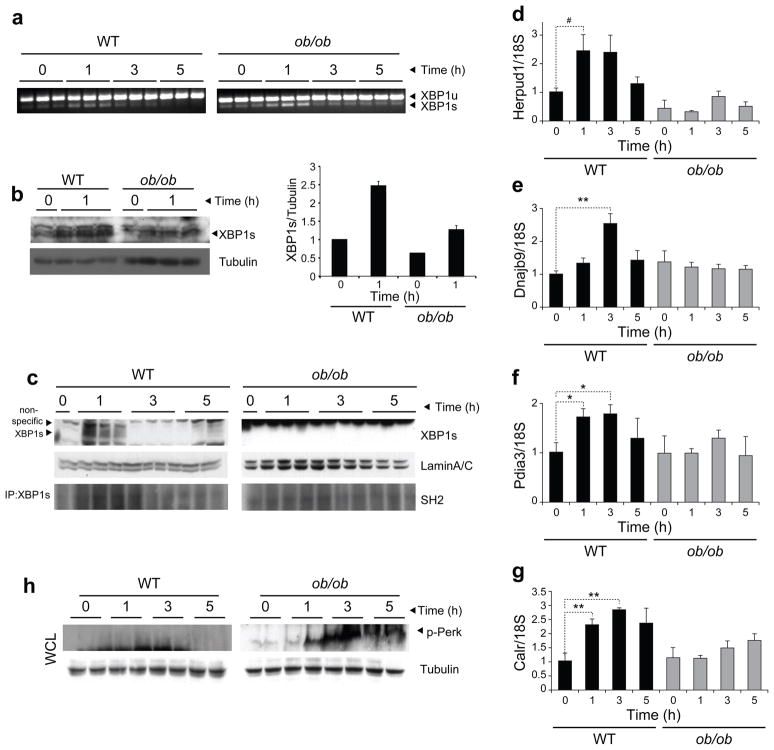
For this reason, we first fasted wild-type (WT) and ob/ob mice for 24 h and then gave them food ad libitum. We collected the liver after fasting and 1, 3 and 5 h after refeeding and then determined the levels of XBP-1s mRNA. Refeeding markedly increased the XBP-1 splicing in the WT mice (Fig. 4a), suggesting that the metabolic load created by refeeding led to the development of ER stress. We obtained similar results in the ob/ob mice (Fig. 4a). The level of XBP-1 splicing was comparable between WT and ob/ob mice at 1 h after refeeding (Fig. 4a). In the WT mice, XBP-1s mRNA levels declined at 3 h and almost completely disappeared after 5 h of refeeding (Fig. 4a). Despite the fact that XBP-1s mRNA levels also declined at the 3-h time point in the liver of ob/ob mice, the presence of considerably higher mRNA levels of XBP-1s 5 h after refeeding indicated that, in the fed state, the obese mice could not resolve the ER stress as well as the WT mice did (Fig. 4a). We also measured the amount of XBP-1s protein in total liver lysates and detected a clear induction of XBP-1s protein expression in WT mice after refeeding (Fig. 4b). We also observed induction of XBP-1s protein expression in ob/ob mice, but to a lower extent relative to the WT mice (Fig. 4b). However, the increased XBP-1s protein amounts from fasting to the refed state were comparable in WT and ob/ob mice (Fig. 4b).
Figure 4.
XBP-1s import to the nucleus is impaired in the ob/ob mice. (a) XBP-1 mRNA splicing in the WT and ob/ob liver during refeeding. (b) XBP-1s protein abundance in total liver lysates. The graph next to the blots depicts the XBP-1s/tubulin ratio at fasting and after 1 h of refeeding. (c) Nuclear XBP-1s protein amounts in WT and ob/ob liver during refeeding (top). Lysates were subjected to immunoprecipitation with XBP-1s–specific antibody and immunoblotted with SH2-specific antibody (bottom). (d–g) Gene expression level of Herpud1 (d), Dnajb9 (e), Pdia3 (f) and Calr (g) at fasting and during refeeding. (h) PERK phosphorylation on Thr980 in the liver of WT and ob/ob mice during refeeding. Experiments were repeated in three independent cohorts. Error bars are means ± s.e.m.; P values were determined by Student’s t test (#P = 0.6, *P < 0.05, **P < 0.01, ***P < 0.001).
