Abstract
Bacteriophage ϕC31 inserts its genome into that of its host bacterium via the integrase enzyme which catalyzes recombination between a phage attachment site (attP) and a bacterial attachment site (attB). Integrase requires no accessory factors, has a high efficiency of recombination, and does not need perfect sequence fidelity for recognition and recombination between these attachment sites. These imperfect attachment sites, or pseudo-attachment sites, are present in many organisms and have been used to insert transgenes in a variety of species. Here we describe the ϕC31 integrase approach to make transgenic Xenopus laevis embryos.
Keywords: Xenopus, ϕC31, integrase, transgenesis, fluorescence
1. Introduction
The frog, Xenopus laevis, has a long history of use for studies in embryonic development. Previously described Xenopus integration techniques often insert multiple copies of a transgene at random sites in the embryos genome (1–4). Though valuable for many experimental designs, these approaches are problematic for researchers who desire transgene expression to approximate endogenous gene expression levels. Rather, a site-directed integration approach that incorporates a regulated single copy of a transgene into the host genome would more closely match endogenous gene expression. The ϕC31 integrase approach is one way to accomplish this aim.
Bacteriophage ϕC31 encodes an integrase enzyme that inserts the phage genome into the genome of various Streptomyces bacteria (5, 6). The integrase protein recognizes a 39-bp-long phage attachment site (attP) in its own genome and a 34-bp-long bacterial attachment site (attB) in the bacterium’s genome and then catalyzes an integration event (7, 8). The integrase enzyme does not require perfect sequence fidelity to recognize the attP site (7). Non-perfect attP sites, or pseudo-attP sites, may have as low as 24% sequence homology to endogenous attP sites and still allow recombination (7) although the recombination efficiency may be decreased.
