Abstract
The Shigella flexneri outer membrane protease IcsP proteolytically cleaves the actin-based motility protein IcsA from the bacterial surface. The icsP gene is monocistronic and lies downstream of an unusually large intergenic region on the Shigella virulence plasmid. In silico analysis of this region predicts a second transcription start site 84 bp upstream of the first. Primer extension analyses and beta-galactosidase assays demonstrate that both transcription start sites are used. Both promoters are regulated by the Shigella virulence gene regulator VirB and both respond similarly to conditions known to influence Shigella virulence gene expression (iron concentration, pH, osmotic pressure, and phase of growth). The newly identified promoter lies upstream of a Shine-Dalgarno sequence and second 5’-ATG-3’, which is in frame with the annotated icsP gene. The use of either translation start site leads to the production of IcsP capable of proteolytically cleaving IcsA. A bioinformatic scan of the Shigella genome reveals multiple occurrences of in-frame translation start sites associated with putative Shine –Dalgarno sequences, immediately upstream and downstream of annotated open reading frames. Taken together, our observations support the possibility that the use of in-frame translation start sites may generate different protein isoforms, thereby expanding the proteome encoded by bacterial genomes.
Introduction
Shigella species are gram-negative intracellular pathogens that cause bacillary dysentery in humans by invading cells of the colonic epithelium (Labrec et al. 1964, Sansonetti 1998). Once inside host cells Shigella move through the cytoplasm and into adjacent cells using actin-based motility. This process is mediated by the Shigella outer membrane protein IcsA (VirG), which polymerizes eukaryotic actin monomers into a tail of tightly bundled filaments on one pole of the bacterium (Bernardini et al. 1989, Goldberg et al. 1993). Shigella flexneri mutants lacking icsA are avirulent in animal models (Makino et al. 1986), demonstrating that actin-based motility is essential for Shigella pathogenicity.
The outer membrane protease IcsP modulates the amount and distribution of IcsA associated with Shigella. The activity of this protease was originally observed when growth medium was found to contain a 95 KDa polypeptide of IcsA after it had supported Shigella growth (Goldberg et al. 1993). Two groups identified IcsP (SopA) as the protease responsible for the cleavage of IcsA (Egile et al. 1997, Shere et al. 1997). Data collected by these two groups throughout five studies demonstrated that IcsP plays a role in the modulation of IcsA and the actin-based motility of Shigella (d'Hauteville et al. 1996, Egile et al. 1997, Shere et al. 1997, Steinhauer et al. 1999,Wing et al. 2005). Although IcsA is localized to the old pole of the bacterium in wild type Shigella (Goldberg et al. 1993), expression of a non-cleavable form of IcsA in Shigella was found to lead to an increase in the circumferential localization of IcsA (d'Hauteville et al. 1996). Similar phenotypes were reported for icsP mutants in vitro (Egile et al. 1997, Shere et al. 1997, Steinhauer et al. 1999). When the intra- and intercellular phenotypes of icsP mutants were analyzed, abnormal actin-based motility and cell-to-cell spread were observed regardless of the serotype examined (Egile et al. 1997, Shere et al. 1997). Furthermore, Shigella cells expressing plasmid-borne icsP were found to lack detectable IcsA on their surfaces the effects of icsP mutation on intercellular movement and plaque formation were also serotype dependent (Steinhauer et al. 1999, Wing et al. 2005). Since dysregulation of the icsP gene generates Shigella phenotypes that are consistent with attenuation of virulence, these studies strongly suggest that the icsP gene and its protein product will be tightly regulated.
Like many of the genes required for virulence of S. flexneri, icsA and icsP are encoded by the large ~230 kb virulence plasmid of S. flexneri. (Jin et al. 2002). This virulence plasmid encodes the transcription factor VirB which positively regulates many genes on the plasmid including icsP (Dorman and Porter 1998, Wing et al. 2004). The VirB-dependent regulation of the icsP promoter requires two distal VirB sites located between positions −1144 and −1130 relative to the annotated transcription start site (TSS) (Castellanos et al. 2009). These binding sites are located within an unusually large (~1.2 kb) intergenic region, which separates the icsP gene and the divergently transcribed ospZ gene.
In Shigella flexneri serotype 2a, coding sequences account for 76.24% of the virulence plasmid (Blattner et al. 1997, Jin et al. 2002). Although the coding density of the Shigella virulence plasmid is lower than the Escherichia coli K12 chromosome (87.8%; 4), the intergenic region upstream of the icsP gene is still abnormally large when compared to the average size of an E. coli K-12 intergenic region (1.2 kb vs. 246 bp, respectively; (Pupo et al. 2000). Furthermore, the remote location of the VirB binding sites that influence icsP expression already implicates this large intergenic region in the transcriptional regulation of the icsP gene.
Based on the role that IcsP plays in maintaining the surface distribution of IcsA and how this ultimately regulates Shigella actin-based motility, we hypothesize that the icsP gene and /or its protein product will be tightly regulated. The aim of this study was to further characterize the regulation of IcsP at both the transcriptional and translational level. To do this, we chose to examine the entire upstream intergenic region for sequence elements involved in the regulation of IcsP.
Results
In silico analyses of the intergenic region upstream of icsP
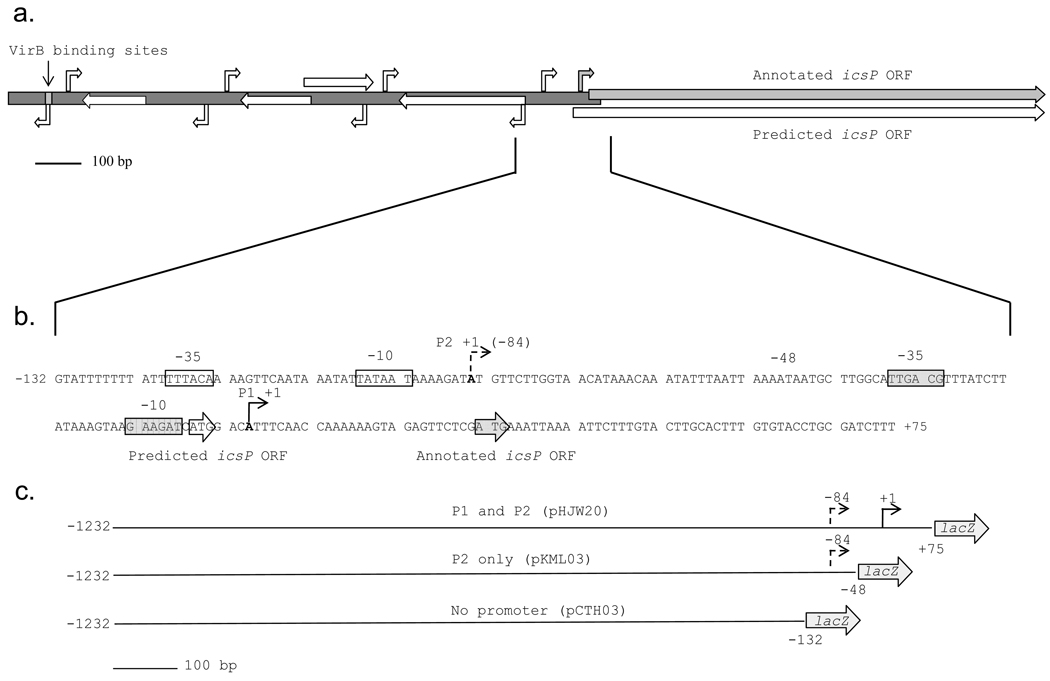
Due to the unusual length of the intergenic region and its involvement in the regulation of icsP, we wanted to further analyze this region for elements contributing to the transcriptional regulation of the icsP gene. To do this, our initial approach was to analyze the entire 1232 bp sequence using in silico tools. To identify putative promoter sequences the intergenic region upstream of icsP was entered into the BPROM program for prediction of promoters regulated by the σ70 subunit of RNA polymerase (http://linux1.softberry.com). The BPROM software identified four putative transcription start sites (TSSs) with associated −10 and −35 sequences at positions −84, −422, −769, and −1106 relative to the previously annotated TSS (Fig. 1a). Interestingly, the BPROM program did not identify the originally annotated promoter. Analysis of the divergent strand predicted four additional promoters approximately 20 bp upstream of each TSS found on the complimentary strand. This is not surprising considering the adenine and thymine composition of the −10 sequence.
