Abstract
Sirtuins function with other biogenic molecules to promote adaptation to caloric restriction in a broad spectrum of eukaryotic species. Sirtuin pathways also converge in the mammalian brain where they appear to protect neurons from nutrient stress. However, few anatomical studies on sirtuins (e.g., SIRT1) are available, particularly those detailing the spatial distribution and sub-cellular localization pattern of SIRT1 in the brain parenchyma. Here we report the characterization of a panel of SIRT1-specific antibodies within rodent (i.e., rat and mouse) and human central nervous systems. Immunocytochemical and Western blot analyses indicate that the sub-cellular localization of SIRT1 is predominantly nuclear throughout the rodent brain and spinal cord. A similar sub-cellular distribution pattern of SIRT1 was detected in human central nervous system material. SIRT1 is ubiquitously present in areas of the brain especially susceptible to age-related neurodegenerative states (e.g., the prefrontal cortex, hippocampus and basal ganglia). Further, we show no apparent species-specific differences in the sub-cellular localization pattern of rodent versus human SIRT1. Finally, we identify the chemical phenotype of SIRT1-containing neurons in a number of brain sites that are strongly compromised by aging. These data provide additional and important anatomical findings for the role of SIRT1 in the mammalian brain, and suggest that SIRT1 pathways are broadly distributed in neurons most susceptible to senescence injury. Activating endogenous sirtuin pathways may therefore offer a therapeutic approach to delay and/or treat human age-related diseases.
Keywords: Basal Ganglia, Hippocampus, Parvalbumin, Prefrontal cortex, Tyrosine hydroxylase
INTRODUCTION
Sirtuins are histone nicotinamide adenosine dinucleotide (NAD)-dependent deacetylases widely recognized for their link to eukaryotic life span (Guarente and Picard, 2005; Michan and Sinclair, 2007). Silent information regulator 1 (SIRT1), the most studied among the seven mammalian sirtuins, increases when situations favor longevity, such as caloric restriction or following treatment with the polyphenol resveratrol. SIRT1 resides primarily in the cell nucleus where it targets several non-histone transcriptional regulators including p53, heat shock factor 1 (HSF1), forkhead transcription factor (FOXO), NF-κB, and peroxisome proliferator-activator receptor-γ coactivator-1α (PGC-1α). These factors promote adaptation to caloric restriction by regulating age-related programs of replicative senescence, protein homeostasis, inflammatory function, and energy metabolism (Vaziri et al., 2001; Motta et al., 2004; Yeung et al., 2004; Nemoto et al., 2005; Rodgers et al., 2005; Li et al., 2007).
SIRT1-regulated pathways also operate in the nervous system where they contribute critically to the proliferation and differentiation of neural progenitors through Notch signaling cues (Prozorovski et al., 2008). Further, cell-based and in vivo rodent models of brain pathology indicate that increased SIRT1 gene dosage reduces β-amyloid deposition typically seen in Alzheimer's disease (AD), and protects against cell necrosis induced by a mutant form of superoxide dismutase I in amyotrophic lateral sclerosis (Qin et al., 2006; Kim et al., 2007). These findings raise the possibility that at least in some forms of neurodegenerative states, SIRT1 (or its agonist resveratrol) could be used to therapeutically mitigate disease progression (Gan and Mucke, 2008). If this is indeed the case, we must first expand our knowledge of SIRT1 protein abundance, distribution, and function in the central nervous system (CNS). In this regard, only three studies have reported the distribution pattern of SIRT1 in the mammalian brain using immunocytochemistry (ICC) and in situ hybridization histochemistry methods (Qin et al., 2006; Michan and Sinclair, 2007; Ramadori et al., 2008). However, there is a discrepancy among these reports on the specificity of anti-sera used for ICC as well as discrepancies in the sub-cellular distribution of SIRT1. To clarify some of these experimental issues, we have carried out detailed ICC and Western blot analyses of the biochemical properties of SIRT1 in rat, mouse, and human brain tissue. In addition, we have characterized the phenotype of certain SIRT1-containing neurons, especially those cells of the brain most susceptible to senescence injury (e.g., neurons in the hippocampus, and substantia nigra).
MATERIALS AND METHODS
Animals
Seven adult male Long-Evans rats (225-265 g; Harlan, Indianapolis, IN) and six C57BL/6J adult male mice (35-45 g; Charles Rivers, Washington, MA) were used in this set of studies. Animals were group-housed, 2-3 per cage under a 12 hr light: dark cycle (lights on 0700) and allowed ad libitum access to food and water. All experiments were performed during the lights on period and were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals and with approval from the NYIT/NYCOM IACUC. All efforts were made to minimize animal stress and to reduce the number of rodents used for the studies detailed below.
