Abstract
Background. The Maraviroc versus Optimized Therapy in Viremic Antiretroviral Treatment-Experienced Patients (MOTIVATE) studies compared maraviroc versus placebo in treatment-experienced patients with CCR5-using (R5) human immunodeficiency virus type 1 (HIV-1), screened using the original Trofile assay. A subset with non-R5 HIV infection entered the A4001029 trial. We retrospectively examined the performance of a genotypic tropism assay based on deep sequencing of the HIV env V3 loop in predicting virologic response to maraviroc in these trials.
Methods. V3 amplicons were prepared from 1827 screening plasma samples and sequenced on a Roche/454 GS-FLX to a depth of >3000 sequences/sample. Samples were considered non-R5 if ≥2% of their viral population scored greater than or equal to −4.75 or ≤3.5 using the PSSMx4/R5 or geno2pheno algorithms, respectively.
Results. Deep sequencing identified more than twice as many maraviroc recipients as having non-R5 HIV, compared with the original Trofile. With use of genotyping, we determined that 49% of maraviroc recipients with R5 HIV at screening had a week 48 viral load <50 copies/mL versus 26% of recipients with non-R5. Corresponding percentages were 46% and 23% with screening by Trofile. In cases in which screening assays differed, median week 8 log10 copies/mL viral load decrease favored 454. Other parameters predicted by genotyping included likelihood of changing to non-R5 tropism.
Conclusions. This large study establishes deep V3 sequencing as a promising tool for identifying treatment-experienced individuals who could benefit from CCR5-antagonist–containing regimens.
Human immunodeficiency virus type 1 (HIV-1) enters and infects a target cell by an interaction of its envelope glycoprotein, gp120, with the cellular CD4 receptor and a co-receptor: CCR5 or CXCR4 [1–4]. CCR5 antagonists, such as maraviroc, inhibit HIV entry via CCR5. These agents work by allosterically altering the conformation of CCR5 at the cell surface, thereby disrupting its interaction with HIV gp120 [1, 5, 6]. However, they have suboptimal activity against viral populations capable of using CXCR4 [7, 8]. Accordingly, before clinical use of CCR5 antagonists, a tropism test is performed to rule out the presence of detectable non–CCR5-tropic (non-R5) virus.
Currently, the most widely used co-receptor tropism tests are the recombinant phenotypic Trofile assay (Monogram Biosciences) [9] and its newer iteration, the Enhanced Sensitivity Trofile assay (ESTA) [10]. Despite their widespread use, there are some practical limitations to these assays, including a long turnaround time, restricted geographic access, and the large sample volume that they require [11]. Genotypic tropism testing is an alternative method [12] that is possible because the sequence of the third variable (V3) loop of HIV gp120 is the principal determinant of tropism [13–18], allowing tropism inference using bioinformatic algorithms, such as PSSMx4/R5 [19] and geno2pheno[coreceptor] (g2p) [20].
However, genotypic assays that are based on standard, population-based V3 sequencing have often had apparently poor sensitivity for detection of non-R5 HIV [21], especially when such species comprise minorities in the viral population below ∼20%, which is the reliable sensitivity of standard sequencing [22, 23]. In comparison, next-generation deep-sequencing approaches have much higher sensitivity and can detect minority HIV variants at much lower levels [24, 25], including minority non-R5 subpopulations [26]. Consequently, this method can capture a detailed cross-section of co-receptor use across a patient's viral population and quantify the prevalence of non-R5 HIV within the patient.
Here, we present an extensive study of deep V3 sequencing as a tool for predicting virologic outcomes on maraviroc-based therapy in treatment-experienced patients in the Maraviroc versus Optimized Therapy in Viremic Antiretroviral Treatment-Experienced Patients (MOTIVATE) 1 and 2 studies. These were placebo-controlled, phase-3 studies of maraviroc in treatment-experienced patients with R5 HIV infection [27, 28]. Patients were originally screened using the original Trofile assay. Of those screened out because of non-R5 HIV, ∼20% (186 of 955) entered the A4001029 trial [8, 28]. We retrospectively tested this method on a total of 1827 blinded screening samples from these 3 clinical trials and assessed its ability to predict virologic responses in maraviroc recipients.
METHODS
Trial Patients, Samples, and Polymerase Chain Reaction (PCR) Methods
Briefly, the V3 loop of HIV gp120 was amplified independently in triplicate by nested reverse-transcriptase PCR (RT-PCR) methods from a total of 1827 screening samples from the 3 trials. These were then sequenced by either standard population-based sequencing [29] or by deep sequencing [30]. The current study focuses on the deep-sequencing data, hereafter referred to as genotyping data.
In total, 1093 of 1827 patients examined in the current study were randomized into the 3 arms of the MOTIVATE (R5) and A4001029 (non-R5) trials (Supplementary data figure 1). Informed consent was obtained from all individuals. Treatment arms were maraviroc once-daily, maraviroc twice-daily, or placebo, plus an optimized background therapy of 3–6 agents, based on treatment history and resistance testing [27, 28]. Note that all phenotypic screening results were obtained using the original Trofile assay (∼10% non-R5 prevalence detection limit [31]) and not the currently used ESTA (0.3% detection limit [10]).