Fig. 1.
Graphical representation of the entire icsP intergenic region and important promoter elements. The entire icsP promoter and gene with the locations of the annotated TSS, ORF, upstream VirB binding sites (grey arrows and box) and predicted TSSs and ORFs (white arrows) (a). Promoter elements of the icsP P1 and P2 promoters (b). P1, solid angled arrow; P2, dashed angled arrow; −10 and −35 sites are boxed and labeled. The P1 and P2 translation start sites are enclosed by arrows and labeled. Schematic representations of the lacZ fusion inserts in pHJW20, pKML03, and pCTH03, respectively (c). The inserts are drawn to scale and the numbers are relative to the P1 TSS.
To examine whether any of the predicted promoters were aligned with potential open reading frames (ORFs), the entire intergenic region was analyzed with the microbial gene finding system Glimmer (NCBI). Only one ORF, identified on the icsP coding strand, lay within 150 bp of a putative TSS found by BPROM. This was the TSS at the −84 position. This ORF begins 33 bp upstream of the annotated icsP gene (Fig. 1b). This would allow for the production of a polypeptide exactly 11 amino acids longer than and yet still in frame with the previously described icsP gene. A common problem with ORF prediction software is that they identify the longest ORFs and frequently miss internally coded start codons (Delcher et al. 1999). This may explain why the Glimmer program did not identify the beginning of the originally annotated icsP gene (Egile et al, 1997).
Identification of two promoters responsible for regulation of the icsP gene
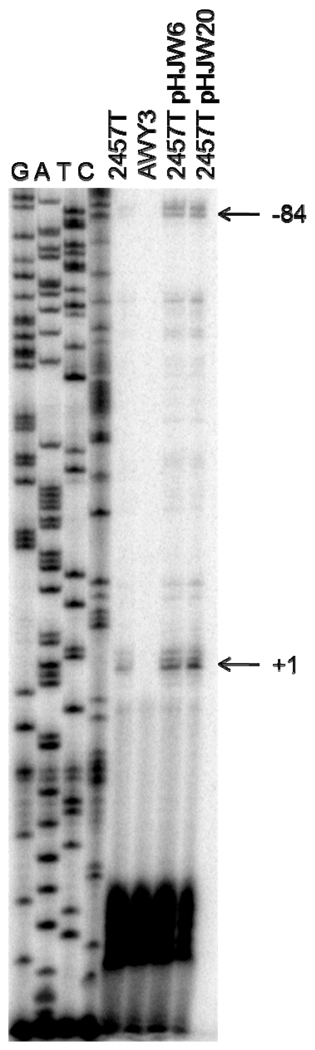
Given the discrepancy between the predicted and annotated TSSs, we wanted to experimentally determine the position of all TSSs using primer extension analysis (Fig. 2). Primer extension analyses were performed on mRNA isolated from the wild type S. flexneri 2a strain (2457T), the isogenic virB mutant (AWY3) and from wild type Shigella carrying either the icsP promoter and gene (pHJW6) or a PicsP-lacZ promoter fusion (pHJW20). Regardless of whether the icsP promoter was carried by the virulence plasmid (2457T) or a low copy cloning vector (pHJW6 & pHJW20), two products were observed, indicating that the icsP gene is transcribed from two promoters. The sizes of these two products indicate that TSSs occur at +1 and −84 relative to the previously annotated icsP TSS (Fig. 2). These data are in agreement with our in silico analyses (Fig. 1b) (Egile et al. 1997). We therefore designated the +1 and −84 TSSs and their accompanying promoter elements P1 and P2, respectively. Quantification of the primer extension products by densitometry indicate that the P2 signal is approximately half that of the P1 signal in all wild type strains. The increased signal intensity of products generated from strains carrying plasmids of the pHJW series was attributed to the copy number of these plasmids. In summary, these analyses identify two promoters involved in the transcriptional regulation of the icsP gene. We next wanted to examine how these two promoters are regulated.
Fig. 2.
Primer extension analysis of the virulence plasmid-encoded icsP gene (2457T), the virB mutant (AWY3), the icsP inducible plasmid (pHJW6), and the icsP-lacZ promoter fusion (pHJW20). The new TSS, P2, is identified with its relative location to the annotated TSS, P1. A sequencing reaction (first four lanes) was used to calibrate the gel. The experiment was repeated three times and representative data are shown.
Both icsP promoters are dependent upon the transcription factor VirB and distal VirB binding sites located over 1 kb upstream of both TSSs
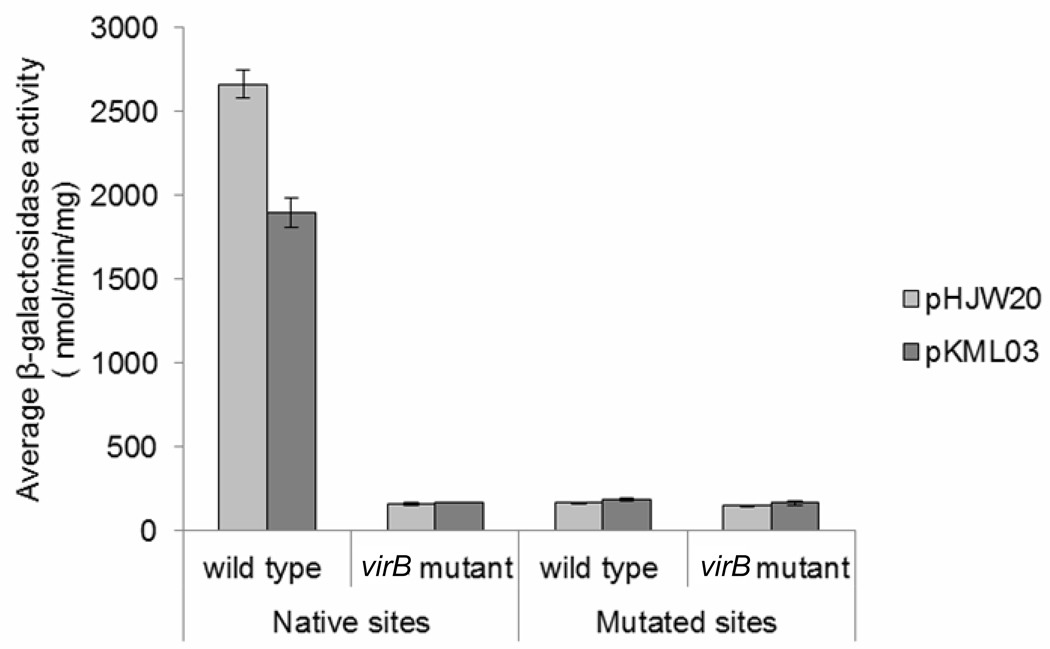
Previous work has demonstrated that icsP is regulated by the Shigella transcriptional regulator VirB (Wing et al. 2004). Two VirB binding sites located over 1 kb upstream of the originally annotated TSS was shown to be required for this VirB-dependent regulation (Castellanos et al. 2009).To determine the effect of VirB-dependent regulation on P1 and P2, β-galactosidase assays were conducted with a series of PicsP-lacZ transcriptional reporter plasmids containing either, P1 and P2 (pHJW20), P2 only (pKML03), or no promoter (pCTH03; Fig.1c), in wild type Shigella or a virB mutant background.