Rodent Brain and Spinal Cord Material
To obtain CNS tissue, rats and mice were injected (IP) with a lethal dose of sodium pentobarbital (50 mg/kg; a dose that euthanizes the animal within minutes) and left undisturbed at room temperature (~23 °C) for 4-5 hr. After this postmortem delay, brains and spinal cords were removed and immediately placed in a fixative buffer (4% paraformaldehyde in sodium phosphate buffer, pH 7.2) for 5 days at 4 °C and then stored for an additional five days in 20% sucrose (dissolved in 0.01 M sodium phosphate buffer). Cut brain and spinal cord sections were collected in a cold cryoprotectant solution (0.05 M sodium phosphate buffer, 30% ethylene glycol and 20% glycerol) and stored at −20 °C until prepared for standard ICC protocols.
Human Brain and Spinal Cord Material
Human tissue material used for ICC was obtained through the generosity of the National Neurological Research Specimen Bank (VAMC). Tissue blocks [e.g., frontal lobe of cortex (central sulcus), inferior colliculus and spinal cord] from adult male subjects with no apparent neurological pathology at the time of death (31-41 years of age) and with a temporal postmortem delay (i.e., autolysis) ranging from 5 to 21.5 hr were processed by the same analytical procedures as described for rodent neural material.
Immunocytochemical Procedures
Rodent and human CNS material was frozen on dry ice, mounted on a sliding microtome and cut into 50-μm coronal sections. To unmask target epitopes, free-floating sections were first washed with 0.5 M potassium phosphate buffer saline (KPBS) and then incubated in a sodium citrate buffer solution (10 mM; pH 9.0; Sigma Chemicals, St. Louis, MO) for 30 min in a water-bath at 80 °C (Jiao et al., 1999; Torres et al., 2004). It should be noted that the above incubation procedure greatly reduces non-specific staining (e.g., endogenous peroxidases) associated with non-perfusion, postmortem procedures (Torres et al., 2004). Following the above antigen retrieval phase, brain and spinal cord sections were incubated for 48 hr at 4 °C with monoclonal and polyclonal IgGs raised against SIRT1 (Millipore, Temecula, CA), parvalbumin (PV; ImmunoStar, Inc. WI) and tyrosine hydroxylase (TH; Sigma, St Louis, MO). Following incubation with appropriate secondary antibodies, brain and spinal cord material was washed in KPBS, mounted onto gelatin-chrom-slipped with DPX mountant (Electron Microscopy Sciences, Ft. Washington, PA). To help identify relevant anatomical structures, brain and spinal cord sections were counterstained with Neutral Red. For double-labeling experiments, we followed a previously published procedure by our group (Horowitz et al., 2003), in which SIRT1 was developed first followed by several rinses in KPBS and then incubated with primary PV and TH. All other subsequent steps were the same as those describe above.
Extraction and Preparation of Rodent Brain Tissue and Human Cells
Brain tissues (cerebellum, prefrontal cortex, basal ganglia, hypothalamus, and hippocampus) were dissected and flash-frozen in liquid nitrogen after rats and mice had been sacrificed by decapitation. Tissues were homogenized in low salt lysing buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM MgCl, 5 mM EDTA, 2 mM EGTA, 1% NP40, and 5% deoxycholate) with protein inhibitors (leupeptin, PMSF, aprotinin, and pepstatin A) included in the buffer solution. The extracts were then centrifuged at 10,000 rpm for 10 min and the supernatant measured for protein content. Aliquots were then mixed with equal amounts of loading buffer containing 200 mM Tris, 8% SDS, 0.4% bromophenol blue, 40% glycerol, and 5% 2-mercaptoethanol. Samples were then heated and immediately loaded onto 8% SDS-polyacrylamide gels, electrophoresed, and transferred to nitrocellulose membranes. Human embryonic kidney cells (HEK-293) and human adult proximal tubular kidney cells (HK-2) were purchased from the American Type Culture Collection (ATCC, Mannasas, VA) and grown to confluence in 80 mm2 cell culture flasks under standard cell culture conditions (Dulbecco's Modified Eagle Medium (DMEM), 10% fetal bovine serum, at 37° C, 5% CO2). Cells were washed 3 X in phosphate buffered saline (PBS), gently scraped off the growing surface, and transferred into a pre-chilled 1.5 ml micro-centrifuge tube. Cell suspensions were centrifuged for 5 min at 60 g rpm in a centrifuge pre-cooled at 4°C and the supernatant discarded. Cell pellets were immediately frozen in liquid nitrogen. Nuclear and cytoplasmic extracts were prepared with the NXTRACT CelLytic nuclear extraction kit according to the manufacturer's instructions (Sigma-Aldrich, St. Louis, MO).