Our main analysis was based on all patients who entered any study (MOTIVATE-1, MOTIVATE-2, or A4001029). Critically, this included all treated patients whose Trofile screening identified them as having non-R5 infection. For additional analyses with respect to tropism assessments by both assays, the patients screened for MOTIVATE-1 (including patients identified by Trofile as having non-R5 infection) but who did not enter a study were also included. However, only patients entering the studies could be examined for virologic responses.
HIV RNA was extracted from 500 μL of plasma per sample using a NucliSENS easyMAG (bioMérieux). Three independent 1-step RT-PCR amplifications were performed with 4 μL of extract per amplification, followed by a second-round amplification using customized primers that included a V3-specific PCR primer and a multiplex barcode (for distinguishing between samples). All primers and thermocycler protocols used are listed in Supplementary Methods.
Emulsion PCR and Pyrosequencing
After PCR amplification, PCR amplicon concentrations were quantified using a Quant-iT Picogreen dsDNA Assay Kit (Invitrogen) and a DTX-880 Multimode Detector (Beckman Coulter). These were combined in equal proportions (2 × 1012 DNA molecules/amplification), purified with Agencourt Ampure PCR Purification beads (Beckman Coulter), and re-quantified. The purified products were then diluted to 2 × 105 molecules/mL and combined at a ratio of 0.6 molecules to 1 emulsion PCR (emPCR) microbead. Oil and emPCR buffer components were shaken with a TissueLyser (Qiagen/Retsch) to allow formation of microreactor micelles around the beads. After emPCR, the beads were washed and enriched for DNA-coated beads in accordance with the manufacturer's instructions. The beads were added onto a picotiter plate at 250,000 beads in each of 4 regions and underwent sequencing with a Genome Sequencer-FLX (Roche/454 Life Sciences).
Bioinfomatic Algorithms for Inferring Tropism from Genotypic Data
Deep sequencing generated read-lengths of ∼250 base-pairs of data in each direction. A typical V3 loop was 105 base-pairs long (35 amino acids). Truncated reads (missing ≥4 bases at either end of V3) were excluded from the analysis, as were samples producing <750 usable reads. Genotyping generated a mean of >3000 V3-sequences/sample. The tropism of each sequence was interpreted by the PSSMx4/R5 or g2p bioinformatic algorithms (optimized cutoff values for non-R5: PSSMx4/R5, greater than or equal to −4.75; g2p, ≤3.5). The overall sample tropism was expressed as the proportion of non-R5 sequences within the sample's viral population. Patients with samples harboring ≥2% non-R5 variants were classified as having non-R5 virus, whereas those with <2% non-R5 variants were classified as having R5 virus. These cutoff values were established by optimizing to week 8 virologic response in a random 75% of the dataset and testing on the remaining 25% [32]. Results for all patients are presented in the text. This 2% cutoff value also approaches the likely level of reproducibility for PCR-based methods. Detailed methodological analyses of this deep-sequencing method will be presented elsewhere.
Population-Based Sequencing as a Comparator
Additional second-round PCR amplifications were also performed prior to standard population-based sequencing on an ABI 3730XL DNA analyzer, according to previously described methods [29]. The cutoff values used were −4.25 for PSSMx4/R5 and 5.75 for g2p [32, 33]. An extensive evaluation of the performance of population-based V3 sequencing will be published elsewhere.
Data Analysis
Where data were missing for patients enrolled in the clinical trials, the previous observation was carried forward, except for the analysis of the proportion of patients with plasma viral load (pVL) <50 copies/mL, where a missing result was considered to be >50 copies/mL. Data from MOTIVATE-1, MOTIVATE-2, and A4001029 were pooled, and both maraviroc arms were combined into a single group. Analyses were restricted to patients with tropism results from both assays. Clinical parameters examined included the following: the median change in log10-transformed HIV pVL from baseline, the proportion of patients with a pVL <50 HIV RNA copies/mL, and time to a tropism switch. The performance of genotyping on these parameters was compared against that of the original Trofile assay. Our results could not be compared with ESTA response rates in this population, because ESTA results were not obtained from patients in these studies.
RESULTS
Tropism Screening by Deep Sequencing Relative to Trofile and Population-Based Sequencing
Overall, genotyping identified 1037 samples (57%) as R5 and 790 (43%) as non-R5 using the PSSMx4/R5 algorithm. PSSM and g2p had ∼90% concordance with one another. For ease of presentation, results will be shown for the PSSMx4/R5 algorithm. When screened with Trofile, 1141 samples (62%) were designated as R5 and 686 (38%) were designated as non-R5 (Dual-/Mixed-tropic or X4). Global concordance of genotyping with Trofile-defined tropism was 82%. Detailed analyses for g2p as well as the PSSMSI/NSI algorithm were largely similar to analyses for PSSMx4/R5 and are presented in Supplementary data table 1.