Many groups have shown that integrase can insert plasmid DNA sequences that contain an attB site into pseudo-attP sites found in a variety of organisms. Utilizing this approach, transgenes have been inserted into the genomes of plant cells (9), mammalian cells (7,10–18), and Drosophila embryos (19). We used the pseudo-attP sites in the Xenopus genome and an attB site containing reporter plasmid to make transgenic Xenopus embryos (20) (Fig. 9.1). Surprisingly, the reporter genes are expressed in some expected tissues but not in others. We recognized this as chromatin position effect and flanked the reporter gene with HS4 insulators. HS4 insulators stop the spread of chromatin silencing and also prevent distant enhancers from acting on a promoter (21, 22). After making transgenic embryos with the new insulated reporter plasmid, we found that the transgenes expressed as expected (20) (Fig. 9.2). The techniques used to generate and recognize ϕC31 integrase mediated transgenic Xenopus embryos are described below.
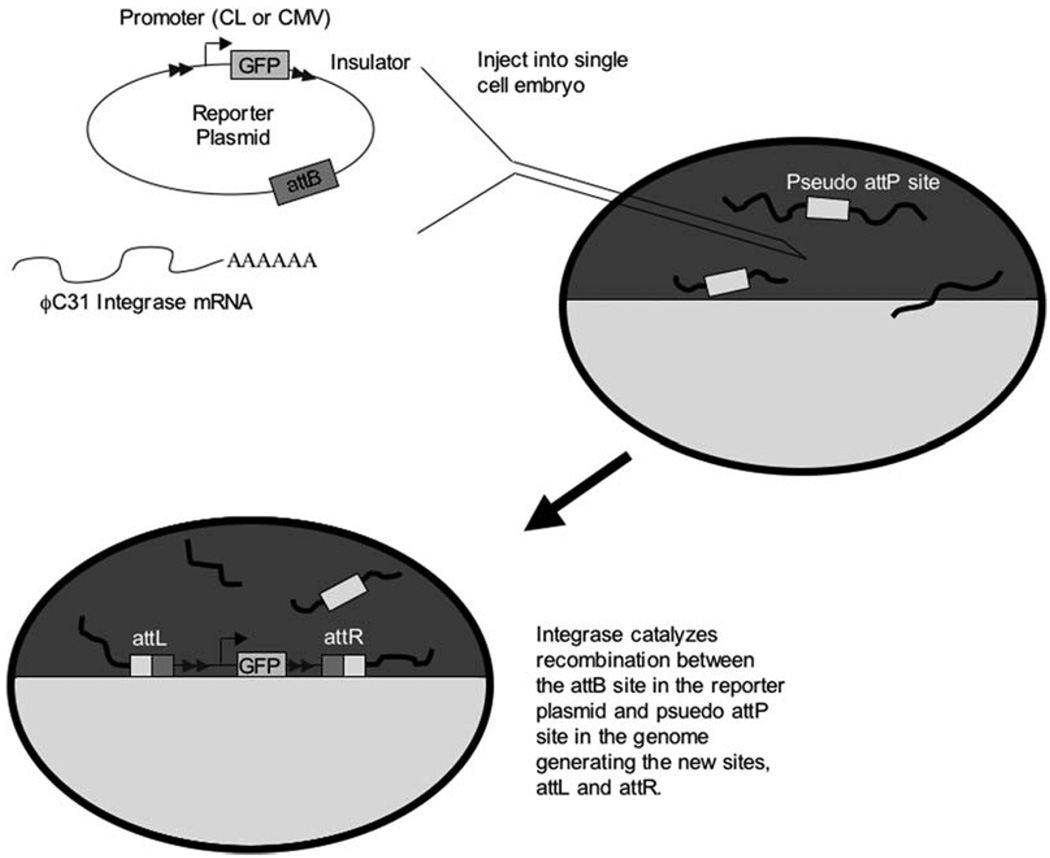
Fig. 9.1.
Representation of ϕC31 integrase-mediated transgenesis in Xenopus laevis. The reporter plasmid containing an attB site and an insulated reporter gene is injected along with integrase mRNA into single-cell embryos. The chromosomes (thick black lines) contain numerous pseudo-attP sites. Inside the single-cell embryo, the integrase protein catalyzes recombination between the attB site in the reporter plasmid (thin black line) and a pseudo-attP site in the embryo’s genome (thick black line). Recombination results in the formation of two new attachment sites, attR and attL flanking the integrated reporter plasmid.
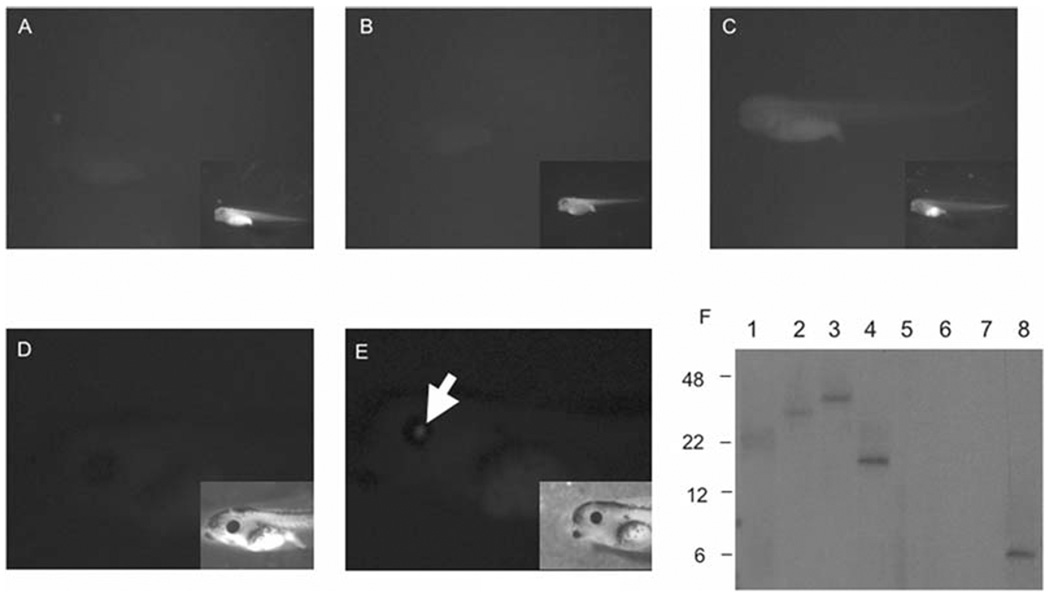
Fig. 9.2.