We also analyzed the protein expression of XBP-1s in isolated nuclear fractions of the liver in the fasting state and at several time points after refeeding. In parallel with the levels of XBP-1s mRNA and total protein, nuclear protein amounts of XBP-1s were increased in the nuclear fractions of the liver of WT mice at 1 h after feeding and were undetectable at 3 and 5 h (Fig. 4c). In contrast, we did not detect any XBP-1s protein in nuclear extracts of the liver of ob/ob mice in three independent experiments (Fig. 4c), even under conditions in which ob/ob samples were loaded at three times higher amounts than WT (data not shown), indicating a severe defect in translocation of XBP-1s to the nucleus in the ob/ob liver.
Next, we performed XBP-1s immunoprecipitation to investigate whether p85 immunoprecipitates with XBP-1s during refeeding conditions. We found that p85 immunoprecipitated with XBP-1s 1 h after refeeding in the WT mice (Fig. 4c), indicating that these two proteins also interact under in vivo conditions. This interaction was markedly reduced in the ob/ob mice (Fig. 4c). Furthermore, when we analyzed gene expression patterns, expression of chaperones such as Herpud1, Dnajb9, Pdia3 and Calr (encoding calreticulin) was considerably upregulated in the WT liver, whereas this chaperone response was totally blunted in the ob/ob mice (Fig. 4d–g).
These results could be due to two different reasons; first, ob/ob mice may have a high-capacity ER that can handle a refeeding-induced metabolic overload without developing substantial ER stress and requiring additional chaperones and UPR activation. Second, despite the fact that ER stress is increased and UPR signaling is activated in ob/ob mice, they may be defective in responding to the increased load owing to the reduced XBP-1s translocation to the nucleus, thus being unable to upregulate the chaperones.
If the lack of ER stress (the first possibility) in the liver of the ob/ob mice during refeeding were the case, PERK signaling would not be triggered, as ER stress would have already been resolved without much upregulation of chaperones. However, with the second possibility, a marked increase in PERK phosphorylation would be expected after refeeding as a result of the lack of a chaperone response and the consequent unresolved ER stress. We therefore analyzed the PERK phosphorylation in WT and ob/ob liver during fasting and refeeding conditions. There was a slight increase in PERK phosphorylation after 1 and 3 h of refeeding in the liver of the WT mice (Fig. 4h); this phosphorylation was rapidly eliminated, leaving no detectable phosphorylated PERK, at 5 h after refeeding (Fig. 4h). These data indicate that the chaperone response was, indeed, enough to block severe activation of UPR in the WT mice. However, refeeding led to marked PERK phosphorylation in ob/ob mouse liver, which was not abated even 5 h after refeeding (Fig. 4h).
p85α or p85β restore glucose homeostasis in ob/ob mice
The lack of p85 binding to XBP-1s, the defective translocation of XBP-1 to the nucleus, the diminished chaperone response and the consequent marked PERK phosphorylation in the ob/ob mice during refeeding suggest that the interaction between p85 and XBP-1s is crucial in the regulation of metabolic homeostasis in the liver. Furthermore, if these postulates are true and if the lack of p85–XBP-1s interaction in the liver of ob/ob mice is essential for downregulation of XBP-1s activity and consequently for development of ER stress, glucose intolerance and type 2 diabetes, then increasing the free levels of p85α or p85β in the liver of ob/ob mice should reinstate the ability of XBP-1s to translocate to the nucleus and, consequently, reduce ER stress and increase glucose tolerance.