Of the 686 samples identified as non-R5 by Trofile, 575 (84%) were also identified as non-R5 by genotyping. An additional 215 samples that were not identified as non-R5 by Trofile were identified as non-R5 by genotyping. Using Trofile as a reference, sensitivity of genotyping was 84%, and specificity was 81%. Using genotyping as a reference, the sensitivity of the original Trofile assay was 73%, and specificity was 89%. Comparing population-based sequencing against deep sequencing, overall concordance was 80%, with 64% sensitivity and 93% specificity. The sensitivity of both Trofile and population-based sequencing was lower when the proportion of non-R5 variants in the viral population was lower according to deep sequencing (Figure 1).
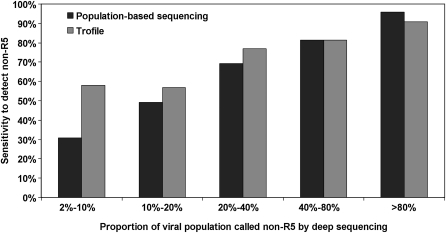
Figure 1.
Sensitivity of population-based V3 sequencing and original Trofile assay with 454 genotyping as the reference. The ability of the population-based V3 sequencing (black bars) and original Trofile (gray bars) to identify screening samples as having non–CCR5-using (non-R5) virus that 454 genotyping had identified as having ≥2% non-R5 virus, stratified by different proportions of non-R5 virus identified in the 454 genotyping result. Both alternative assays seemed to have decreased sensitivity for non-R5 human immunodeficiency virus (HIV) when such variants were present at lower proportions of the viral population. When non-R5 virus was present at 2%–10% according to 454 genotyping, 31% (69 of 224) and 58% (130 of 224) of the samples were also identified as having non-R5 virus by population-based sequencing and Trofile, respectively. These were 49% (57 of 116) and 57% (66 of 116) in the 10%–20% group; 69% (81 of 117) and 77% (90 of 117) in the 20%–40% group; 81% (118 of 145) and 81% (118 of 145) in the 40%–80% group; and 96% (180 of 188) and 91% (171 of 188) in the >80% group.
Of interest, deep sequencing detected at least some non-R5 HIV in >90% of patients (1700 of 1827 patients), regardless of tropism classification. Samples with R5 HIV by Trofile had a median of 0.1% non-R5 variants at screening according to deep-sequencing results. However, non-R5 levels <2% likely have low reproducibility and should be considered with caution. The Trofile non-R5 group excluding dual/mixed samples (ie, only “pure” X4 by Trofile; n = 39) had a median of 93% non-R5 virus.
Patients with R5 virus by Trofile but non-R5 virus by genotyping had 12% non-R5 HIV present at screening (n = 167), which was much higher than results obtained when both assays indicated R5: 0.1% (n = 926). This suggests that the original Trofile assay did not reliably detect patients with low-level non-R5 variants present at screening (Figure 1), consistent with the finding that 8% of patients had non-R5 results at baseline [27], and consistent with ESTA results for the Maraviroc versus Efavirenz Regimens as Initial Therapy (MERIT) trial of maraviroc [34].
Of the 1827 patients screened, 1093 actually entered the maraviroc (n = 851) or placebo (n = 242) arms of the trials. Baseline characteristics of patient groups screened by both methods are presented in Table 1. The R5 groups by either method were similar in terms of baseline pVL and CD4+ cell count, as were the non-R5 groups. Among maraviroc recipients, genotyping identified over twice as many patients as being unlikely to respond to maraviroc, compared with Trofile (n = 240 vs 111).
Table 1.
Baseline Characteristics of Treated Population, Stratified by Tropism Status by Genotype and Trofile
| Variable | Geno R5(n = 775) | Trofile R5 (n = 925) | Geno non-R5(n = 318) | Trofile non-R5(n = 168) |
| Baseline pVL, median log10 HIV RNA copies/mL | 4.85 | 4.88 | 5.04 | 5.07 |
| CD4+ cell count, median cells/mm3 | 177 | 168 | 72 | 54 |
| Non-R5 variants in deep-sequencing screening result, median % (IQR) | 0.1(0.0–0.2) | 0.1(0.0–0.7) | 19(7.0–54.0) | 28(7.0–63.0) |
NOTE. Geno non-R5, identified as having non-R5 virus by genotyping; Geno R5, identified as having R5 virus by genotyping; HIV, human immunodeficiency virus; IQR, interquartile range; pVL, plasma viral load; R5, CCR5-using virus; Trofile non-R5, identified as having non-R5 virus by Trofile assay; Trofile R5, identified as having R5 virus by Trofile assay.
Early Virologic Response to Maraviroc
Screening genotype was a predictor of response to maraviroc-based antiretroviral therapy in treatment-experienced patients. Maraviroc recipients found to have R5 HIV by genotyping had consistently better virologic outcomes than did those found to have non-R5 HIV. Using a number of parameters, genotyping performed similarly to or marginally out-performed the original Trofile assay in predicting virologic response. Virologic performance was slightly better in the maraviroc twice-daily arm, compared with the maraviroc once-daily arm (data not shown), but these arms have been pooled to simplify the presentation of the results.