Our data show that P2 alone contributes approximately 70% of the total promoter activity in wild type cells. In contrast, in the virB mutant background the activity of both promoters is significantly decreased (Fig. 3), indicating the activity of both promoters is dependent upon VirB. To further test the role of the distal VirB binding sites on P1 and P2 regulation, base pair substitutions that completely abolish the previously annotated VirB binding sites (Castellanos et al. 2009) were introduced into each of the reporter plasmids and promoter activity was measured in a wild type and virB mutant background. In wild type cells both constructs carrying the mutated binding sites exhibited a significant reduction in icsP promoter activity, and this activity was similar to that observed in the virB mutant background (Fig. 3). Furthermore, our primer extension analysis revealed no detectable primer extension products in the Shigella virB mutant lane (Fig. 2, AWY3). Taken together, these data indicate that VirB acts as a transcriptional regulator for both icsP promoters and that both promoters require the presence of the two distal VirB binding sites to mediate this effect.
Fig. 3.
Activity of the wild type icsP promoter or a promoter carrying substitutions in the distal VirB binding sites (centered at – 1137, with respect to P1) in wild type S. flexneri (2457T) and the virB mutant (AWY3). Assays were run in triplicate and the means and standard deviations are shown.
Both promoters respond similarly to changes in phase of growth, iron concentration, pH, and osmotic pressure
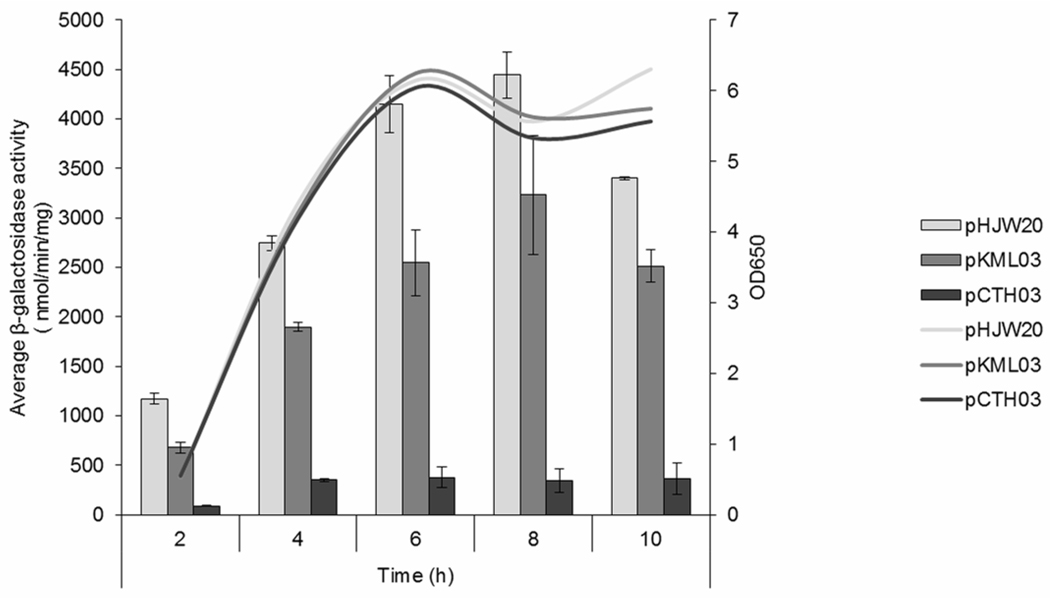
Multiple promoters often allow for the differential regulation of a single gene product in response to bacterial growth or environmental stimuli (Erickson et al. 1987, Raina et al. 1995). In Shigella, many virulence genes are regulated at the level of transcription by changes in temperature, pH, osmolarity, and iron concentration (Mitobe et al. 2009, Murphy and Payne 2007, Porter and Dorman 1997). The presence of two promoters upstream of the icsP gene raises the possibility that these promoters respond differently to growth phase and/or environmental cues. Although expression of VirB is known to be regulated by these environmental signals and VirB is required for transcription from both promoters as demonstrated here, we wanted to investigate whether changes in environmental conditions experienced by Shigella during colonization would allow for more refined control of icsP transcription through capitalization of the two-promoter architecture. Therefore, we examined the change in activities of the two promoters following a decrease in either, pH, osmolarity, or iron concentration using the pHJW20 and pKML03 constructs. A decrease in iron concentration, pH, or osmotic pressure all seemed to affect each promoter similarly, with P2 contributing between approximately 60–75% of the combined promoter activity (Table 2). Since icsA expression is regulated in a growth phase-dependent manner (Goldberg et al. 1994), we also conducted a time course assay to determine whether the relative activity of the two promoters varies with phase of growth. Our data show that both icsP promoters are maximally active during stationary phase and that P2, represented by pKML03, contributes approximately 59% (2 h) to 76% (10 h) of the combined promoter activity, represented by pHJW20 (Fig. 4). These data indicate that the relative contribution of P1 and P2 to overall activity of the icsP promoters remains constant under a variety of conditions, suggesting that the two icsP promoters do not appear to be differentially regulated, at least under the conditions tested here.
Table 2.
Contribution of P2 promoter activity to total icsP promoter activity
| Environmental parameter | Specific condition in Luria-Bertani medium | Percent contributiona |
|---|---|---|
| pH | 7.4 | 75.54 ± 6.96 |
| 5.5 | 61.88 ± 19.55 | |
| Iron concentration | Normal (0 µg ml-1 EDDA) | 69.86 ± 1.12 |
| Reduced (15 µg ml-1 EDDA) | 74.81 ± 5.48 | |
| Osmotic pressure | Normal NaCl concentration (LB) | 72.32 ± 0.07 |
| Half NaCl concentration (LO) | 67.20 ± 3.96 |
Average percent contributions are based upon the promoter activity of P2 (pKML03) compared to the total promoter activity (pHJW20). Assays were run in triplicate and the means and standard deviations are shown.
Fig. 4.
Activity of the icsP promoter constructs in response to growth phase. pHJW20 is a construct carrying both promoters, pKLM03 carries the P2 promoter alone, pCTH03 carries neither icsP promoter. Promoter activities were measured throughout growth (0–10 h) and optical densities of the cultures carrying each promoter construct are shown. The data is normalized using pMIC21, the promoterless lacZ gene. Assays were run in triplicate and the means and standard deviations are shown.