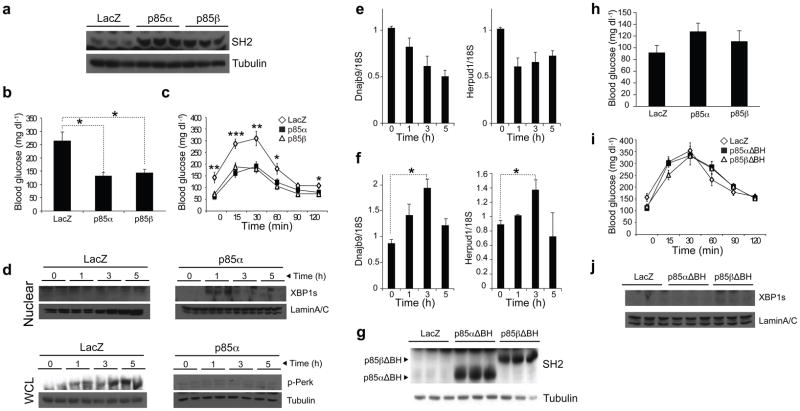
To test this hypothesis, we overexpressed p85α or p85β in the liver of ob/ob mice by tail vein injection of Ad-p85α or Ad-p85β and Ad-LacZ as a control (Fig. 5a). After 4 d of injection, we fasted the mice for 6 h and measured blood glucose concentrations. Blood glucose concentrations of the Ad-p85α– or Ad-p85β–injected group were at euglycemia levels and were dramatically lower than those in the Ad-LacZ– injected group (Fig. 5b). Glucose tolerance test (GTT) at day 6 revealed a major improvement in the glucose disposal rate in the p85α– or p85β–overexpressing groups (Fig. 5c).
Figure 5.
Overexpression of p85α and p85β in the liver of ob/ob mice increases glucose tolerance and establishes euglycemia. (a–f) Eight-week-old, male ob/ob mice were injected with Ad-LacZ (2.5 · 109 plaque-forming units (PFU) per mouse) (n = 7) or Ad-p85α (3 · 109 PFU per mouse) (n = 7) or Ad-p85β (2.5 · 109 PFU per mouse) (n = 7) through the tail vein. (a) Liver p85 abundance on day 8 after the injections. (b) Blood glucose concentrations (mg dl–1) after 6-h fasting on day 4 of the injections. (c) GTT on day 6 after injection. (d) XBP-1s nuclear protein amounts and PERK phosphorylation on Thr980 at fasting, and 1, 3 and 5 h after refeeding. (e,f) mRNA levels of Dnajb9 and Herpud1 in the liver of Ad-LacZ–injected (e) or Ad-p85α–injected (f) ob/ob mice during refeeding. (g–j) Eight-week-old, male ob/ob mice were injected with Ad-LacZ (2.5 · 109 PFU per mouse) (n = 6), Ad-p85αΔBH (2.5 · 109 PFU per mouse) (n = 6) or Ad-p85βΔBH (2.5 · 109 PFU per mouse) (n = 6) through the tail vein. (g) Western blotting for SH2 at day 8 after injection. (h) Blood glucose concentration (mg dl–1) after 6-h fasting on day 4 of the injections. (i) GTT on day 6 after injection. (j) XBP-1s nuclear protein amounts and Lamin A/C control levels in the liver at fasting and 1, 3 and 5 h after refeeding. Each experiment was repeated independently three times. Error bars are means ± s.e.m.; P values were determined by Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001).
In another experiment, we overexpressed p85α in the liver of ob/ob mice by tail vein injection of Ad-p85α and then subjected the mice to the fasting and refeeding regimen starting on day 6 after injection. The overexpression of p85α restored the ability of XBP-1s to migrate to the nucleus (Fig. 5d) and, in fact, blocked development of ER stress during refeeding; PERK phosphorylation on Thr980 was markedly blocked during refeeding in the ob/ob mice (Fig. 5d). In parallel to these observations, refeeding-induced chaperone upregulation was also significantly increased (Fig. 5e,f).
We next overexpressed p85αΔBH or p85βΔBH in the liver of ob/ob mice by tail vein injection of Ad-p85αΔBH or Ad-p85βΔBH and Ad-LacZ as a control (Fig. 5g). We measured the blood glucose concentrations at the 6-h fasted state at day 4 after injection; there was a slight increase in the blood glucose concentrations of the p85αΔBH– and p85βΔBH–overexpressing groups, but these alterations were not significant (Fig. 5h). GTT at day 6 after injection showed that glucose disposal curves after glucose injection were not different among the groups (Fig. 5i). These results show that the BH domain is necessary for p85 to act on XBP-1s. Indeed, migration of XBP-1s was still defective in the Ad-p85αΔBH– or Ad-p85βΔBH– injected ob/ob mice (Fig. 5j).