The median week 8 pVL change from baseline was examined to minimize the number of patients who had discontinued the study because of reasons such as treatment failure or loss-to-follow-up but who were receiving the drug for a sufficient time to measure the efficacy of maraviroc. Maraviroc recipients who were found to have R5 virus by genotyping had a combined median week 8 decrease in pVL from baseline of 2.4 log10 copies/mL (interquartile range [IQR], 1.7–2.9 log10 copies/mL; n = 611), which was approximately twice as large as the 1.4 log10 copies/mL decrease (IQR, 0.2–2.7 log10 copies/mL; n = 240) for patients classified as having non-R5 virus by genotyping. Results in which missing patients were censored or where data were restricted to only those screened for MOTIVATE-1 were largely similar (data not shown).
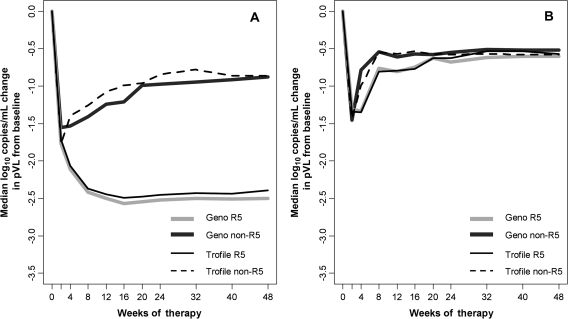
Using Trofile, the corresponding results were similar: 2.4 log10 copies/mL (IQR, 1.3–2.8 log10 copies/mL; n = 740) for patients with R5 virus versus 1.3 log10 copies/mL (IQR, 0.3–2.7 log10 copies/mL; n = 111) for patients with non-R5 virus. For placebo recipients, the week 8 pVL decreases were modest (0.5–.8 log10 copies/mL) and similar regardless of genotypic tropism. Median pVL responses for patients who received maraviroc and those who received placebo over the course of the studies are shown in Figures 2A and 2B, respectively.
Figure 2.
A, Median change in plasma viral load (pVL) from baseline in the maraviroc arms. Patients identified at screening as having CCR5-using (R5) virus by either genotyping or Trofile had much larger median decreases in pVL from baseline, relative to patients identified at screening as having non-R5 virus. Light gray and dark gray lines correspond to deep-sequencing R5 (n = 611) and non-R5 (n = 240) groups, respectively, whereas thinner solid black and dotted black lines correspond to Trofile R5 (n = 740) and non-R5 (n = 111) groups. B, Median change in pVL from baseline in the placebo arms. The pVL decreases from baseline for patients receiving placebo were similar to those for maraviroc-receiving patients identified as having non-R5 virus, and were small regardless of screening tropism or assay used. Light gray and dark gray lines correspond to deep sequencing R5 (n = 164) and non-R5 (n = 78) groups, respectively, whereas thinner solid black and dotted black lines correspond to Trofile R5 (n = 185) and non-R5 (n = 57) groups.
Longer-Term Virologic Efficacy
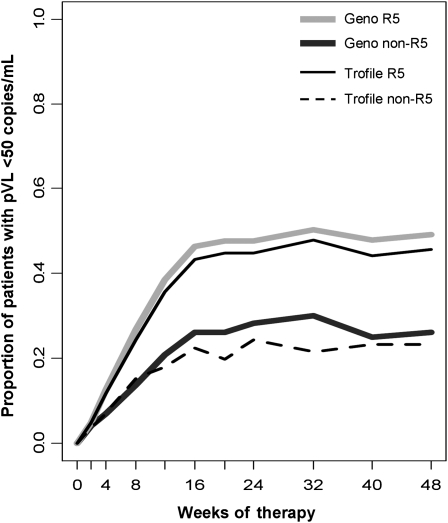
The efficacy of maraviroc in patients identified by genotyping as having R5 virus was sustained to week 48 (Figure 3). These patients were more likely to achieve a pVL <50 copies/mL at week 48, compared with the non-R5 group. In total, 49% (301 of 611) of the R5 group and 26% (62 of 240) of the non-R5 group had virologic suppression at week 48 when screened by genotyping. By Trofile, these were 46% (337 of 740) and 23% (26 of 111), respectively.
Figure 3.
Percentage of patients with plasma viral load (pVL) <50 human immunodeficiency virus (HIV) RNA copies/mL in maraviroc arms. A higher proportion of patients identified at screening by either method as having CCR5-using (R5) virus had a pVL <50 HIV RNA copies/mL, compared with the patients with non-R5 virus. Light gray and dark gray lines correspond to deep-sequencing R5 (n = 611) and non-R5 (n = 240) groups, respectively, whereas thinner solid black and dotted black lines correspond to Trofile R5 (n = 740) and non-R5 (n = 111) groups. Geno, genotypic tropism result by deep sequencing.
The genotypic non-R5 group could be divided roughly in half, with 127 patients having low-prevalence (2%–20%) non-R5 virus and 113 patients having >20% non-R5 virus. The group of patients with 2%–20% non-R5 virus according to deep sequencing had minority non-R5 variants that were not reliably detected by standard population-based sequencing methods (Figure 1). Importantly, this group of patients had poor response to maraviroc, with 27% (34 of 127) of the patients achieving virologic suppression at week 48, similar to the non-R5 group as a whole (26%) and to patients with >20% non-R5 virus (25%; 28 of 113). The rate of virologic response among placebo recipients was similar to that among maraviroc recipients identified as having non-R5 HIV, ranging from 17% to 23% depending on tropism or assay.