ϕC31 integrase-mediated transgenesis of insulated reporter plasmids generate Xenopus embryos with tissue-appropriate expression. In every case, the insert shows a brightfield image of the embryo. (A) Non-injected stage 42 embryo. (B) Stage 42 embryo injected with 5 pg of CMV-EGFP-DI-attB reporter plasmid. (C) Stage 42 embryo injected with 5 pg of CMV-EGFP-DI-attB reporter plasmid and 1 ng of integrase mRNA. (D) Stage 44 embryo injected with 5 pg of CL-EGFP-DI-attB plasmid alone. (E) Stage 44 embryo injected with 5 pg of CL-EGFP-DI-attB plasmid and 1 ng of integrase mRNA. GFP expression is indicated with the white arrow. (F) Southern blot demonstrating single integration events. Lanes 1 and 2 contains DNA harvested from single stage 46 CMV-EGFP-DI-attB transgenic embryos that expressed GFP uniformly. Lanes 3–4 contain DNA harvested from single stage 46 CL-EGFP-DI-attB transgenic embryos that expressed GFP in the lens of the eye. Lane 5 contains stage 46 non-injected DNA. Lane 6 contains DNA from a stage 46 embryo injected with CMV-EGFP-DI-attB plasmid alone. Lane 7 contains DNA from a stage 46 embryo injected with CL-EGFP-DI-attB plasmid alone. Lane 8 contains 10 pg of CMV-DI-EGFP-attB plasmid linearized with BamHI. Markers in kilobase pairs are indicated to the left of the membrane.
2. Materials
2.1. Plasmids and Plasmid Preparation Reagents
Plasmids: The integrase plasmid pET11-phiC31poly(A) (23) can be obtained from Dr. Michelle Calos (calos@stanford.edu), and the attB reporter plasmids CMV-EGFP-DI-attB and CL-EGFP-DI-attB (20) can be obtained from Dr. Daniel Weeks (daniel-weeks@uiowa.edu). Permission to use HS4 insulator sequences and HS4 insulator constructs needs to be obtained from Dr. Gary Felsenfeld (gary.felsenfeld@nih.gov) (see Note 1).
Plate and culture reagents: Bacto-Yeast Extract, Bacto-Tryptone powder, and Bacto-agar (BD Biosciences, Sparks, MD).
Antibiotics: Ampicillin and Kanamycin (Sigma Aldrich, St. Louis, MO).
Plasmid Purification Kits: Qiaprep Spin Miniprep Kit, HiSpeed Plasmid Maxi Kit (Qiagen, Valencia, CA).
In vitro RNA transcription: T7 mMessage machine (Ambion, Austin, TX).
Agarose (RPI, Mt. Prospect, IL).
10X TAE: 0.4-M Tris-acetate, 0.01-M EDTA.
10X Non-denaturing DNA loading buffer: 50% glycerol, 60-mM EDTA, 1% SDS, and 0.05% bromophenol blue.
1% agarose, 2.2-M formaldehyde MOPS gel.
10X Denaturing RNA loading buffer.
Gel electrophoresis apparatus for flat-bed agarose gel electrophoresis (Owl Separation Systems, Portsmouth, NH).
Spectrophotometer: Nanodrop ND1000 (Nanodrop Technologies, Wilmington, DE).
UV-light transilluminator.
2.2. Xenopus Injection
Xenopus laevis: May be obtained from either Xenopus I (Dexter, MI) or Nasco (Fort Atkinson, WI).
Human Chorionic Gonadotropin (Sigma Aldrich).
Tricaine (3-aminobenzoic acid ethyl ester) (Sigma Aldrich).