Defective XBP-1s translocation in Pik3r1−/−;Pik3r2−/− mice
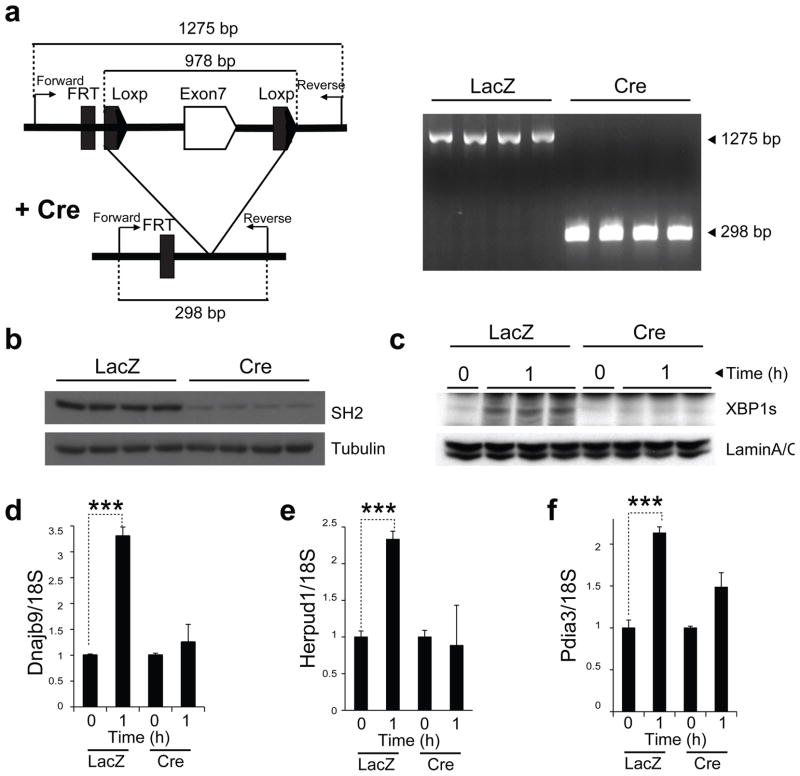
We also sought to investigate whether depletion of p85α and p85β in the liver of lean mice influences fasting and refeeding–induced translocation of XBP-1s to the nucleus. For this purpose, we used the Pik3r1f/f;Pik3r2−/− mouse model. At 7 weeks of age, we split male Pik3r1f/f;Pik3r2−/− mice into two groups. The first group was injected with Ad-LacZ and the second was injected with Cre recombinase–expressing adenovirus (Ad-Cre) through the tail vein. To determine whether Cre-mediated recombination was complete, we isolated DNA from the liver of the Ad-LacZ– and Ad-Cre–injected mice. Expression of Cre led to complete recombination of the Pik3r1 allele in the liver of Pik3r1f/f;Pik3r2−/− mice (Fig. 6a). Analysis of protein amounts in the liver samples of Pik3r1f/f;Pik3r2−/− mice injected with Ad-Cre revealed a marked reduction in the amount of p85 protein (Fig. 6b). We starved both groups for 24 h at 14 d after injection and then refed them for 1 h. Refeeding-induced XBP-1s translocation to the nucleus was lost in the Cre-injected group (Fig. 6c). In parallel to the blunted XBP-1s migration to the nucleus, chaperone response in the liver of Ad-Cre–injected Pik3r1f/f;Pik3r2−/− mice was significantly reduced when compared with the Ad-LacZ– injected group (Fig. 6d–f).
Figure 6.