Gel Electrophoresis and Western Blotting
For Western blot analysis, 5 μg of total protein from all animal brain tissue and human cells were separated on 4-20% HEPES acrylamide gels (Pierce, Rockford, IL) and transferred to nitrocellulose-supported membranes. Membranes were blocked with 5% milk and incubated with primary antibodies overnight at 4°C. Primary antibodies were used at the following dilutions in 5% milk in Tris-buffered saline with 0.1% Tween-20: Rabbit polyclonal α-SIRT1, (A310-204A; AbVantage Pack; Bethyl Laboratories, Montgomery, Tx) at 1:2000; mouse monoclonal α-beta-tubulin (T0198; Sigma, St. Louis, MO) at 1:1000. After primary antibody incubation, membranes were incubated with a horseradish peroxidase-coupled secondary antibody (HRP-conjugated goat IgG; 31430 or 31460; Pierce, Rockford, IL) for 1 hr at room temperature and processed with the ECL Plus chemiluminescence system (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). Luminescence signals were recorded on clear blue X-ray film (Pierce, Rockford, IL), scanned and evaluated for appropriate signal.
Data Analysis
Total number of positive nuclear or cytoplasmic immunolabeling was counted using the dissector principle as previously described (Torres et al., 2005) on a one to six series of 50 μm coronal sections. Cell and blood vessel distribution was computed from neural tissue observed at magnifications of 10X, 20X, and 40X. Adjacent coronal sections were stained with Neutral Red to enable precise anatomical delineation of the rodent prefrontal cortex, hippocampus, substantia nigra, and spinal cord regions. Images were obtained from an Olympus microscope and then processed with a video-based digital image analysis technique (Torres et al., 2005). Data are reported as means ± SEM. Student's t-tests were performed with the assumption of unequal variance to test for differences between group means. Statistically significant differences were defined as P ≤ 0.05.
RESULTS
Overall distribution pattern of SIRT in the rodent brain
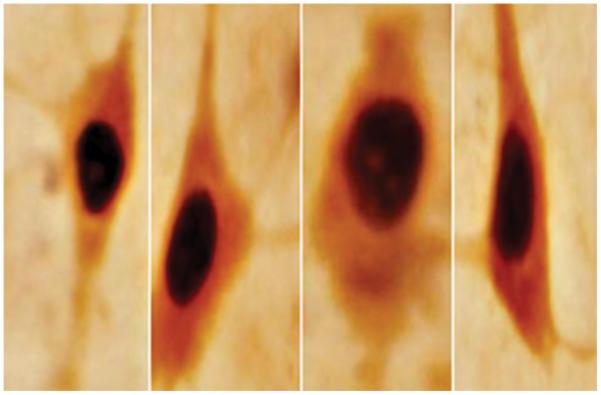
Immunocytochemical analysis of rat and mouse brain using anti-SIRT1 clone 2G1/F7 (mouse monoclonal IgG1, see Fig. 1 for details), showed SIRT1 to be localized throughout the brain parenchyma. In every brain region tested (50 μm thick coronal sections, n = 6/brain region), the sub-cellular distribution of SIRT1-immunoreactivity was predominantly nuclear. No apparent species-dependent differences in nuclear labeling or anatomic distribution were observed between rat and mouse brains, thus indicating species cross-reactivity properties of the anti-SIRT1 2G1/F7 mouse monoclonal IgG1 (Fig. 2). To obtain convergent data regarding validation of sub-cellular distribution and anatomical landscape of (C57BL/6J) mouse SIRT1, we compared our labeling findings to those of the Allen Brain Atlas; www.brainatlas.org. In general, we found that the anatomical distribution of SIRT1 derived with the aforementioned antibody coincided well with the mRNA expression patterns depicted online for the mouse frontal cortex, hippocampus and mid-brain. Further, our SIRT1-immunoreactivity findings complemented those previously reported by Qin et al (2006) and Michan and Sinclair (2007) in mouse frontal cortex, hippocampus and striatum. It should be noted that in our current studies, rat and mouse brains were not fixed with formaldehyde or glutaraldehyde solutions thus minimizing the possibility of epitope modification. Finally, the specificity of our ICC method was determined by negative controls (i.e., replacing the primary SIRT1 antibody with serum). Under these experimental conditions, SIRT1 labeling in the cell nucleus was completely absent throughout the rat and mouse brain parenchyma. Thus, convergent data showing brain regions expressing mRNA encoding the target antigen, sub-cellular localization of the transcription factor in the cell nucleus, and specificity controls for ICC methods, provide confirmation of the anti-SIRT1 2G1/F7 mouse monoclonal IgG1 signal.
Fig. 1.
Schematic diagram of human SIRT1 and specific domains (1-letter abbreviation) used to raise the anti-sera, anti-SIRT1 (2G1/F7) for ICC. aa = amino acids. Amino acid similarities between rodent and human SIRT1 epitopes are shown in red. Both rat and mouse SIRT1 epitopes, share approximately 67% similarity with that of human.
Fig. 2.