10X Marc’s Modified Ringers Solution (MMR): 1-M NaCl, 20-mM KCl, 20-mM CaCl2, 10-mM MgCl2, 50-mM HEPES at pH 7.4.
Injection buffer: 88-mM NaCl, 10-mM HEPES.
0.3X MMR with 3% Ficol Type 400 (Sigma Aldrich).
2% Cysteine (RPI) made in dH2O, pH 7.8–7.9 with NaOH. Should be made on the day of injection.
Microinjection needle puller.
Micromanipulator: Singer MK-1 (Singer instrument company, Somerset, England) or similar instrument.
Microinjector: Inject+Matic (Geneva, Switzerland) or similar instrument.
Glass capillary tubes (Singer).
18°C incubator.
2.3. Microscopy
Dissecting microscope: Nikon SMZ (Nikon Instruments, Melville, NY) and Zeiss Stemi SV 11 fluorescent dissecting microscope (Zeiss MicroImaging, Thornwood, NY).
Compound microscope with fluorescence: Zeiss Axioplan 2 (Carl Zeiss MicroImaging) or similar instrument.
Camera for photo documentation: SPOT camera (Diagnostic Instruments, Sterling Heights, MI), Zeiss Axiocam (Carl Zeiss MicroImaging), or similar instrument.
2.4. Southern Blotting
Genomic DNA purification: Qiagen DNeasy (Qiagen).
Restriction enzymes: multiple suppliers.
Ethidium bromide (Sigma Aldrich).
Hybond-N+ nylon membrane (GE Healthcare, Chicago, IL).
Rediprime II DNA Labeling System (GE Healthcare).
Redivue α-P32 deoxycytidine (GE Healthcare).
RapidHyb solution (Ambion).
X-ray film, Kodak Biomax XAR (Eastman Kodak Co, Rochester, NY).
20X SSC: 3.0-M NaCl and 0.3-M sodium citrate at pH 7.0.
Depurination buffer: 0.25-M HCl.
Denaturation buffer: 1.5-M NaCl with 0.5-M NaOH.
Neutralization buffer: 1-M Tris-HCl, pH 8.0, 1.5-M NaCl.
Hybridization Oven: Hybaid (Thermo Corporation, Waltham, MA).
3. Methods
3.1. Preparation of Plasmids and ϕC31 Integrase mRNA
Transform the pET11-phiC31poly(A) plasmid and the attB reporter plasmids (CMV-EGFP-DI-attB and CL-EGFP-DIattB) into Escherichia coli and plate onto antibiotic selective plates using standard techniques. The pET11-phiC31poly(A) plasmid contains an ampicillin resistance gene and the attB reporter plasmids contain a kanamycin resistance gene. We have found that insulated attB reporter plasmids frequently undergo rearrangements and thus recommend using Stbl2 cells (Invitrogen) which are designed to prevent recombination.
Select individual colonies on each plate and grow 3-ml cultures at 37°C overnight with agitation. Use 1 ml for mini-preps (Qiaprep mini-prep kit) and save the other 2 ml of culture at 4°C. Using the mini-prep-generated DNA, confirm plasmid size and insulator orientation by restriction enzyme digestion and gel analysis. Once plasmid size and insulator orientation are confirmed, inoculate 1 l of LB broth containing the appropriate antibiotic with the remaining 2 ml of culture. Grow at 37°C with agitation overnight.
Perform maxi-preps on the 1-l cultures following the manufacturer’s instructions (Hi Speed Plasmid Maxi Kit). Do not add RNase to buffer P1.
Confirm maxi-prep-generated plasmid sequences by restriction enzyme digestion and gel analysis. Store the maxi-prep-generated DNAs at −20°C.
Linearize 5 µg of pET11-phiC31poly(A) maxi-prep DNA with either BamHI or EcoRI restriction enzymes. Run approximately one-tenth of the digestion on a 1% agarose gel to ensure linearization. Heat inactivates the restriction enzyme in the remaining portion of digestion by heating reaction to 65°C for 20 min. Precipitate the DNA by adding one-tenth the volume of 5-M NH4 acetate and 2 volumes of ethanol. Place solution at −20°C for 15 min and centrifuge at 10,000 g for 15 min. Remove the supernatant and resuspend the pellet in 10 µl of RNase-free TE buffer.