Interestingly, the virologic outcomes for maraviroc recipients showed a general inverse relationship with the percentage of non-R5 virus present at screening according to genotyping. Patients with 0% non-R5 virus had the greatest success, showing a median week 8 pVL decrease of 2.6 log10 copies/mL, with 65% (58 of 89) of the patients having week 48 virologic suppression. Patients with 0%–1% non-R5 virus had slightly poorer outcomes, with a median week 8 pVL decrease of 2.4 log10 copies/mL and a 48% rate of virologic suppression (234 of 491 patients). This decreased again in patients with 1%–2% non-R5 virus, who had a median decrease of 2.1 log10 copies/mL and a 29% rate of virologic suppression (9 of 31 patients). Patients with >2% non-R5 virus (ie, the group identified as having non-R5 virus by genotyping) all showed similar low virologic responses, as detailed above.
Changes in Viral Tropism
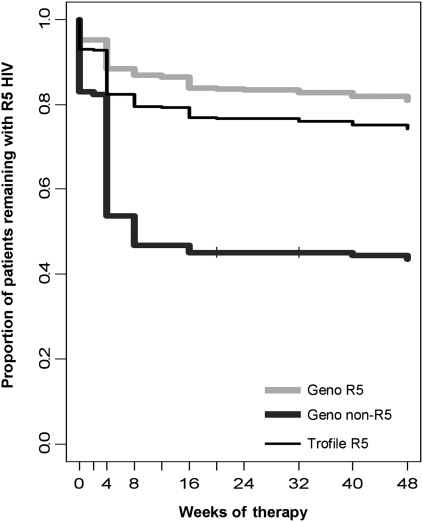
As a separate endpoint, patients were analyzed according to whether they experienced a change in their Trofile result from R5 to non-R5 (a tropism “switch”) over the course of the studies (Figure 4). This parameter is both clinically relevant for maraviroc-based therapy and functioned as a measure separate from changes in pVL. Among those patients originally identified as having R5 virus by Trofile screening, those identified as having non-R5 virus by genotyping were almost twice as likely to have non-R5 HIV emerge by week 24, compared with patients identified as having R5 virus by both methods. A total of 40% (72 of 180) of the maraviroc recipients who switched tropism were identified by genotyping as having ≥2% non-R5 virus at screening. Tropism switches occurred in 18% (111 of 612) of the patients identified as having R5 virus by genotyping, which was lower than the rate among those identified as having R5 virus by Trofile alone (25%; 180 of 724 patients).
Figure 4.
Time to change in tropism from CCR5-using (R5) to dual/mixed (DM) or X4 virus in maraviroc arms. The change in tropism in the maraviroc arms, where all patients had R5 virus identified at screening by Trofile and switched tropism to DM or X4 over the course of the studies, according to the Trofile assay. Light gray and dark gray lines correspond to genotyping R5 (n = 605) and non-R5 (n = 135) groups, respectively, whereas the thinner solid black line corresponds to the Trofile R5 (n = 740) group. HIV, human immunodeficiency virus. Geno, genotypic tropism result by deep sequencing.
Among patients who switched tropism, maraviroc recipients classified as having R5 virus by Trofile but as having non-R5 virus by genotyping were treated for a mean of 4.6 weeks before Trofile gave a non-R5 result, which was more than twice as quickly as for patients for whom both tests indicated R5 virus (9.7 weeks).
Response Stratified by Background Drug Activity
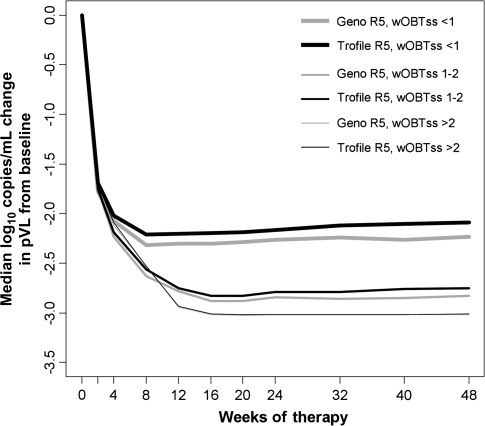
Patients were also classified according to a weighted optimized background therapy susceptibility score (wOBTss)—in general, the number of active drugs in the patient's background regimen at baseline, with nucleoside reverse-transcriptase inhibitors scoring 0.5 [35]. Genotyping was predictive of virologic success for patients who received maraviroc-based therapy, regardless of wOBTss (Figure 5).
Figure 5.
Median change in plasma viral load (pVL) from baseline in patients with CCR5-using (R5) virus stratified by weighted optimized background therapy susceptibility score (wOBTss). Maraviroc-treated patients identified as having R5 virus at screening by genotyping (light gray lines) or Trofile (black lines). Patients identified at screening at having R5 virus by either method who also had wOBTss >2 (n = 68 and n = 80, respectively) showed the largest pVL decreases from baseline. Patients with wOBTss 1–2 (n = 151 and n = 197) showed intermediate pVL decreases, and patients with wOBTss ≤1 (n = 392 and n = 463) showed poorer changes in pVL. The wOBTss >2, 1–2, and ≤1 groups are indicated by thin, intermediate, and thick lines, respectively.