Transcription from the two icsP promoters allows for translation initiation from two sites
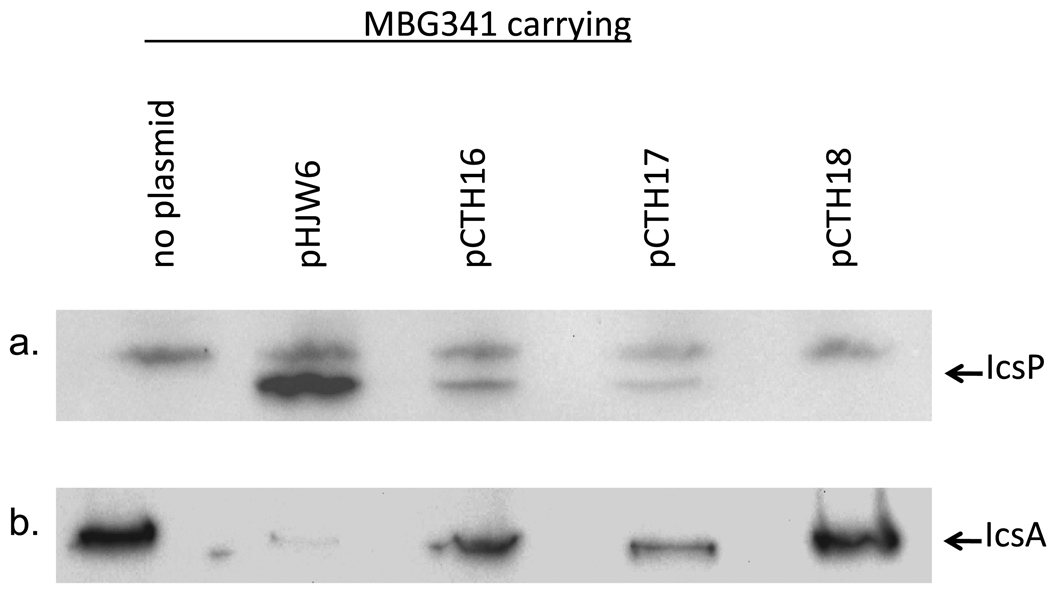
Our in silico analysis revealed the potential for an additional translation start site upstream and yet still in frame with the annotated translation start site (Fig. 1b). This second translation start site lies downstream of P2, raising the possibility that it is unique to P2 regulated transcripts. To examine whether this translation start site is used in the production of the mature, secreted IcsP protein, IcsP levels were measured in an icsP mutant carrying a low copy plasmid containing either both translation start sites (pHJW6), a single translation start site (pCTH16/downstream only or pCTH17/upstream only) or no translation start sites (pCTH18). Western blot analysis of whole cell protein preparations harvested from cells carrying each of these constructs, show that levels of the mature form of IcsP decreases when either the upstream or downstream translation start sites are eliminated (pHJW6 compared to pCTH16 and pCTH17 respectively) and are undetectable when translation from both sites is prevented (pCTH18). These data indicate that both translation start sites can be used to produce the mature form of IcsP (Fig. 5a). Furthermore, since IcsP is an outer membrane protease, which is secreted across the inner membrane via the general secretion pathway, our detection of mature IcsP in cells which lack either one translation start site or the other, strongly suggests that two isoforms of nascent IcsP are made and that each form is rapidly processed to a single mature form during secretion. This idea is supported by the signal peptide prediction program Signal P 3.0 (http://www.cbs.dtu.dk/services/SignalP) which predicts a single signal peptide cleavage site for each of the predicted, nascent IcsP isoforms (data not shown).
Fig. 5.
Amount and activity of IcsP produced from either the upstream or the downstream translation start site. IcsP levels measured in the icsP mutant (MBG341) or the icsP mutant carrying either pHJW6, pCTH16, pCTH17 or pCTH18 (a). Proteolytic activity of IcsP generated from either the upstream or the downstream translation start site, as judged by loss of IcsA. IcsA levels measured in the icsP mutant (MBG341) or the icsP mutant carrying either pHJW6, pCTH16, pCTH17 or pCTH18 (b). The same whole cell protein extracts were used for both the IcsP and IcsA blots. The experiment was repeated three times and representative data are shown.
Translation initiation from either start codon produces IcsP capable of proteolytically cleaving IcsA
Having established that two translation start sites are used in the production of IcsP, we next wanted to test whether the resulting IcsP proteins differed in their proteolytic activity, as judged by their cleavage of IcsA. To do this we probed the same protein cell extracts utilized for the IcsP analysis with an IcsA antibody. Our data show a reduction of the IcsA signal in all lanes except those containing cell extracts from the icsP mutant (MBG341) and 2457T pCTH18; a construct which lacks both translation start sites (Fig. 5b). This observed decrease in full length IcsA protein levels is consistent with cleavage of IcsA by IcsP. These data therefore suggest that regardless of which translation start site is used to make the IcsP protein, the resulting protease is capable of cleaving IcsA.
Putative translation start sites are commonly found around start codons of annotated open reading frames of the S. flexneri chromosome and virulence plasmid
Our studies of the icsP promoter and gene had revealed that two in frame translation start sites can each lead to the production of a mature IcsP protein. To address how frequently additional translation start sites are found associated with S. flexneri ORFs, we conducted a genome-wide scan of the S. flexneri genome. For this analysis we assumed that translation start sites consist of two main elements: the start codon and Shine-Dalgarno sequence or ribosome binding site (RBS).
First, a position specific scoring matrix (PSSM) for the RBS associated with annotated ORFs on the Shigella flexneri genome (both plasmid and chromosome) was generated. We decided to use a PSSM instead of a consensus, because these matrices capture commonly observed variations within the motif. The reverse complement to the consensus of the generated PSSM is 100% identical to the 3’ end of the Shigella flexneri 16S rRNA 5-'CTCCTT-3' and similar to the consensus SD sequence of E. coli, although offset by 1 nt: E. coli 5’–AGGAGG-3’; Shigella flexneri 5’ -AAGGAG-3’. Consequently, the generated logo (Online resource 1) is likely to be an accurate representation of the consensus RBS used in Shigella flexneri.
Second, to identify alternative translation start codons, fifty in-frame codons up and downstream of start codons of all annotated ORFs (both plasmid and chromosome) of S. flexneri 2a strain 301 were examined for presence of potential start codons (Jin et al. 2002, Wei et al. 2006). The position-specific scoring matrix (PSSM), specific for the RBS in S. flexneri 2a strain 301 (Online Resource 1), was then applied to 20 nucleotide stretches of sequence upstream of every potential start codon, to determine whether the identified potential alternative start codons were associated with RBS sequences,
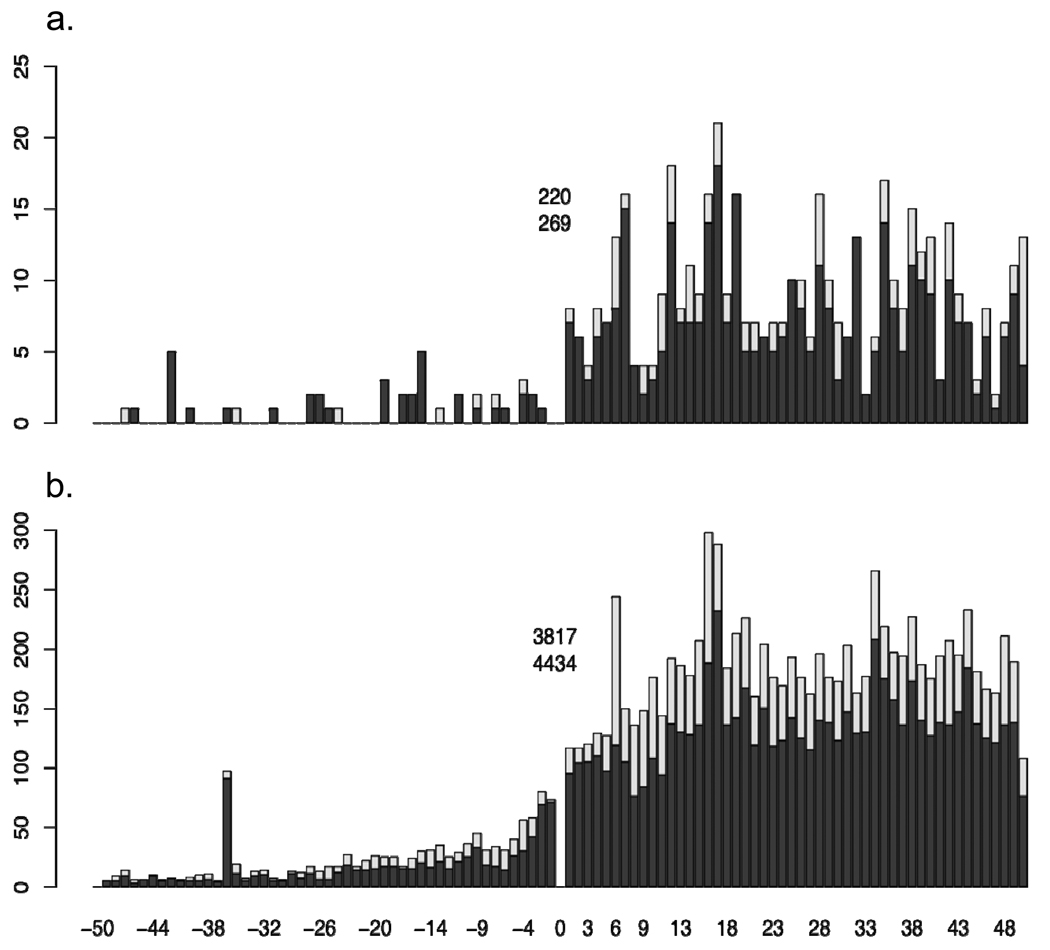
Our analysis revealed that out of the 4705 analyzed ORFs, 4165 have at least one additional putative start codon within a fifty codon range, up and downstream of the annotated start codon (raw data available upon request). In each case, the alternative translation start site lies in frame with the originally annotated start site and is not interrupted by a stop codon. For 2070 of the ORFs at least one of these alternative start codons is associated with a putative RBS (with a P value < 0.04). Figure 6A and B shows the distribution of alternative start codons and alternative translation start sites within the examined range of genomic DNA. The trend within the distribution is better defined among ORFs on the chromosome, because of the larger number of ORFs examined. The number of detected start codons quickly declines within intergenic spaces compared to those within the ORFs, due to present of in-frame stop codons, however local maximums of alternative translation start sites are located at codon 10, and between 35 and 40 codons upstream of the annotated translation start site. Within the coding regions, local maximums are found between codon 18–20 and at codon 35. The distribution of p-values for RBSs associated with annotated and alternative start codons are shown in Online Resource 2. It is clear from these values that some alternative translation start sites are associated with much stronger RBSs than the annotated translation starts. I still think we need a better conclusion here.