Synthesize the ϕC31 integrase mRNA using the T7 mMessage machine following the manufacturer’s instructions. Because the protocol includes a DNase treatment step, the DNase needs to be removed or inactivated. We routinely follow the LiCl precipitation protocol described in the manufacturer’s instructions. Resuspend the integrase mRNA in RNase-free water at a concentration of 1 mg/ml.
Run 1 µg of ϕC31 integrase mRNA on a formaldehyde MOPS gel in 1X MOPS buffer to ensure that the transcript is approximately 1.9 kilobases long.
Store ϕC31 integrase mRNA (for up to 1 month) at −80°C until it is needed.
3.2. Injection of Xenopus Embryos
Obtain approval from host institution to house and care for Xenopus adult frogs and embryos. If Xenopus experience is minimal, the book, Early Development of Xenopus laevis – A Laboratory Manual (24), may be a useful reference to consult.
Induce Xenopus females to lay eggs by injecting 1 ml (1000 IU) of human chorionic gonadotropin into the dorsal lymph sac the night before desired day of egg collection.
The next morning, the cloaca on the injected females should be swollen and there may be eggs in the water tank holding the frogs.
After eggs are present in the tank, inject a lethal dose of Tricaine (1 ml of a 10% solution) into the dorsal lymph sac of a Xenopus male and then surgically remove the testes. Store the testes in 1X MMR.
Induce egg laying into a dry petri dish by spreading apart the female’s legs and gently squeezing and rubbing the pelvic region. Immediately fertilize the eggs by rubbing a small piece of testis (~one-sixth of a testis) through the eggs. Then crush the testis in ~1 ml of 0.3X MMR and spread this solution over the eggs. Let the eggs sit in the sperm solution for about 1 min and then flood the eggs with 0.1X MMR.
Successful fertilization can be determined if the eggs align with the animal pole (pigmented half) facing up. Embryos should be spherical, uniform size, and have smooth even pigmentation of the animal hemisphere. At 30 min after fertilization, remove the 0.1X MMR and place the embryos in 2% cysteine, pH 7.8–7.9 for 2 min to remove the jelly coats.
Remove the cysteine and wash the embryos in three 5-ml washes of 0.3X MMR.
Place the embryos in 0.3X MMR, 0.3% ficol.
Inject single-cell embryos into the center of the animal hemisphere with either 10 nl of injection solution, 10 nl containing 5 pg of reporter plasmid resuspended in injection solution, or 10 nl containing 5 pg of reporter plasmid + 1 ng of ϕC31 integrase mRNA also resuspended in injection solution.
3.3. Monitoring Developing Embryos for Transgenesis
Allow injected embryos to develop at 18°C in 0.3X MMR, 0.3% Ficol for approximately 6–8 h after injection and then transfer to 0.3X MMR.
Remove delaminating or dead embryos and provide fresh 0.3X MMR at least twice a day for the first 3 days.
On the second day after fertilization, begin to monitor the embryos for GFP expression using a fluorescent microscope optimized to detect green fluorescence (see Note 2). Embryos injected with 1 ng integrase mRNA and 5 pg of CMV-EGFPDI-attB should express GFP uniformly (Fig. 9.2C) while embryos injected with 1-ng integrase mRNA and 5 pg of CL-EGFP-DI-attB should express GFP only in the lens of the eye (Fig. 9.2E). Embryos injected with 5 pg of reporter plasmid alone rarely give GFP expression (Fig. 9.2B). Digital photography with long exposure times may detect fluorescence before it is visible to the eye.
Once fluorescence is detected, photograph the embryos using a fluorescent microscope with a digital microscope. If embryo movement prevents photography, consider anesthetizing the embryos with 0.02% Tricaine. Embryos may be exposed to the Tricaine solution for approximately 5 min; longer exposure times may lead to the demise of the embryos. Embryos should be placed back into 0.3X MMR after photography.