Maraviroc was successful in either of the R5 groups where the wOBTss was between 1 and 2. The proportions of these patients with a week 48 pVL <50 copies/mL were 58% (179 of 311) and 53% (205 of 389) when screened by genotyping and Trofile, respectively. The predictive ability of genotyping was more pronounced at more-compromised background regimens (Figure 5). The proportions of patients with undetectable viral loads were 33% (81 of 232) and 29% (78 of 271) of R5-classified patients with wOBTSS <1 by genotyping or by Trofile, respectively.
Assay Discordance
Where screening assay results differed, virologic outcomes for patients who received maraviroc slightly favored genotyping. Among the discordant patients, in cases in which Trofile indicated R5 but genotyping identified 2% non-R5 virus (n = 135), median log10 copies/mL decreases in pVL at week 8 were lower, at 1.8 log10 copies/mL, as was the decrease in the concordant non-R5 group (1.2 log10 copies/mL; n = 105). In cases in which genotyping screening identified patients as having R5 virus but Trofile screening identified them as having non-R5 virus, the median week 8 pVL decrease was 2.6 log10 copies/mL (n = 6), which was similar to the decrease in the concordant R5 group (2.4 log10 copies/mL; n = 605).
Either or both assays indicating non-R5 virus was a poor prognostic indicator of longer-term maraviroc response. At week 48, the proportions of patients with suppressed pVLs were 27% (36 of 135) for the Trofile R5 and genotyping non-R5 group and 0% (0 of 6) for the Trofile non-R5 and genotyping R5 group. The rate of viral suppression at week 48 was twice as high in the concordant R5 group than in the concordant non-R5 group: 50% (301 of 605) versus 25% (26 of 105).
In the Trofile R5 and genotyping non-R5 group, 55% (74 of 135) of the maraviroc recipients changed tropism, which was a much higher rate than that in the concordant R5 group (18%; 111 of 605). Most patients identified by Trofile as having non-R5 virus continued to have non-R5 virus over the course of the study period, regardless of concordance with genotyping.
Finally, an analysis was performed to approximate ESTA screening data by combining the screening and baseline Trofile results. If either result indicated non-R5 virus, the patient was classified as being in the Trofile non-R5 group and likely had non-R5 HIV fluctuating near the limits of detection of Trofile. In this analysis, the combined Trofile assays performed similarly to deep sequencing at screening alone, with similar virologic responses among the discordant groups (data not shown).
DISCUSSION
This study represents, to our knowledge, the largest clinical application of next-generation sequencing technology to date. Deep V3 sequencing was able to detect and quantify low prevalence sub-populations of CXCR4-using HIV within a large set of clinical isolates. This method was predictive of virologic response to a maraviroc-containing regimen and matched or surpassed the predictive ability of the original Trofile assay on a number of parameters, including the proportion of patients who achieved a pVL of <50 copies/mL and the likelihood of switching tropism while receiving maraviroc.
Retrospectively screening with genotyping led to more than twice as many maraviroc recipients being identified as having non-R5-HIV, compared with the number identified by Trofile. Patients identified as having R5 virus by Trofile who were identified as having non-R5 virus at screening by genotyping were more likely to change their Trofile result to dual/mixed or X4 during the trials, suggesting earlier non-R5 detection by genotyping, compared with the original Trofile assay. Among maraviroc recipients who experienced tropism switches, genotyping would have identified 40% as having non-R5 virus. Thus, the high sensitivity of deep sequencing was able to account for a substantial portion of tropism switches as being attributable to the presence of low-level non-R5 variants that were not detected by Trofile.
The emergence of minority non-R5 variants detectable by deep sequencing but not by Trofile after treatment with maraviroc has been shown previously [26]. In the MERIT trial of maraviroc in treatment-naive patients, low X4 sensitivity of the original Trofile assay was determined to be a primary reason that maraviroc failed to demonstrate noninferiority to efavirenz [34]. When this trial was retrospectively re-analyzed with ESTA, more patients were identified as harboring non-R5 virus [34].
A potential benefit of deep sequencing over standard population-based sequencing is that the latter cannot reliably detect variants present below ∼20% of the viral population [22, 23], whereas deep sequencing can reliably detect quasi-species present at much lower levels, as shown here. Only 37% of maraviroc recipients with 2%–20% non-R5 variants were identified by population-based V3 sequencing (58% were identified by Trofile), yet these patients still showed suboptimal virologic responses. Thus, like ESTA, deep sequencing may represent an enhanced sensitivity tropism test, able to detect minority non-R5 variants. However, the added clinical benefit of capturing low-prevalence non-R5 variants should be weighed against the accessibility and relative affordability of standard sequencing. Importantly, population-based sequencing had >80% concordance with deep sequencing in this same dataset.