Nuclear translocation of XBP-1s is impaired in the liver-specific Pik3r1−/−;Pik3r2−/− mice. Seven-week-old, male Pik3r1f/f;Pik3r2−/− mice were injected with Ad-LacZ (n = 6) (7.5 · 109 PFU per mouse) and Ad-Cre (n = 6) (7.5 · 109 PFU per mouse) via tail vein. (a) Schematic characterization of Cre-mediated recombination of Pik3r1 allele (left) and PCR products after recombination (right). (b) p85 protein amounts at day 15 after injection. (c) Nuclear protein amounts of XBP-1s at fasting and 1 h after refeeding. (d–f) mRNA levels of Dnajb9 (d), Herpud1 (e) and Pdia3 (f) at fasting and after 1 h refeeding. Experiments were repeated in three independent cohorts. Error bars are means ± s.e.m.; P values were determined by Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001).
Taken together, our results indicate that p85α and p85β are crucial partners of XBP-1s and mediate translocation of XBP-1s to the nucleus.
DISCUSSION
XBP-1 is a b-ZIP transcription factor that belongs to the ATF–cAMP response element–binding protein family of transcription factors. Together with ATF6, it orchestrates the UPR in ER stress conditions2,3. To date, understanding of the factors that modulate the activity of XBP-1 or its binding partners is fragmentary. Our results here have identified a previously unknown function of p85α and p85β and have shown that, in addition to being regulatory subunits for p110, p85α and p85β also function as regulatory subunits for XBP-1s. We showed that both p85α and p85β bind XBP-1 and increase its nuclear translocation. The absence of p110 in the XBP-1–p85 complex strongly supports the notion that regulation of XBP-1s activity by p85α or p85β is not related to the known function of class I PI3K. Furthermore, treatment with either a P13K inhibitor (LY-204002) or a P13K-mammalian target of rapamycin inhibitor (BEZ235) led to an increase in XBP-1s translocation in MEFs that were infected with Ad-XBP-1s (data not shown), which indicates that increased p110 catalytic activity cannot be responsible for p85-mediated nuclear translocation of XBP-1s.
In addition, by demonstrating that the interaction between p85 and XBP-1 is stimulated by insulin signaling, and that insulin increases the nuclear concentration of XBP-1s by disrupting the heterodimerization of p85α and p85β and thus promoting their association with XBP-1s, we have not only uncovered a previously unknown interaction between p85 and XBP-1s but also identified a new pathway for signaling via the insulin receptor (Supplementary Fig. 7).
More noteworthy, we showed that this mechanism is defective in obese and insulin-resistant ob/ob mice, which are unresponsive to ER stress during refeeding as marked by reductions in XBP-1s nuclear translocation and consequent chaperone upregulation. Although ER stress was resolved in WT mice without much PERK phosphorylation during refeeding, obese mice developed severe UPR, which was not resolved at the postprandial states owing to their inability to activate the necessary XBP-1s–derived chaperone machinery. Our experiments have led to an unexpected discovery that when the amount of p85α or p85β is increased in the liver of obese and diabetic ob/ob mice, glucose tolerance is substantially increased and blood glucose concentrations are reduced to euglycemia. Furthermore, in p85-overexpressing liver XBP-1s, translocation to the nucleus was reinstated, the chaperone response was restored and PERK phosphorylation during refeeding was greatly reduced. The lack of these responses after overexpression of BH-mutant p85α, which cannot bind XBP-1s, strongly indicates that the interaction between XBP-1s and p85α is crucial for the observed effects of p85 in ob/ob mice. These results indicate that increasing the availability of free p85 in obese and diabetic states could provide a new therapeutic strategy for treatment of type 2 diabetes.
In complete agreement with the gain-of-function experiments, silencing of p85α and p85β expression led to severe defects in tunicamycin-induced XBP-1s translocation to the nucleus and increased the vulnerability of these cells to developing ER stress. Furthermore, depletion of p85α in the liver of Pik3r1f/f;Pik3r2−/− mice with Ad-Cre infections completely blocked XBP-1s translocation to the nucleus during fasting and refeeding experiments. These results show that p85α and p85β are prerequisites for XBP-1s to migrate to the nucleus during metabolic overloading.