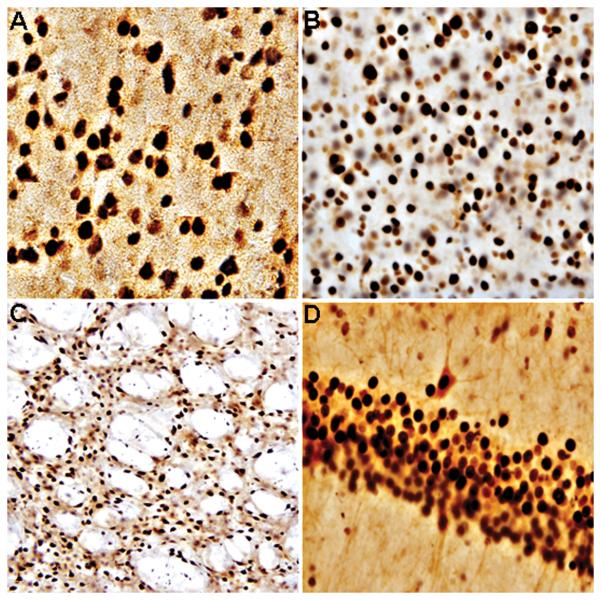
Representative bright-field photomicrographs depicting characteristic SIRT1-immunoreactivity in rodent brain. A = rat; prefrontal cortex. B = mouse; prefrontal cortex. C = rat; striatum. D = rat; hippocampus. SIRT1 was localized to the cell nucleus with intense and diffuse labeling in all cases. A and B = 40X magnification. C and D = 20X magnification.
Specificity of antibodies and Western blot analyses
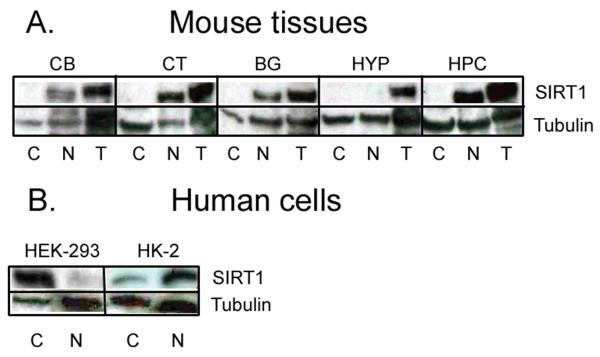
To confirm further the specificity of anti-sera used in these studies, we conducted Western blot analyses using brain extracts from Long-Evans rats and C57BL/6J mice, as well as extracts from human kidney cells (stability of proteins in postmortem human brain tissue was not optimal for Western blots). Surprisingly, the anti-SIRT1 2G1/F7 mouse monoclonal IgG1 failed to detect protein bands with molecular masses corresponding to that of native SIRT1 (data not shown). In sharp contrast, other commercially purchased anti-SIRT1 antibody (anti-SIRT1 A310-204A rabbit polyclonal IgG1) detected protein bands that matched those reported for mammalian SIRT1 in prefrontal cortex, hippocampus, basal ganglia, and cerebellum, but not hypothalamic extracts (Fig. 3A). As previously seen in ICC material, mouse SIRT1 protein bands were predominantly detected in nuclear as opposed to cytoplasmic homogenates. The specificity of anti-SIRT1 A310-204A rabbit polyclonal IgG1 was also attested with human lysates generated from HEK-293 human embryonic kidney cells. This analysis revealed the anti-SIRT1 A310-204A rabbit polyclonal IgG1 successfully detected SIRT1 in lysates of human embryonic kidney (Fig. 3B). However, SIRT1-immunoreactivity was predominantly detected in the cytoplasm as opposed to cell nucleus material. To control for possible developmental differences in SIRT1 expression, we probed adult kidney cells with the same anti-SIRT A310-204A rabbit polyclonal IgG1 antibody. Intriguingly, proximal tubule cells showed a predicted nuclear sub-cellular localization of the sirtuin protein. In general, therefore, the anti-SIRT1 2G1/F7 mouse monoclonal IgG1 was found to be only suitable for ICC, whereas the anti-SIRT1 A310-204A rabbit polyclonal IgG1 was suitable for Western blotting. In addition, we uncovered a developmental divergence in the sub-cellular localization pattern of SIRT1 in human kidney cells.
Fig. 3.
Representative Western blots of SIRT1 in rodent (A) CB = Cerebellum; CT = Prefrontal cortex; BG = Basal ganglia; HYP = Hypothalamus; HPC = Hippocampus. Note the relative absence of SIRT1 in mouse hypothalamus. Western blots of SIRT1 in human embryonic kidney cells (B). C = Cytoplasm; N = Nucleus; T = Total (i.e., both cytoplasmic and nuclear extracts). In human embryonic kidney (HEK-293) cells, SIRT1 is predominantly confined to the cytoplasm. This cytoplasmic expression, however, is reversed to the nucleus in adult human kidney (HK-2) cells. Tubulin was used to verify equal loading of proteins.