Fig. 6.
Distribution of putative start codons positioned up and downstream of the annotated start codon (0) in the S. flexneri virulence plasmid (a) and chromosome (b). The total number of predicted start codons are shown for each coordinate (light grey), the total number of predicted start codons associated with predicted Shine-Dalgarno sequences (dark grey). The inset values indicate the total number of annotated translation start sites (bottom) and annotated translation start sites associated with a predicted Shine-Dalgarno sequence (top).
Discussion
This work has identified a second TSS for the Shigella icsP gene. Transcription from this site ultimately allows translation to proceed from a newly identified translation start site located 33 bp upstream of the originally annotated translation start site. Our work demonstrates that regardless of which translation start site is used, a mature form of IcsP is made that is capable of proteolytically cleaving the Shigella actin based motility protein IcsA. It remains unclear whether the complex organization of the icsP regulatory region simply allows for transcriptional and translational redundancy or whether this organization allows for the exquisite control over icsP transcription and subsequent protein production in response to cellular and/or environmental cues.
Regulation from multiple promoters has been well documented in other bacterial systems and is usually found to contribute to the differential regulation of a single gene. While our experiments did not allow us to identify conditions which lead to the differential regulation of the two icsP promoters, we can now eliminate decreases in pH, osmolarity and iron concentration from other environmental conditions encountered in the human host, which have the potential to differentially regulate the two icsP promoters. Our data show that both icsP promoters are most active during stationary phase cultures. This pattern of expression is in agreement with the clearance of IcsA from the bacterial cell surface in stationary phase cultures (Goldberg et al. 1994) and the model for IcsP activity during pathogenesis, which proposes a build-up of IcsA on one pole of the bacterial outer membrane, maintenance of a tight polar cap of IcsA by removing IcsA that diffuse away from the pole through the activity of IcsP, and finally clearance of IcsA from the bacterial surface by IcsP (Goldberg et al. 1994, Steinhauer et al. 1999).
Another way multiple promoters can be differentially regulated is by the use of different transcription factors. Many virulence genes in Shigella are commonly regulated by the transcription regulator, VirB. Previous work, by us and others, has demonstrated that transcription of icsP is positively regulated by VirB (Castellanos et al. 2009, Le Gall et al. 2005, Wing et al. 2004). The work presented here reveals that both icsP promoters are positively regulated by VirB and that this regulation is mediated by a VirB binding site located over 1 kb upstream of the originally annotated TSS, because site directed mutagenesis of these binding sites reduce promoter activity of both P1 and P2 to level observed in virB mutant. VirB functions at Shigella promoters, including the icsP promoter, by alleviating transcriptional repression mediated by the nucleoid structuring protein H-NS, rather than by activating the promoter per se. Since the activity of P1 and P2 increases in the presence of VirB, both promoters are likely to be repressed by H-NS. Whether or not other DNA binding proteins interact with the long intergenic region upstream of the icsP promoter, and what role this has in the regulation of the two icsP promoters remains unclear at this stage, but this is an avenue of research investigation in our laboratory.
In bacterial genomes, alternative sigma factors sometimes allow the differential regulation of multiple promoters associated with a single gene. In our study the BPROM software failed to identify the previously annotated TSS in our analysis. This might indicate the possible use of an alternative sigma factor, even though both icsP promoters contain −35 sequences closely resembling the consensus for σ70-dependent regulation. Since the activity of both icsP promoters is maximal in stationary phase cultures, the most likely alternative sigma factor to be used to control icsP expression is the stationary phase sigma factor, σS. Despite maximal activity of the two icsP promoters in stationary phase cultures, neither P1 nor P2 contain the 5’-TGTGC-3’consensus sequence immediately upstream of the −10 element, which is a feature of promoters capable of binding σS (Gaal et al. 2001).
In all of our β-galactosidase assays the relative contribution of each icsP promoter to total icsP promoter activity remained similar, regardless of the conditions used; P2 promoter activity contributed approximately 60–75% of the total promoter activity. Nevertheless, in our primer extension analysis, the P2 generated transcripts contribute to approximately 33% of the total signal intensity from both P1 and P2. Interestingly, the reduced level of P2 versus P1 transcripts in our primer extension analyses is consistent with the decreased amount of IcsP protein when the upstream translation start is used versus the downstream translation start. Therefore it seems likely that the apparent discrepancy between the primer extension analysis and the β-galactosidase assay results is most likely caused by differences in the mRNA leader of the lacZ reporter constructs, although this has yet to be confirmed.
The organization of the icsP promoter region means that if transcription occurs from P1, translation can only occur from the downstream translation start site, whereas if transcription occurs from P2 then translation has the potential to start from either the upstream of downstream translation start site. This raises the question whether both translation start sites are used in a P2 transcript. Recent studies on polysome organization (Brandt et al. 2009) indicate that the distance between the two translation start sites in a P2 transcript (11 codons) is unlikely to allow simultaneous ribosome binding. This, along with the fact that our bioinformatics studies reveal no good match to the consensus Shine-Dalgarno sequence associated with the downstream translation start site, strongly suggests that the upstream translation start site would be favored in P2 transcripts, at least in the absence of accessory translation initiation factors.
Our work demonstrates that two translation start sites can be used to generate the mature and active form of IcsP. Since IcsP is secreted using the general secretion pathway, the two nascent isoforms must include an amino terminal signal sequence consisting of positively charged amino acids involved in protein targeting to the inner membrane for secretion (Fekkes and Driessen 1999). Interestingly, analyses of the two nascent isoforms of IcsP using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP), predicts that each isoform is likely to be cleaved at the same position, to release two different signal peptides, but the same mature IcsP protein. The extended positively charged amino terminus of the longer nascent IcsP protein, could allow for enhanced protein processing and translocation similar to the enhanced secretion of outer membrane proteins observed in E. coli (Akita et al. 1990). More efficient processing and translocation of the longer IcsP product may explain why the shorter IcsP protein product is expressed at a higher level but contributes to less IcsA cleavage as indicated by densitometry analyses of our IcsP and IcsA western blots.