What could be the advantage of this mechanism? Insulin levels increase sufficiently to activate a potent downstream intracellular signaling response when organisms are exposed to an excess of nutrient availability. Activation of insulin receptor signaling increases protein synthesis and nutrient storage and promotes differentiation and growth20,21,23. This metabolic processes can lead to ER stress. Phosphorylation of PERK during UPR reduces global translation initiation to reduce the load of protein folding in the ER and resolve stress6,7. However, under conditions where more protein synthesis is required, such as during insulin and other growth factor signaling, early activation of PERK can lead to premature cessation of protein synthesis, and increased IRE1 kinase activity can block insulin receptor signaling. At this point, we presume that active insulin receptor signaling increases the efficiency of XBP-1s by promoting its association with p85, leading to faster and higher upregulation of the chaperones. This mechanism could be highly beneficial for the cell to handle the increasing load in the ER, without activating the three arms of the UPR that would otherwise block protein synthesis and also inhibit IRS activity.
This delicate crosstalk between the PI3K system and the UPR might have evolved to ensure the utilization of nutrients at the highest level of their availability. The UPR is a key part of the cellular antiviral armamentarium33. During viral infections, especially infections by enveloped viruses, there is a heavy demand for membrane proteins and lipids for morphogenesis. The cell uses UPR activation to reduce protein synthesis via PERK phosphorylation. This is an antidefense mechanism for the cell, as a reduction in protein synthesis limits the replication of the invading viruses33. It is possible that signaling systems that activate PI3K act as a signal for the cell to differentiate the ER stress caused by metabolic processes from other ER stress–causing pathological conditions and help the cell to continue with the required metabolic machinery.
Indeed, in WT mice, where insulin receptor signaling is intact, p85 binds XBP-1s and increases its nuclear translocation. Concomitant with XBP-1s transport to the nucleus, chaperones are upregulated and ER stress is resolved without considerable activation of PERK. However, in an insulin-resistant condition, such as in the ob/ob mice, refeeding and metabolic overload cannot induce association of p85 with XBP-1s, and there is a severe defect in XBP-1s translocation to the nucleus. Consequently, chaperones are not upregulated and the ER cannot cope with the increasing demand, which, in turn, leads to severe PERK phosphorylation and probably premature cessation of protein synthesis and augmentation of insulin resistance through the kinase activity of IRE1, which might contribute to the development of postprandial hyperglycemia and insulin resistance.
The mechanism presented in our current work shows that the intact response of p85 to insulin receptor signaling (that is, the loss of heterodimerization by p85α and p85β and their subsequent interaction with XBP-1s) is necessary for efficient functioning of the ER and for the necessary chaperone response during metabolic overload. Interference with this insulin-induced pathway may lead to a vicious cycle that accelerates the progression of insulin resistance to a more severe degree, ultimately leading to type 2 diabetes. More notably, our results further indicate that circumventing this vicious cycle by increasing the free amount of either isoform of p85 might provide a therapeutic approach for treatment of type 2 diabetes.
METHODS
Dithiobis[succinimidyl propionate] crosslinking and immunoprecipitation
We dissolved DSP in DMSO to a final concentration of 250 mg ml–1. We washed cells with ice-cold PBS and incubated with 5 ml of PBS containing 1 mg ml–1 DSP for 7 min at 25 °C. We quenched the reaction by addition of 500 μl of 1 M Tris (pH 8.5) for 1 min at 25 °C. We washed the cells with ice-cold PBS, and lysed them with 750 μl RIPA buffer (50 mM Tris-HCl (pH 7.5), 2 mM EGTA, 0.3% CHAPS, 100 mM NaF, 10 mM sodium pyrophosphate, 10 μg ml–1 leupeptin, 10 μg ml–1 aproptonin, 2 mM PMSF and 20 nM okadaic acid). We purchased CHAPS and okadaic acid from CalBiochem, and we purchased all other reagents from Sigma-Aldrich. We incubated cell lysates with antibody (0.3–0.6 μg) overnight at 4 °C with gentle rotation. We added protein A– or protein G–Sepharose beads (80 μl or in a 1 in 5 dilution) (GE Healthcare Bioscience) to the tubes and rotated at 4 °C for 2 h. We precipitated beads by centrifugation at 16,000g for 30 s and washed them three times with cold RIPA buffer containing 150 mM NaCl. We resuspended the pellets with beads in 2× Laemmli buffer and incubated the solutions at 100 °C for 5 min.