Sub-cellular localization of SIRT1 to human CNS
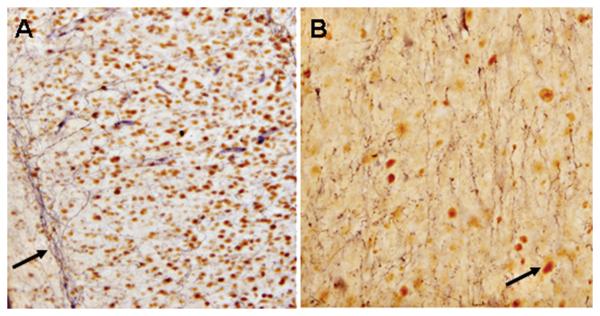
Because neurochemical insights from animal studies are ultimately extrapolated to human brain function, we examined the sub-cellular distribution of SIRT1 to normal human frontal pole cortex (50 μm thick coronal sections, n = 4) and spinal cord (sacral level; 50 μm thick coronal sections, n = 6). This enabled us to characterize and compare the biochemical properties of SIRT1 in the human CNS with those of rat and mouse tissue. Using the same anti-SIRT1 2G1/F7 mouse monoclonal IgG1, we found SIRT1-positive labeling in the adult human frontal pole cortex and spinal cord. The sub-cellular localization of SIRT1 was confined predominantly to the nucleus of as-yet unknown neurons (Fig. 4). Postmortem intervals of up to 21.5 hr did not have a significant effect on SIRT1-immnoreactivity, as shorter postmortem intervals of 5 hr showed a similar labeling profile of the transcription factor (data not shown). In this context, it should be noted that in our current studies, rats and mice were also exposed to a postmortem interval of 4-5 hr, thus resembling some of the agonal states experienced by humans (Torres et al., 1992). Under these postmortem interval conditions, SIRT1-immunoreactivity in the spinal cord was identical in both rodent (sacral level; 50 μm thick coronal sections, n = 6) and human sample material (Fig. 5). In general, then, the anti-SIRT1 2G1/F7 mouse monoclonal IgG1, was able to detect endogenous SIRT1 in the human CNS. As SIRT1 is thought to modulate chromatin structure by deacetylating specific lysine residues in histones H1, H3 and H4 (Michan and Sinclair, 2007), our findings of SIRT1 in the cell nucleus supports function of the target protein and its logical sub-cellular localization within the neuron.
Fig. 4.
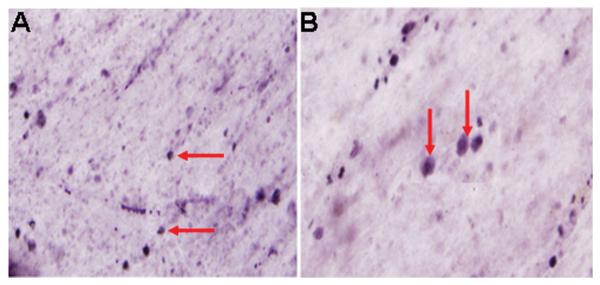
Representative bright-field photomicrographs depicting characteristic SIRT1-immunoreactivity in human brain. Sub-cellular distribution of the NAD-dependent histone deacetylase is predominatly nuclear in frontal pole cortical neurons. Arrows point to cell nuclei labeled positive for SIRT1. A = 20X magnification. B = 40X magnification.
Fig. 5.
Representative bright-field photomicrographs depicting characteristic SIRT1-immunoreactivity in rat (A) and human (B) spinal cord. SIRT1 was localized to the cell nucleus with intense and diffuse labeling in both mammalian samples. Arrows point to cell nuclei labeled positive for SIRT1. A and B = 20X magnification
Identification of SIRT1-containing neurons in the rodent brain
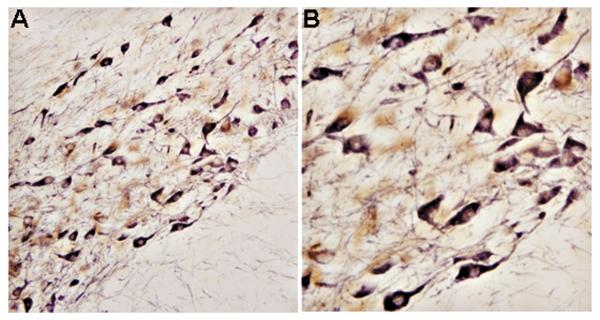
SIRT1 modulates fundamental mechanisms in cell longevity and can alleviate cell pathology when up-regulated. However, the identity or phenotype of most neurons containing SIRT1 is unknown, especially those groups of cells most affected by senescence injury or neurodegenerative states. To this end, we performed double-labeling ICC in the hippocampus, and substantia nigra of rat and mouse sample material. The following specific, well-characterized primary antibodies were used: PV for the hippocampus and TH for the substantia nigra. PV is a Ca2+-binding protein that also serves as a phenotypic marker for GABA-containing neurons in the CNS, whereas TH is the rate-limiting enzyme for dopamine (DA) synthesis. Against this background, we found SIRT1-immunoreactivity confined to hippocampal (CA1) neurons labeled positive for PV (24.5 ± 4.8 mm2; Bregma −3.14 mm; Paxinos and Watson, 1986). As expected, PV was present predominantly in neuronal somata and their processes, especially proximal dendrites (Fig. 6). This double-labeling pattern was also observed in the rodent cerebellum, where nuclear SIRT1 and cytoplasmic PV were co-localized to the same (Purkinje) neuron (data not shown).