3.4. Southern Blot Analysis to Confirm Integration
Isolate DNA from single-stage 46 embryos using the Qiagen DNeasy kit following the manufacturer’s instructions. One-stage 46 embryo should yield approximately 50 µg of DNA. DNA should be collected from non-injected embryos, embryos injected with reporter plasmid alone, and transgenic embryos determined by green fluorescence.
Digest 5–10 µg of harvested DNA and 10–100 pg of reporter plasmid with a restriction enzyme that cuts the reporter plasmid at a single site overnight (for CL-EGFP-DI-attB or CMV-DI-EGFP attB, BamHI is a suitable enzyme).
Run digested DNA on a 0.6% agarose gel. To allow for good separation, use a gel that is 10–15-cm long and run the gel until the bromophenol blue dye front is approximately 2 cm from the bottom of the gel. Stain the gel with ethidium bromide and then photograph the gel while on a UV transilluminator. Place a UV light visible ruler next to the gel while taking the photograph as it will be useful to approximate the size of the insertions.
Transfer the DNA onto a positively charged nylon membrane using standard southern blot protocols.
Following the transfer, rinse the membrane in 5X SSC and prehybridize in Ambion RapidHyb supplemented with 0.1 µg/ml herring sperm DNA at 45°C for 1–2 h with agitation.
Generate a P32-labeled EGFP probe. We digest the CLEGFP-DI-attB plasmid or CMV-EGFP-DI-attB plasmid with BamHI and AgeI, run the digested DNA on a 1% agarose gel, and then isolate the GFP fragment using the Qiagen Gel Extraction Kit. We then label the EGFP fragment using Rediprime II Random Prime Labeling System following the manufacturer’s instructions. Free nucleotides are removed using the Qiagen nucleotide removal kit.
After prehybridizing the membrane in Ambion RapidHyb for 1–2 h, boil the labeled probe for 5 min and then place the probe on ice for 5 min. dd denatured probe to prehybridization solution and hybridize at 45°C overnight.
After hybridization, rinse the blot with 2X SSC, 0.1% SDS at room temperature and then wash the blot with two 68°C 100-ml washes of 2X SSC, 0.1% SDS (5 min each), two 100-ml washes using 1X SSC, 0.1% SDS (10 min each), and four 100-ml washes using 0.1X SSC, 0.1% SDS (30–60 min each).
Sandwich washed blot in plastic wrap and expose to Kodak MS film. We recommend using film cassettes with intensifying screens at −80°C. Develop the first exposure 12 h after placing the film. If the signal is weak, additional exposure time may be needed (48–120 h).
Analyze the southern blot for evidence of genomic integration. Southern blot insertion sites have ranged in size from 10 kb to 50 kb in size (Fig. 9.2F).
Acknowledgements
We would like to thank Professor Michele Calos for providing the pET11phiC31poly(A) plasmid, Professor Gary Felsenfeld for providing the HS4 insulator sequences, and Paul Kreig for providing the gamma crystallin lens promoter. This work was supported by funding from the NIH (GM069944 and DC007481). Bryan Allen is a student in the Medical Scientist Training Program at the Roy J. and Lucille A Carver College of Medicine, University of Iowa.
Footnotes
MTA agreements are required from Dr. Gary Felsenfeld (NIH) to use the HS4 insulator sequences and Dr. Daniel Weeks (Iowa) for the CMV-EGFP-DI-attB and CL-EGFP-DI-attB plasmids. An MTA is also required to obtain the plasmid pET11phiC31poly(A) from Dr. Michele Calos (Stanford).