XBP-1s-Flag immunoprecipitation and silver staining
To identify the XBP-1s–associating proteins, we cultured MEFs in 150-mm cell culture dishes and infected them with Ad-XBP-1s. After 16 h of incubation in medium containing 1% FBS, we crosslinked cells with DSP as described above and lysed them with RIPA buffer without NaCl. We washed Flag M2 affinity resins (Sigma-Aldrich) three times with RIPA buffer. We then added 50 μl of a 50% slurry of the resins to cell lysates that had previously been cleared and incubated with rotation for 6 h at 4 °C. Finally, we washed the beads three times with lysis buffer containing 150 mM NaCl. To elute Flag-tagged proteins, we incubated beads in elution buffer (50 mM HEPES (pH 7.4), 500 mM NaCl, 0.5% CHAPS and 50 μg μl–1 Flag peptide) for 30 min at 30 °C. We combined the eluates from five dishes and loaded them in a single well of a 16 × 28 cm SDS-PAGE gel (8%). We performed silver staining to the manufacturer’s instructions (Bio-Rad).
Analysis of dissociation of p85α and p85β after insulin stimulation
We optimized the doses of p85α-Flag– and p85β-HA– expressing adenoviruses to a dosage at which association and dissociation of p85α and p85β could be detected. Low amounts of exogenous p85α and p85β were necessary to see the insulin effect on the p85α-p85β complex. We infected cells with adenoviruses in 1% FBS containing medium for 16 h and then treated them with insulin (500 nM) for 10, 30 and 45 min. We gently rocked the dish every 3 min during the treatment to evenly distribute the insulin. After the stimulation, we washed the cells with ice-cold PBS and crosslinked proteins with DSP as described above. We pulled down p85α-Flag with 1 μl of Flag-specific antibody (Sigma-Aldrich) at 4 °C overnight. We added protein G–Sepharose beads to the lysate-antibody mixture and further rotated the mixture for 2 h. Subsequently, we washed the beads three times with cold RIPA buffer containing 150 mM NaCl. After resolving the proteins in SDS-PAGE, we immunoblotted with HA-specific antibody (Santa Cruz Biotechnology).
Lysis of liver tissue
For direct immunoblotting, we homogenized liver tissue (200 mg) in 4 ml of ice-cold tissue lysis buffer (25 mM Tris-HCl (pH 7.4), 10 mM Na3VO4, 100 mM NaF, 50 mM Na4P2O7, 10 mM EGTA, 10 mM EDTA, 1% NP-40, 10 μg ml–1 leupeptin, 10 μg ml–1 aproptonin, 2 mM PMSF and 20 nM okadaic acid) in 50 ml round-bottom tubes. We centrifuged samples at 8,000g for 20 min at 4 °C and transferred the supernatants into tubes. We centrifuged the supernatants for 30 min at 16,000g. We removed the lipid layer, transferred the lysates into new tubes and centrifuged for 30 min at 16,000g at 4 °C. We denatured the protein in 1× Laemmli buffer by boiling at 100 °C for 5 min. For XBP-1s–p85 coimmunoprecipitation, we homogenized liver tissue (200 mg) in 4 ml of coimmunoprecipitation lysis buffer (40 mM HEPES pH 7.4, 2 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 0.3% CHAPS, 10 μg ml–1 leupeptin, 10 μg ml–1 aproptonin, 2 mM PMSF and 20 nM okadaic acid) in 50-ml round-bottom tubes. After clearing the lysate with centrifugation as described above, we added primary antibody to the lysate and incubated overnight at 4 °C with gentle rotation. The next day, we added 40 μl of protein A–Sepharose CL–4B beads (GE Healthcare) to the lysate-antibody mixture and rotated for 2 h. Subsequently, we washed the beads three times with lysis buffer containing 150 mM NaCl. We denatured the immunoprecipitated by addition of 30 μl of 2× Laemmli buffer and boiling for 5 min.
Statistical analyses
We used Student’s t test to determine P values for statistical significance. Error bars represent the s.e.m. (*P < 0.05, **P < 0.01, ***P < 0.001).