Fig. 6.
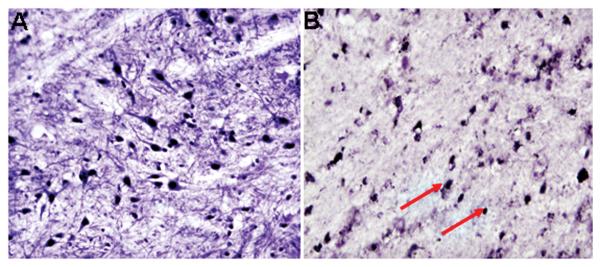
Representative bright-field photomicrographs depicting characteristic SIRT1 and PV double-labeling ICC in rat hippocampus. As predicted, SIRT1 was localized to the cell nucleus, whereas PV was confined to the cytoplasmic compartment. 40X magnification.
Immunocytochemical analysis of the substantia nigra, a region in the mid-brain that is crucial for the initiation of movement, showed relatively high levels of SIRT1 (see above). We next determined whether SIRT1-labeling material corresponded to specific groups of neurons bearing the signature for TH. Indeed, double-labeling ICC in the rodent substantia nigra revealed a significant number of SIRT1-containing neurons to be positive for TH as well (37.2 ± 2.7 mm2; Bregma −5.20 mm; Paxinos and Watson, 1986). There was a strong TH labeling of neuronal somata and their proximal dendrites in the substantia nigra (Fig. 7). These dendritic processes, in turn, were observed to directly terminate in receptive synaptic fields of the fundus striati and nucleus accumbens (Fig. 8). The spatial distribution pattern of TH in the mid-brain corresponded well with that previously reported in the literature (Lee and Price, 2001). TH is a valid phenotypic marker for the neurotransmitter DA, and DA neurons of the mid-brain are progressively lost in Parkinson's disease (PD). Thus, we have confirmed here the anatomical presence of SIRT1 in vulnerable DA neurons of the substantia nigra. A more vexing issue, which begins to encroach on the relevance of this study to PD, is whether SIRT1 in the mid-brain acts to protect DA neurons from pathogenic protein (e.g., α-synuclein) assemblies. Regardless, SIRT1 appears to be well positioned to rescue vulnerable neurons from aging and progressive cell death.
Fig. 7.
Representative bright-field photomicrographs depicting characteristic SIRT1 and TH double-labeling ICC in rat substantia nigra. As predicted, SIRT1 was localized to the cell nucleus, whereas TH was confined to the cytoplasmic compartment. A = 20X magnification. B = 40X magnification.
Fig. 8.
Representative bright-field photomicrographs depicting characteristic SIRT1-immunoreactivity in the cell nucleus and TH-labeling of neuronal fibers terminating within receptive DA fields of the rat fundus striate (A; Bregma 0.70 mm; Paxinos and Watson, 1986) and nucleus accumbens (B; Bregma 0.70 mm; Paxinos and Watson, 1986). Arrow in A points to neuronal fibers, whereas arrow in B points to cell nucleus labeled positive for SIRT1. A and B = 20X magnification.
DISCUSSION
Our results demonstrate that the deacetylase SIRT1 is ubiquitously found across the entire breadth of rodent and presumably human CNS. The distinct sub-cellular localization of SIRT1 was predominantly nuclear with no apparent immunolabeling detected in diffuse cytoplasmic compartments or neuronal processes of various cell types. These microscopic observations are consistent with previous findings of SIRT1 mRNAs and proteins occurring in mature rat and mouse brains (Qin et al., 2006; Michan and Sinclair, 2007; Ramadori et al., 2008). In addition, we report for the first time the sub-cellular localization of SIRT1 protein to human cortical and sacral neurons. Thus, the repetitive cytology across the six layers of the mammalian neocortex contains a significant number of SIRT1-containing neurons where they have been implicated in DNA stability and cell proliferation (Gan and Mucke, 2008). This anatomical analysis also highlights the presence of SIRT1 in all four major divisions of the mammalian spinal cord (i.e., cervical, thoracic, lumbar and sacral columns). At present, however, very little is known about the specific role of SIRT1 or other sirtuins in the spinal cord.