Most patients identified as having R5 virus by genotyping still had low levels (<2%) of detectable non-R5 virus. Despite this, good virologic responses to maraviroc were seen in this population. It is possible that the background antiretrovirals were able to suppress the non-R5 variants in these patients. Alternatively, it may be the case that a minimum threshold of non-R5 HIV must be surpassed before treatment with CCR5-antagonists is compromised; our data may indicate that this threshold is ∼2% of the viral population. The overall activity of the background regimen is also likely to be a major factor.
Some limitations of this study and the deep sequencing method in general should be acknowledged. A major limitation is the current relatively high cost of deep sequencing. Also, the labor involved in preparing samples for pyrosequencing is intensive and complex. Third, there may be correlates of tropism outside V3 [36, 37], which this method does not capture. This study was retrospective in nature, and a randomized clinical trial with exclusion of patients identified as having non-R5 virus at screening by deep sequencing, instead of by Trofile, may have yielded different results. This deep-sequencing approach has also not been published in a treatment-naive population.
Surprisingly, there is no ESTA data available for these studies. This is a concern, because maraviroc is primarily prescribed for treatment-experienced patients, but ESTA, the most commonly used tropism assay, has not been formally validated in MOTIVATE. This is a major limitation for the interpretation of the results of the current study. However, a preliminary comparison has been performed between deep sequencing and ESTA [38]. The prescreening of patients with Trofile also limited the number of treated patients with non-R5 virus who were examined in this study. These limitations, coupled with the good performance of our method, support a prospective trial evaluating the relative merits of genotypic and phenotypic approaches, which would also establish the true sensitivities and specificities of both approaches.
Overall, despite the study's limitations, deep sequencing showed good performance in predicting a variety of clinical parameters, including pVL decreases, the likelihood of achieving virologic suppression, and time to a tropism switch. This large study establishes deep-sequence analysis of the HIV envelope V3 loop as an extremely promising tool for identifying treatment-experienced individuals who could receive clinical benefit from CCR5-antagonist–containing therapy regimens.
Supplementary Material
References
- 1.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 2.Soriano V, Geretti AM, Perno CF, Fatkenheuer G, Pillay D, et al. Optimal use of maraviroc in clinical practice. AIDS. 2008;22:2231–2240. doi: 10.1097/QAD.0b013e3283136d95. [DOI] [PubMed] [Google Scholar]
- 3.Briz V, Poveda E, Soriano V. HIV entry inhibitors: mechanisms of action resistance pathways. J Antimicrob Chemother. 2006;57:619–627. doi: 10.1093/jac/dkl027. [DOI] [PubMed] [Google Scholar]
- 4.Esté J, Telenti A. HIV entry inhibitors. Lancet. 2007;370:81–88. doi: 10.1016/S0140-6736(07)61052-6. [DOI] [PubMed] [Google Scholar]
- 5.Dorr P, Westby M, Dobbs S, et al. Maraviroc (UK-427,857), a potent, orally bioavailable, selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother. 2005;49:4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter NJ, Keating GM. Maraviroc. Drugs. 2007;67:2277–2288. doi: 10.2165/00003495-200767150-00010. [DOI] [PubMed] [Google Scholar]
- 7.Westby M, Lewis M, Whitcomb J, et al. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol. 2006;80:4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saag M, Goodrich J, Fätkenheuer G, et al. A double-blind, placebo-controlled trial of maraviroc in treatment-experienced patients infected with non-R5 HIV-1. J Infect Dis. 2009;199:1638–1647. doi: 10.1086/598965. [DOI] [PubMed] [Google Scholar]
- 9.Whitcomb JM, Huang W, Fransen S, et al. Development characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51:566–575. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reeves JD, Coakley E, Petropoulos CJ, Whitcomb JM. An enhanced-sensitivity trofile assay. J Viral Entry. 2009;3(3):94–102. [Google Scholar]
- 11.Rose JD, Rhea AM, Weber J, Quinones-Mateu ME. Current tests to evaluate HIV-1 co-receptor tropism. Curr Opin HIV AIDS. 2009;4:136–142. doi: 10.1097/COH.0b013e328322f973. [DOI] [PubMed] [Google Scholar]
- 12.Sierra S, Kaiser R, Thielen A, Lengauer T. Genotypic coreceptor analysis. Eur J Med Res. 2007;12:453–462. [PubMed] [Google Scholar]