While this study adds to our understanding of the icsP intergenic region and the role of this region plays in the regulation of the Shigella outer membrane protease, further investigation is needed to understand whether additional regulatory elements exist within this intergenic region and how these elements affect the production of IcsP and ultimately Shigella actin-based motility. Although the purpose of the second promoter and second translation start site remains unclear, our results suggest that production of the outer membrane protease IcsP may be more intricately regulated than previously thought.
The genome-wide screen for alternative translation start sites conducted within the present study, along with our observations at made at the icsP promoter, provides the first evidence that functional alternative in-frame translation start sites in the genome of S. flexneri 2a strain 301 may be a general phenomenon rather than something specific for icsP gene. Whether our observation is restricted to the genome of S. flexneri 2a strain 301 or is the general property of microbial genomes will require additional studies. It should be noted that recent work by Tucker and Escalante-Semerena (Tucker and Escalante-Semerena 2010), demonstrates that two translation start sites allow the production of two isoforms of CobB to be made from a single gene in Salmonella enterica and that these two isoforms have different biological activities. Our findings, in conjunction with these studies, imply that the use of alternative translation start sites may increase the size of the proteome and, in some instances, lead to a larger range of physiological functions being encoded by the bacterial genome than was previously acknowledged.
Materials and Methods
Bacterial strains, plasmids, and media
The bacterial strains and plasmids used in the present study are listed in Table 1. E. coli strains were grown at 37 °C in Luria-Bertani (LB) broth with aeration or on LB agar (LB broth containing 1.5% [wt/vol] agar). S. flexneri were grown at 37 °C in Trypticase Soy Broth (TSB) with aeration or on Trypticase Soy Agar (TSA) (TSB containing 1.5% [wt/vol] agar). Where appropriate, chloramphenicol was added at a final concentration of 25 µg ml−1. To ensure that Shigella strains had maintained the large virulence plasmid during manipulation, Congo red binding was tested on TSA plates containing 0.01% (wt/vol) Congo red (Sigma Chemical Co., St. Louis, Mo.).
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| S. flexneri | ||
| 2457T | S. flexneri serotype 2a | (Labrec et al. 1964) |
| AWY3 | 2457T virB:Tn5; Knr | (Wing et al. 2004) |
| MBG341 | 2457T icsP::Ampr | (Shere et al. 1997) |
| Plasmids | ||
| PicsP-lacZ reporters | ||
| pHJW20 | icsP promoter region transcriptionally fused to lacZ in pACYC184 Cmr; carries 1232 bp of wild-type sequence upstream of the icsP transcription start site and unique XbaI site upstream of lacZ gene | (Castellanos et al. 2009) |
| pHJW36 | pHJW20 lacking P2 promoter elements | (Castellanos et al. 2009) |
| pMIC18 | pHJW20 carrying 14 bp substitutions in the two upstream VirB-VirBbinding sites | (Castellanos et al. 2009) |
| pMIC21 | pHJW20 lacking all icsP promoter sequences | (Castellanos et al. 2009) |
| pKML03 | pHJW20 lacking previously annotated promoter elements | This work |
| pCTH02 | pKML03 carrying 14 bp substitutions in the two upstream VirB-binding sites | This work |
| pCTH03 | pHJW20 lacking P1 and P2 sequences | This work |
| PicsP-icsP reporters | ||
| pHJW6 | icsP promoter and gene cloned into pACYC184 | (Wing et al. 2004) |
| pCTH16 | pHJW6 lacking P2 specific promoter elements | This work |
| pCTH17 | pHJW6 with 4 bp substitutions in the downstream translation start site | This work |
| pCTH18 | pCTH17 lacking P2 specific promoter elements | This work |
Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Knr, kanamycin resistance.
Plasmid construction
The starting point for this work was the PicsP-lacZ reporter plasmids pHJW20 and pMIC18 (described in (Castellanos et al. 2009); Table 1). pHJW20 carries 1232 bps upstream of the TSS of the icsP promoter annotated by Egile et al. (Egile et al. 1997), the first 48 bp of the icsP coding region cloned upstream of a translation stop site, and a unique XbaI site upstream of a promoterless lacZ gene, so the expression of lacZ is directly regulated by the icsP promoter. pMIC18 is identical to pHJW20, but carries a 14 bp substitution that destroys the two upstream VirB-binding sites that are required for the VirB–dependent regulation of icsP (Castellanos et al. 2009).
To create pKML03, a truncated icsP promoter fragment was amplified from pHJW20 using oligonucleotides W93 (5’-TGGGTTGAAGGCTCTCAAGGGC-3’) and W123 (5’-TATTTTGCTCTAGATTTTAATTAAATATTTGTTTATGTTACC-3’). The PCR fragment was digested with PstI and XbaI, and the resulting DNA fragment was ligated into pHJW20 previously digested with PstI and XbaI. The resulting construct lacked the P1 TSS, and its −10 and −35 promoter elements, due to a 48 bp truncation at the 3’ end of the icsP promoter region. To create pCTH02, mutated VirB binding sites from pMIC18 were isolated on a PstI and PacI restriction fragment and introduced into pKML03 previously digested with PstI and PacI. The resulting construct therefore carried mutated, instead of wild-type VirB binding sites. To create pCTH03, a truncated icsP promoter fragment was amplified from pHJW20 using oligonucleotides W93 and W167 (5’-TATTTTGCTCTAGACCTCATTGTGCGAATAAAGTAACGG-3’). The PCR fragment was digested with BglII and XbaI, and the resulting DNA fragment was ligated into pHJW20 previously digested with BglII and XbaI. The resulting construct therefore lacked both P1 and P2, due to a 132 bp truncation at the 3’ end of the icsP promoter region.
To measure IcsP production and IcsP protease activity, the plasmid pHJW6 and its derivatives were used (described in (Wing et al. 2004); Table 1). pHJW6 is identical to pHJW20, but instead of carrying a PicsP-lacZ fusion this plasmid carries the full icsP coding region downstream of the icsP promoter region.
To create pCTH16, the sequence encoding the icsP gene was isolated from pHJW6 using PacI and BamHI restriction enzymes and used to replace the lacZ gene in pHJW36. pHJW36 lacks the −35 and part of the −10 promoter elements for P2 and this has been demonstrated to result in an inactive P2, as evidenced by i) primer extension analysis (unpublished data) and ii) the drop in total icsP promoter activity (Castellanos et al. 2009), consequently the newly formed construct pCTH16 could be used to measure IcsP protein production generated from P1 specific transcripts and hence the downstream translation start site. To create pCTH17, regions encoding a portion of the downstream Shine-Dalgarno sequence and the downstream methionine were mutated by introducing base pair substitutions in both sites using a QuikChange Lightning site-directed mutagenesis kit from Agilent Technologies and oligonucleotides W259 (5’-GTGCAAGTACAAAGAATTTTAATTTGAGCGAGAACTCGACTTTTTTGGTTGAAATGTCCATGA-3’) and W260 (5’-TCATGGACATTTCAACCAAAAAAGTCGAGTTCTCGCTCAAATTAAAATTCTTTGTACTTGCAC-3’). The substitutions used to disrupt the downstream translation start site were chosen to minimize the effect on the upstream translated protein product. Specifically the Shine-Dalgarno sequence was mutated from AAGTAG to AAGTCG, this resulted in the substitution of a valine codon for another valine codon, and the methionine codon ATG was mutated to a leucine codon CTC. The resulting amino acids have similar biochemical properties. To create pCTH18, pCTH17 was digested with PacI and BamHI to obtain the mutated sequence eliminating the downstream translation start site, and the resulting DNA fragment was ligated into pCTH16 previously digested with PacI and BamHI. The resulting construct consequently lacked the upstream translation start site and carried a mutated downstream Shine-Dalgarno and translation start site.