SIRT1-immunoreactivity was also detected in the rodent hippocampus (cell layers of Ammon's horn from CA1 to CA4), basal ganglia circuits (e.g., substantia nigra), brain stem (e.g., raphe nucleus), and cerebellum (e.g., pontine reticular nucleus). Collectively, these results support and extend the hypothesis that SIRT1 proteins are found throughout the adult brain parenchyma where they are likely to deacetylate both histone and non-histone targets. The ubiquitous expression pattern of SIRT1 in rodent brain indicates that most neurons have NAD-dependent histone deacetylases that protect them from various metabolic stressors. We suspect therefore that cell-specific SIRT1 exists to modulate tissue-specific transcriptional consequences to stress. One physical stressor that the mammalian brain must continuously contend with is the progressive aggregation of pathogenic protein assemblies. For instance, α-synucleins, amyloid β-peptides, and polyglutamines accumulate over the course of a life-span and must periodically be removed before they reach threshold levels of toxicity as exemplified in PD, AD and Huntington's disease, respectively (Taylor et al., 2002). The existence of such potentially toxic, aggregation-prone proteins has put considerable evolutionary pressure on the brain for evolving (counter) protective measures that resist aggregated structural states of pathology. Sirtuin pathways fulfill this protective role as they modulate fundamental mechanisms of cell repair, and cell survival (Torres et al., 2008). Consistent with this hypothesis, sirtuin expression is intimately linked to the cell cycle and in regulating metabolism in response to dietary changes (Michan and Sinclair, 2007). Consequently, it is conceivable that by implementing nutritious caloric restriction paradigms during specific cell cycles, sirtuin pathways could theoretically forestall or prevent altogether many late-onset neurodegenerative diseases (Guarente and Picard, 2005; Baur and Sinclair, 2006).
We also detected SIRT1 in rodent tissue and human cell extracts using Western blot procedures with different commercially available antibodies. Our results show the presence of protein bands closely matching predictive sizes of ~85 kDa for rodents and ~120 kDa for humans. This prediction is predicated on our recent analysis of the rodent and human proteome (NCBI; www.ncbi.nlm.nih.gov/) showing that the average SIRT1 protein length is rats is 657 amino acid residues, whereas for the mouse is 737, and for the human is 757 amino acid residues. Nevertheless, the anti-SIRT1 2G1/F7 mouse monoclonal IgG1 recognized SIRT1 in both rodent and human brain extracts by ICC procedures, whereas the anti-SIRT1 A310-204A rabbit polyclonal IgG1 was only suitable for the detection of mammalian SIRT1 by Western blot methods. However, both commercially available antibodies recognized SIRT1 to be predominantly sub-localized to the nucleus, at least in adult organisms. These data are consistent with the idea that SIRT1, SIRT6, and SIRT7 are found mainly in the mammalian cell nucleus, although SIRT1 also performs critical biological functions in the cytoplasm as well. The remaining sirtuins have been described as mitochondrial sirtuins (i.e., SIRT3-SIRT5) or cytoplasmic sirtuins (i.e., SIRT2) (Michan and Sinclair, 2007).
In the current study, we also reveal the identity of certain SIRT1-containing neurons in the rodent hippocampus and substantia nigra. Neurons in these two brain areas appear to sub-serve cognitive (hippocampus) or movement (substantia nigra) functions, and although their structural and physiological differences are beginning to be understood, their vulnerability differences to aging or late-onset neurodegenerative diseases are largely unknown. In the rodent hippocampus, we found that SIRT1 and PV were diffusely arranged in the nucleus and cytoplasm respectively, and were therefore co-localized to the same nerve cell. Neurons expressing the Ca2+-binding protein constitute an abundant sub-population of GABA cells in the cerebral cortex and the hippocampal formation. In these brain regions, PV-containing cells are the major perisomatic-targeting interneurons where they play a key role in neuronal network activity, including the generation of electrical oscillations at particular frequencies (Karson et al., 2009). In general, our results suggest an as-yet unknown role of SIRT1 in PV and GABA cells with active neurogenesis cycles, at least in the hippocampus.
Our anatomical data also confirm the co-localization of SIRT1 with TH to neurons of the substantia nigra. In this regard, vulnerable DA cells of the substantia nigra are lost during PD progression, whereas other neurons are spared (Lee and Price, 2001). This is a puzzling aspect of late-onset neurodegenerative diseases. Why are pathological processes not always evenhanded across all neurons? And more importantly, why do DA cells die despite having SIRT1 complexes that normally protect neurons from aberrant senescence? A possible answer to these questions is the exaggerated propensity for DA to oxidize and form multiple reactive oxygen species as well as the protein-modifying DA quinone (Dauer and Przedborski, 2003). It is conceivable that during the spontaneous oxidation of DA, reactive oxygen species and/or quinone molecules attack one or more sites of the SIRT1 protein eventually silencing its deacetylase activity. The subsequent loss of SIRT1-dependent protection would result in the enhanced and selective demise of DA cells. There is still considerable work to be done, particularly in animal models of PD, to determine the role of DA oxidation and subsequent SIRT1 deacetylation within nigro-striatal neurons.