- 13.Shioda T, Levy JA, Cheng-Mayer C. Macrophage T cell line-tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 14.De Jong JJ, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouchier RA, Brouwer M, Broersen SM, Schuitemaker H. Simple determination of human immunodeficiency virus type 1 syncytium-inducing V3 genotype by PCR. J Clin Microbiol. 1995;33:906–911. doi: 10.1128/jcm.33.4.906-911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan S, Speck R, Power C, Gaffen S, Chesebro B, Goldsmith MA. V3 recombinants indicate a central role for CCR5 as coreceptor in tissue infection by HIV type 1. J Virol. 1999;73:2350–2358. doi: 10.1128/jvi.73.3.2350-2358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman NG, Seillier-Moiseiwitsch F, Ahn J, Walker JM, Swanstrom R. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J Virol. 2002;76:3852–3864. doi: 10.1128/JVI.76.8.3852-3864.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen MA, Li FS, van 't Wout AB, et al. Improved coreceptor usage prediction genotypic monitoring of R5-to-X4 transition by motif analysis of HIV-1 env V3 loop sequences. J Virol. 2003;77:13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sing T, Low AJ, Beerenwinkel N, et al. Predicting HIV co-receptor usage based on genetic clinical covariates. Antivir Ther. 2007;12(7):1097–1106. [PubMed] [Google Scholar]
- 21.Low AJ, Dong W, Chan D, et al. Current V3 genotyping algorithms are inadequate for predicting X4 co-receptor usage in clinical isolates. AIDS. 2007;21:F17–F24. doi: 10.1097/QAD.0b013e3282ef81ea. [DOI] [PubMed] [Google Scholar]
- 22.Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol. 1999;37:2291–2296. doi: 10.1128/jcm.37.7.2291-2296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer S, Kearney M, Maldarelli F, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Droege M, Hill B. The Genome Sequencer FLXTM System–longer reads, more applications, straight forward bioinformatics more complete data sets. J Biotechnol. 2008;136:3–10. doi: 10.1016/j.jbiotec.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 25.Bushman FD, Hoffman C, Ronen K, et al. Massively parallel pyrosequencing in HIV research. AIDS. 2001;22:1411–1415. doi: 10.1097/QAD.0b013e3282fc972e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Archer J, Braverman MS, Taillon BE, et al. Detection of low-frequency pretherapy chemokine (CXC motif) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. AIDS. 2009;23:1209–1218. doi: 10.1097/QAD.0b013e32832b4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gulick RM, Lalezari J, Goodrich J, et al. Maraviroc for previously treated patients with R5 HIV-1 infection. N Eng J Med. 2008;359:1429–1441. doi: 10.1056/NEJMoa0803152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fätkenheuer G, Nelson M, Lazzarin A, et al. Subgroup analyses of maraviroc in previously treated R5 HIV-1 infection. N Engl J Med. 2008;359:1442–1455. doi: 10.1056/NEJMoa0803154. [DOI] [PubMed] [Google Scholar]
- 29.Harrigan PR, McGovern R, Dong W, et al. In: Program and abstracts of the 5th IAS Conference on HIV Pathogenesis, treatment Prevention. Cape Town, South Africa: Screening for HIV tropism using population-based V3 genotypic analysis: a retrospective virological outcome analysis using stored plasma screening samples from MOTIVATE-1 [abstract WELBA101] pp. 19–22. July 2009. [Google Scholar]
- 30.Swenson LC, Moores A, Low AJ, et al. Improved detection of CXCR4-using HIV by V3 genotyping: application of population-based and ‘deep’ sequencing to plasma RNA and proviral DNA. J Acquir Immune Defic Syndr. 2010;54(5):506–510. doi: 10.1097/QAI.0b013e3181d0558f. [DOI] [PubMed] [Google Scholar]
- 31.Trinh L, Han D, Huang W, et al. Technical validation of an enhanced sensitivity Trofile HIV coreceptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5. Antivir Ther. 2008;13(suppl 3):A128. [Google Scholar]
- 32.Harrigan PR MOTIVATE Tropism Study Group. Optimization of clinical cutoffs for determining HIV co-receptor use by population and “deep” sequencing methods. Philadelphia, PA: Infectious Diseases Society of America; 29 October–1 November 2009. [Google Scholar]
- 33.McGovern RA, Dong W, Mo T, et al. In: Program and abstracts of the 12th European AIDS Conference. Cologne, Germany: Optimization of clinically relevant cut-points for the determination of HIV co-receptor usage to predict maraviroc responses in treatment experienced (TE) patients using population V3 genotyping [abstract PE3.4/8] pp. 11–14. November 2009. [Google Scholar]
- 34.Cooper DA, Heera J, Goodrich J, et al. Maraviroc versus efavirenz, both in combination with zidovudine/lamivudine, for the treatment of antiretroviral-naïve subjects with CCR5-tropic HIV-1. J Infect Dis. 2010;201:803–813. doi: 10.1086/650697. [DOI] [PubMed] [Google Scholar]
- 35.Valdez H, Lewis M, Delogne C, et al. In: Program and abstracts of the 48th Annual ICAAC/IDSA 46th Annual Meeting. Washington, DC: Weighted OBT susceptibility score (wOBTss) is a stronger predictor of virologic response at 48 weeks than baseline tropism result in MOTIVATE 1 and 2; pp. 25–28. October 2008. [Google Scholar]
- 36.Nabatov AA, Pollakis G, Linnemann T, Kliphius A, Chalaby MIM, Paxton WA. Intrapatient alterations in the human immunodeficiency virus type 1 gp120 V1V2 V3 regions differentially modulate coreceptor usage, virus inhibition by CC/CXC chemokines, soluble CD4, and the b12 and 2G12 monoclonal antibodies. J Virol. 2004;78:524–530. doi: 10.1128/JVI.78.1.524-530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho MW, Lee MK, Carney MC, Berson JF, Doms RW, Martin MA. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swenson L, Dong W, Mo T, et al. Program and abstracts of 17th Conference on Retroviruses Opportunistic Infections. San Francisco, CA: Large-scale application of deep sequencing using 454 technology to HIV tropism screening; pp. 16–19. February 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.