Quantification of icsP promoter activity using PicsP-lacZ reporters
Activity of the icsP promoters were determined by measuring β-galactosidase activity (as described previously (Castellanos et al. 2009) using the Miller protocol (Miller 1972)) in strains carrying pHJW20 or derivatives. Routinely, transcription was analyzed in three independent transformants in early stationary phase cultures. Cells were routinely back-diluted 1:100 and grown for 5 h in TSB, to ensure icsP expression. To measure the effects of growth phase on promoter activity, cells were grown for 2 to 10 h in 2 h intervals. To measure the effects of pH on promoter activity, cells were grown in LB with a pH of 5.5 buffered with a final concentration of 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) or a pH of 7.4 buffered with 100 mM 3-(N-morpholino)propanesulfonic acid (MOPS). To measure the effects of osmotic pressure on promoter activity, cells were grown in either LB or LO (Porter and Dorman 1994). To measure the effects of iron concentration on promoter activity, cells were grown in either LB or LB supplemented with 15 µg ml−1 EDDA to chelate iron. Optical densities were measured using a DU 520 general purpose UV/Vis spectrophotometer (Beckman Coulter). Promoter activity was normalized using pMIC21, the promoterless lacZ reporter construct.
Transcription start site mapping of the icsP gene
Transcription start sites of the icsP gene were identified through RNA extraction and primer extension analysis procedures as described previously (Wing et al., 1995) using a protocol adapted from Aiba (Aiba et al. 1981, Wing et al. 1995). Total cellular RNA was extracted using the hot-phenol method from 109 cells harvested from early stationary phase cultures (Aiba et al. 1981). Residual DNA within samples was digested with DNase I (Qiagen) at 37°C for 1 h in DNase I buffer according to Ambion instructions (Ambion 2001). Integrity of total RNA was checked by formaldehyde gel electrophoresis and ethidium bromide staining as described by Sambrook (Sambrook and Russell 2001). The oligonucleotide primer W183 (5’-AAAGTGCAAGTACAAAG-3’) was 5’-end-labeled with [γ-32P]ATP by using T4 polynucleotide kinase (Promega). One picomole of 32P-labeled primer and 5 µg of total RNA were lyophilized and redissolved in 30 µl of hybridization buffer (Aiba et al. 1981). The reaction was incubated at 75°C for 15 min followed by a cooling and incubation at 37°C for a total of 3 h. Following an ethanol precipitation, reverse transcription was completed using Superscript II reverse transcriptase (Invitrogen) according to manufacturer’s instructions. Remaining RNA was degraded with 10 mg/ml RNase A (Sigma) for 30 min at 37°C and the reaction terminated by ethanol precipitation. The precipitate was dissolved in 5 µl of loading dye (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol) and electrophoresed on a 6% glycerol tolerant polyacrylamide gel containing 7 M urea. Following electrophoresis, the gel was transferred to Whatman paper and then vacuum dried before overnight exposure to a phosphorescent screen. The screen was visualized the following morning using a Typhoon 9410 variable mode imager (Amersham). The sequencing ladder generated from pBluescript KSII+ (Stratagene) and a M13 reverse primer (5’-GAGCGGATAACAATTTCACACAGG-3’) with the Sequenase 2.0 kit (usb) according to manufacturer’s instructions, was used to size the primer extension products. Densitometry analysis was conducted using Vision Works LS image acquisition and analysis software (UVP).
Quantification of IcsP production and IcsA cleavage in Shigella
IcsP production and activity (IcsA cleavage) was measured by western blot analysis. Cells from early stationary phase cultures were harvested and whole-cell protein extracts were prepared as described previously (Steinhauer et al. 1999). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a 12.5% SDS-PAGE gel. Equivalent amounts of protein were loaded by normalizing the volume to cell density. Western blot analyses were performed with an affinity-purified IcsP or IcsA rabbit antiserum. The IcsP and IcsA antisera were raised against peptides sequences predicted to fall in surface exposed regions of the two proteins, the L3 loop of IcsP (based on the model of OmpT; (Vandeputte-Rutten et al. 2001) and the α-domain of IcsA, which is proteolytically cleaved from the surface of Shigella by IcsP. Each antibody was ultimately detected by chemiluminescence using a UVP BioSpectrum imaging system and accompanying software. Densitometry analysis was conducted as previously described.
In silico analyses of the icsP gene, its protein product and the position of translation start sites in Shigella open reading frames
Throughout our work sequences files were accessed and analyzed using the software program “Clone Manager 9 Basic Edition” (Scientific and Educational Software). Transcription start site predictions were performed using the BPROM program (http://linux1.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb). This algorithm predicts potential transcription start positions regulated by σ70 promoters (major E. coli promoter class). The linear discriminant function combines characteristics describing functional motifs and oligonucleotide composition of these sites. BPROM has 80% accuracy for E. coli σ70-dependent promoter recognition. Open reading frame predictions were performed using Glimmer (NCBI), which is a system for finding genes in microbial DNA using interpolated Markov models (IMMs) to identify the coding regions and distinguish them from noncoding DNA (Delcher et al. 1999). To predict the presence and location of proteolytic cleavage sites within nascent IcsP the SignalP 3.0 Server program (http://www.cbs.dtu.dk/services/SignalP/) was used (Emanuelsson et al. 2007). This algorithm predicts potential signal peptides and cleavage sites based on a combination of several artificial neural networks and hidden Markov models.
Bioinformatics screen for alternative translation start sites and associated Shine-Dalgarno sequences in S. flexneri 2a str. 301
Translation start sites were assumed to consist of two main components: a start codon and a Shine-Dalgarno sequence (Ribosome Binding Site, RBS). The full genome sequence of S. flexneri 2a str. 301 (chromosome and plasmid) as well as the most recent annotation was downloaded from GenBank FTP site (ftp://ftp.ncbi.nih.gov/genbank/genomes/Bacteria/Shigella_flexneri_2a/).
To identify position-specific scoring matrix (PSSM) for Shine-Dalgarno sequence specific for S. flexneri 2a str. 301, 20 nucleotides of sequence data upstream of the start codon of each annotated open reading frame (ORF) were extracted from the genome (chromosome and plasmid). Then retrieved regions of DNA were searched for overrepresented motifs using locally installed MEME program (Multiple EM for Motif Elicitation). The identified RBS motif was truncated to include only highly conserved positions (Online Resource 1). A motif sequence logo was created using an online program (http://weblogo.berkeley.edu/logo.cgi).
To identify presence of alternative translation start sites in genome of S. flexneri 2a str. 301, fifty codons upstream and downstream of start codon of every annotated ORF was tested for presence of possible in-frame start codon (ATGs or GTGs only), if an in-frame stop codon (TAG, TAA or TGA) was detected between annotated and alternative start codons further searches around this particular annotated start codon was terminated. 20 nucleotide sequences upstream every possible alternative start codon were collected, and then searched for presence of RBS using the program MAST (Motif Alignment & Search Tool) and determined before PSSM for RBS. Distance form annotated start site, p-value for motif presence (if RBS was found) and exact sequence of start codon were extracted. Data were manipulated, managed and graphically represented using custom R and Perl scripts.
Supplementary Material
Acknowledgements
We thank N. Ward and C. Ross for their assistance with this project. This study was supported by the National Institutes of Health (NIH) grant P20 RR-016464 from the IDeA Networks of Biomedical Research Excellence Program of the National Center for Research Resources grant, by the NIH grant R15 AI090573-01 and by UNLV start-up funds to H.J.W. K.M.L. was a 2009 Recipient of a Barry Goldwater Scholarship & C.T.H. is currently supported in part by the Post-9/11 GI Bill.