In conclusion, we have provided information on the sub-cellular localization and anatomical distribution of SIRT1 in rodent and human nervous systems. Additionally, we have characterized the phenotype of several SIRT1-containing neurons of the hippocampus and substantia nigra. Finally, we have provided information on the specificity of several commercially available monoclonal and polyclonal antibodies raised against SIRT1. Taken together, this information should provide us with further experimental insights for examining the neurobiology of SIRT1 in the CNS, especially as it relates to age-related neurodegenerative diseases.
Acknowledgments
This work was supported in part by NIH grant #R15MH64513-02A1 (JMH).
LITERATURE CITED
- Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nature Reviews. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- Dauer W, Przedborski S. Parkinson's disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- Gan L, Mucke L. Paths of Convergence: Sirtuins in aging and neurodegeneration. Neuron. 2008;58:10–14. doi: 10.1016/j.neuron.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Picard F. Calorie restriction-the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Horowitz JM, Goyal A, Ramdeen N, Hallas BH, Horowitz AT, Torres G. Characterization of fluoxetine plus olanzepine treatment in rats: a behavior, endocrine, and immediate early gene expression analysis. Synapse. 2003;50:353–364. doi: 10.1002/syn.10276. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Sun Z, Lee T, Fusco FR, Kimble TD, Meade CA, Cuthbertson S, Reiner A. A simple and sensitive antigen retrieval method for free-floating and slide-mounted tissue sections. J. Neu. Meth. 1999;39:149–162. doi: 10.1016/s0165-0270(99)00142-9. [DOI] [PubMed] [Google Scholar]
- Karson M, Tang A, Milner T, Alger B. Synaptic cross talk between perisomatic-targeting interneuron classes expressing cholecystokinin and parvalbumin in hippocampus. J. Neurosci. 2009;29(13):4140–54. doi: 10.1523/JNEUROSCI.5264-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO. J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Price DL. Advances in genetic models of Parkinson's disease. Clinical Neurosci. Res. 2001;1:456–466. [Google Scholar]
- Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem. J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1 alpha. J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 2nd Ed. Academic Press; San Diego: 1986. [Google Scholar]
- Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schroter F, Ninnemann O, Siegart E, Bendix I, Brustle O, Nitsch R, Zipp F, Aktas O. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nature Cell Biology. 2008;10 doi: 10.1038/ncb1700. PAGE #s. [DOI] [PubMed] [Google Scholar]
- Qin W, Yang T, Ho L, Zhao Z, Wang J, Chen L, Zhao W, Thiyagarajan M, MacGrogan D, Rodgers JT, Puigserver P, Sadoshima J, Deng H, Pedrini S, Gandy S, Suave AA, Pasinetti GM. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J. Biol. Chem. 2006;281:21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- Ramadori G, Lee CE, Bookout AL, Lee S, Williams KW, Anderson J, Elmquist JK, Coppari R. Brain SIRT1: Anatomical Distribution and Regulation by Energy Availability. J. Neurosci. 2008;28(40):9989–9996. doi: 10.1523/JNEUROSCI.3257-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1 alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1994. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- Torres G, Frisella PD, Yousuf SJ, Sarwar S, Baldinger L, Zakhary SM, Leheste JR. A ChIP-cloning approach linking SIRT1 to transcriptional modification of DNA targets. Biotechniques. 2008;44:7. doi: 10.2144/000112748. [DOI] [PubMed] [Google Scholar]
- Torres G, Lee S, Rivier C. Immunocytochemical and in situ hybridization detection of hypothalamic neuropeptides from postmortem unfixed rat brains. Neuroscience Letters. 1992;146:96–100. doi: 10.1016/0304-3940(92)90181-6. [DOI] [PubMed] [Google Scholar]
- Torres G, Hallas BH, Vernace VA, Jones C, Gross KW, Horowitz JM. A neurobehavioral screening of the ckr mouse mutant: implications for an animal model of schizophrenia. Brain Res. Bulletin. 2004;62:315–326. doi: 10.1016/j.brainresbull.2003.09.020. [DOI] [PubMed] [Google Scholar]
- Torres G, Hallas BH, Lorig EN, Strauss J, Horowitz JM. Dynamic expression of molecular chaperones in structurally and functionally intact endothelial cell networks. Preclinica, 2004;2:197–203. [Google Scholar]
- Torres G, Meeder BA, Hallas BH, Spernyak JA, Mazurchuk R, Jones C, Gross KW, Horowitz JM. Ventricular size mapping in a transgenic model of schizophrenia. Dev. Brain Res. 2005;154:35–44. doi: 10.1016/j.devbrainres.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E., Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2 (SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]