References
- 1.Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- 2.Sparrow DB, Latinkic B, Mohun TJ. A simplified method of generating transgenic Xenopus. Nucleic Acids Res. 2000;28:E12. doi: 10.1093/nar/28.4.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogino H, McConnell WB, Grainger RM. Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mech. Dev. 2006;123:103–113. doi: 10.1016/j.mod.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Pan FC, Chen Y, Loeber J, Henningfeld K, Pieler T. I-SceI meganuclease-mediated transgenesis in Xenopus. Dev. Dyn. 2006;235:247–252. doi: 10.1002/dvdy.20608. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Jeppesen I, Nielsen K, Jensen TG. Phi c31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Ther. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]
- 6.Kuhstoss S, Rao RN. Analysis of the integration function of the streptomycete bacteriophage phi C31. J Mol Biol. 1991;222:897–908. doi: 10.1016/0022-2836(91)90584-s. [DOI] [PubMed] [Google Scholar]
- 7.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol.& Cell. Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorpe HM, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Nat. Acad. Sci. USA. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lutz KA, Corneille S, Azhagiri AK, Svab Z, Maliga P. A novel approach to plastid transformation utilizes the phiC31 phage integrase. Plant J. 2004;37:906–913. doi: 10.1111/j.1365-313x.2004.02015.x. [DOI] [PubMed] [Google Scholar]
- 10.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc. Nat. Acad. Sci. USA. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalberg TW, Genise HL, Vollrath D, Calos MP. phiC31 integrase confers genomic integration and long-term transgene expression in rat retina. Invest. Ophthalmol. Vis. Sci. 2005;46:2140–2146. doi: 10.1167/iovs.04-1252. [DOI] [PubMed] [Google Scholar]
- 12.Thomason LC, Calendar R, Ow DW. Gene insertion and replacement in Schizosaccharomyces pombe mediated by the Streptomyces bacteriophage phiC31 site-specific recombination system. Mol. Gen. & Genomics: MGG. 2001;265:1031–1038. doi: 10.1007/s004380100498. [DOI] [PubMed] [Google Scholar]
- 13.Olivares EC, Hollis RP, Chalberg TW, Meuse L, Kay MA, Calos MP. Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nat. Biotechnol. 2002;20:1124–1128. doi: 10.1038/nbt753. [DOI] [PubMed] [Google Scholar]
- 14.Held PK, Olivares EC, Aguilar CP, Finegold M, Calos MP, Grompe M. In vivo correction of murine hereditary tyrosinemia type I by phiC31 integrase-mediated gene delivery. Mol. Ther. 2005;11:399–408. doi: 10.1016/j.ymthe.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nat. Biotechnol. 2003;21:321–324. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- 16.Keravala A, Portlock JL, Nash JA, Vitrant DG, Robbins PD, Calos MP. PhiC31 integrase mediates integration in cultured synovial cells and enhances gene expression in rabbit joints. J. Gene Med. 2006;8:1008–1017. doi: 10.1002/jgm.928. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa Y, Tanaka N, Murakami K, Uchiyama T, Kumaki S, Tsuchiya S, Kugoh H, Oshimura M, Calos MP, Sugamura K. Phage phiC31 integrase-mediated genomic integration of the common cytokine receptor gamma chain in human T-cell lines. J. Gene Med. 2006;8:646–653. doi: 10.1002/jgm.891. [DOI] [PubMed] [Google Scholar]
- 18.Bertoni C, Jarrahian S, Wheeler TM, Li Y, Olivares EC, Calos MP, Rando TA. Enhancement of plasmid-mediated gene therapy for muscular dystrophy by directed plasmid integration. Proc. Natl. Acad. Sci. USA. 2006;103:419–424. doi: 10.1073/pnas.0504505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen BG, Weeks DL. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat. Methods. 2005;2:897–898. doi: 10.1038/nmeth814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 2003;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 22.West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 23.Hollis RP, Stoll SM, Sclimenti CR, Lin J, Chen-Tsai Y, Calos MP. Phage integrases for the construction and manipulation of transgenic mammals. Reprod. Biol. Endocrinol. 2003;1:79. doi: 10.1186/1477-7827-1-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sive HL, Grainger RM, Harland RM. Early Development of Xenopus laevis : A Laboratory Manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]