Footnotes
Online Resource 1 Position-specific scoring matrix for ribosome binding sites associated with the S. flexneri 2a strain 301 virulence plasmid pCP301 (NC_004851.1) and chromosome (NC_004337.1). Six nucleotide-long motif overrepresented in immediate upstream regions of annotated start codons identified by MEME (http://meme.sdsc.edu) and used to search for Shine-Dalgarno sequences upstream of predicted additional start codons.
Online Resource 2 Distribution of p-values for predicted ribosome binding sites with respect to both the annotated translation start sites (black) and the predicted additional translation start sites (red).
References
- Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 [PubMed] [Google Scholar]
- Akita M, Sasaki S, Matsuyama S, Mizushima S. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J Biol Chem. 1990 [PubMed] [Google Scholar]
- Ambion . TechNotes Newsletter. DNase I Demystified. [Google Scholar]
- Bernardini ML, Mounier J, d'Hauteville H, Coquis-Rondon M, Sansonetti PJ. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc Natl Acad Sci U S A. 1989 doi: 10.1073/pnas.86.10.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997 doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, Baumeister W. The native 3D organization of bacterial polysomes. Cell. 2009 doi: 10.1016/j.cell.2008.11.016. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- Castellanos MI, Harrison DJ, Smith JM, Labahn SK, Levy KM, Wing HJ. VirB Alleviates H-NS Repression of the icsP Promoter in Shigella flexneri from Sites More Than One Kilobase Upstream of the Transcription Start Site. J Bacteriol. 2009 doi: 10.1128/JB.00313-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999 doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Hauteville H, Dufourcq Lagelouse R, Nato F, Sansonetti PJ. Lack of cleavage of IcsA in Shigella flexneri causes aberrant movement and allows demonstration of a cross-reactive eukaryotic protein. Infect Immun. 1996 doi: 10.1128/iai.64.2.511-517.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman CJ, Porter ME. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol Microbiol. 1998 doi: 10.1046/j.1365-2958.1998.00902.x. [DOI] [PubMed] [Google Scholar]
- Egile C, d'Hauteville H, Parsot C, Sansonetti PJ. SopA, the outer membrane protease responsible for polar localization of IcsA in Shigella flexneri. Mol Microbiol. 1997 doi: 10.1046/j.1365-2958.1997.2871652.x. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007 doi: 10.1038/nprot.2007.131. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Erickson JW, Vaughn V, Walter WA, Neidhardt FC, Gross CA. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1987 doi: 10.1101/gad.1.5.419. [DOI] [PubMed] [Google Scholar]
- Fekkes P, Driessen AJ. Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Biol Rev. 1999 doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MB, Theriot JA, Sansonetti PJ. Regulation of surface presentation of IcsA, a Shigella protein essential to intracellular movement and spread, is growth phase dependent. Infect Immun. 1994 doi: 10.1128/iai.62.12.5664-5668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MB, Barzu O, Parsot C, Sansonetti PJ. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. Infect Agents Dis. 1993 [PubMed] [Google Scholar]
- Jacques N, Dreyfus M. Translation initiation in Escherichia coli: old and new questions. Mol Microbiol. 1990 doi: 10.1111/j.1365-2958.1990.tb00679.x. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yuan Z, Xu J, Wang Y, Shen Y, Lu W, Wang J, Liu H, Yang J, Yang F, Zhang X, Zhang J, Yang G, Wu H, Qu D, Dong J, Sun L, Xue Y, Zhao A, Gao Y, Zhu J, Kan B, Ding K, Chen S, Cheng H, Yao Z, He B, Chen R, Ma D, Qiang B, Wen Y, Hou Y, Yu J. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 2002 doi: 10.1093/nar/gkf566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrec EH, Schneider H, Magnani TJ, Formal SB. Epithelial Cell Penetration as an Essential Step in the Pathogenesis of Bacillary Dysentery. J Bacteriol. 1964 doi: 10.1128/jb.88.5.1503-1518.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall T, Mavris M, Martino MC, Bernardini ML, Denamur E, Parsot C. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology. 2005 doi: 10.1099/mic.0.27639-0. doi: 10.1099/mic.0.27639-0. [DOI] [PubMed] [Google Scholar]
- Makino S, Sasakawa C, Kamata K, Kurata T, Yoshikawa M. A genetic determinant required for continuous reinfection of adjacent cells on large plasmid in S. flexneri 2a. Cell. 1986 doi: 10.1016/0092-8674(86)90880-9. [DOI] [PubMed] [Google Scholar]
- Miller J. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Mitobe J, Morita-Ishihara T, Ishihama A, Watanabe H. Involvement of RNA-binding protein Hfq in the osmotic-response regulation of invE gene expression in Shigella sonnei. BMC Microbiol. 2009 doi: 10.1186/1471-2180-9-110. doi: 10.1186/1471-2180-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy ER, Payne SM. RyhB, an iron-responsive small RNA molecule, regulates Shigella dysenteriae virulence. Infect Immun. doi: 10.1128/IAI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Dorman CJ. Differential regulation of the plasmid-encoded genes in the Shigella flexneri virulence regulon. Mol Gen Genet. 1997 doi: 10.1007/s004380050550. [DOI] [PubMed] [Google Scholar]
- Porter ME, Dorman CJ. A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J Bacteriol. 1994 doi: 10.1128/jb.176.13.4187-4191.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A. 2000 doi: 10.1073/pnas.180094797. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 1995 doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning : a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sansonetti PJ. Molecular and cellular mechanisms of invasion of the intestinal barrier by enteric pathogens. The paradigm of Shigella. Folia Microbiol (Praha) 1998 doi: 10.1007/BF02818608. [DOI] [PubMed] [Google Scholar]
- Shere KD, Sallustio S, Manessis A, D'Aversa TG, Goldberg MB. Disruption of IcsP, the major Shigella protease that cleaves IcsA, accelerates actin-based motility. Mol Microbiol. 1997 doi: 10.1046/j.1365-2958.1997.4681827.x. [DOI] [PubMed] [Google Scholar]
- Steinhauer J, Agha R, Pham T, Varga AW, Goldberg MB. The unipolar Shigella surface protein IcsA is targeted directly to the bacterial old pole: IcsP cleavage of IcsA occurs over the entire bacterial surface. Mol Microbiol. 1999 doi: 10.1046/j.1365-2958.1999.01356.x. [DOI] [PubMed] [Google Scholar]
- Tucker AC, Escalante-Semerena JC. Biologically Active Isoforms of CobB Sirtuin Deacetylase in Salmonella enterica and Erwinia amylovora. J Bacteriol. 2010 doi: 10.1128/JB.00874-10. doi: 10.1128/JB.00874-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte-Rutten L, Kramer RA, Kroon J, Dekker N, Egmon MR, Gros P. Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. EMBO J. 2001 doi: 10.1093/emboj/20.18.5033. doi: 10.1093/emboj/20.18.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Yang J, Zhu J, Zhang X, Leng W, Wang J, Xue Y, Sun L, Li W, Wang J, Jin Q. Comprehensive proteomic analysis of Shigella flexneri 2a membrane proteins. J Proteome Res. 2006 doi: 10.1021/pr0601741. doi: 10.1021/pr0601741. [DOI] [PubMed] [Google Scholar]
- Wing HJ, Goldman SR, Ally S, Goldberg MB. Modulation of an outer membrane protease contributes to the virulence defect of Shigella flexneri strains carrying a mutation in the virK locus. Infect Immun. 2005 doi: 10.1128/IAI.73.2.1217-1220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing HJ, Yan AW, Goldman SR, Goldberg MB. Regulation of IcsP, the outer membrane protease of the Shigella actin tail assembly protein IcsA, by virulence plasmid regulators VirF and VirB. J Bacteriol. 2004 doi: 10.1128/JB.186.3.699-705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing HJ, Williams SM, Busby SJ. Spacing requirements for transcription activation by Escherichia coli FNR protein. J Bacteriol. 1995 doi: 10.1128/jb.177.23.6704-6710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.