Supplementary Material
Acknowledgments
We thank members of the Ozcan laboratory for their contributions to this project. We are grateful to L. Cantley (Harvard Medical School) for kindly providing us with the Pik3r1f/f;Pik3r2−/−mice. We would like to thank the Children’s Hospital Boston Proteomics Core Facility and H. Steen for the tandem mass spectroscopy analysis. This study was supported by junior faculty start-up funds provided to U.O. by Children’s Hospital Boston, Translational Research Award, an RO1 grant (R01DK081009) provided to U.O., and Timothy Murphy funds provided to Division of Endocrinology, Children’s Hospital Boston.
Footnotes
Author Contributions
S.W.P. came up with the hypothesis, designed and performed the experiments, analyzed the data and wrote the manuscript. Y.Z., J.L., A.L., C.S. and J.C. performed the experiments. K.U. came up with the hypothesis and provided reagents. U.O. came up with the hypothesis, designed and performed the experiments, analyzed the data and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 2.Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 3.Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 4.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 5.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum–resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 7.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 8.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 9.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 10.Urano F, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 11.Nishitoh H, et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 13.Lee K, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 15.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sriburi R, et al. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)–induced endoplasmic reticulum biogenesis. J Biol Chem. 2007;282:7024–7034. doi: 10.1074/jbc.M609490200. [DOI] [PubMed] [Google Scholar]
- 17.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozcan U, et al. Endoplasmic reticulum stress links obesity, insulin action and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 19.Ozcan L, et al. Endoplasmic reticulum stress plays a central role in development of leptin resistance. Cell Metab. 2009;9:35–51. doi: 10.1016/j.cmet.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Saltiel AR, Pessin JE. Insulin signaling pathways in time and space. Trends Cell Biol. 2002;12:65–71. doi: 10.1016/s0962-8924(01)02207-3. [DOI] [PubMed] [Google Scholar]
- 21.Nandi A, Kitamura Y, Kahn CR, Accili D. Mouse models of insulin resistance. Physiol Rev. 2004;84:623–647. doi: 10.1152/physrev.00032.2003. [DOI] [PubMed] [Google Scholar]
- 22.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 23.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 24.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Y, Bagrodia S, Cerione RA. Activation of phosphoinositide 3-kinase activity by Cdc42Hs binding to p85. J Biol Chem. 1994;269:18727–18730. [PubMed] [Google Scholar]
- 26.Okkenhaug K, Vanhaesebroeck B. New responsibilities for the PI3K regulatory subunit p85α. Sci STKE. 2001;2001:pe1. doi: 10.1126/stke.2001.65.pe1. [DOI] [PubMed] [Google Scholar]
- 27.Furuya F, Ying H, Zhao L, Cheng SY. Novel functions of thyroid hormone receptor mutants: beyond nucleus-initiated transcription. Steroids. 2007;72:171–179. doi: 10.1016/j.steroids.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuboi N, et al. The tyrosine phosphatase CD148 interacts with the p85 regulatory subunit of phosphoinositide 3-kinase. Biochem J. 2008;413:193–200. doi: 10.1042/BJ20071317. [DOI] [PubMed] [Google Scholar]
- 29.Déléris P, Gayral S, Breton-Douillon M. Nuclear Ptdlns(3,4,5)P3 signaling: an ongoing story. J Cell Biochem. 2006;98:469–485. doi: 10.1002/jcb.20695. [DOI] [PubMed] [Google Scholar]
- 30.Sephton CF, Mousseau DD. Dephosphorylation of Akt in C6 cells grown in serum-free conditions corresponds with redistribution of p85/PI3K to the nucleus. J Neurosci Res. 2008;86:675–682. doi: 10.1002/jnr.21516. [DOI] [PubMed] [Google Scholar]
- 31.Dai Y, et al. Haloperidol induces the nuclear translocation of phosphatidylinositol 3′-kinase to disrupt Akt phosphorylation in PC12 cells. J Psychiatry Neurosci. 2007;32:323–330. [PMC free article] [PubMed] [Google Scholar]
- 32.Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2α enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin JH, Walter P, Yen TS. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.