Abstract
Background. Pandemic H1N1 (pH1N1) surveillance data showed lower attack rates but higher risk of severe outcomes with advanced age. We explored immuno-epidemiologic correlates of surveillance findings including humoral and cell-mediated immunity (CMI).
Methods. In an age-based design, ∼100 banked/residual sera per 10-year age stratum were assessed by hemagglutination inhibition (HI) and microneutralization (MN) assays for preexisting antibody to pH1N1 and recent seasonal H1N1 and H3N2 strains. In a separate birth cohort design defined by childhood influenza A/subtype priming (1919–1929: H1N1; 1945–1949: H1N1; 1958–1960: H2N2; 1969–1970: H3N2; 1978–1989: H3N2/H1N1), whole blood was collected from up to 50 volunteers per birth cohort. The ratio of Th1(IFN-γ):Th2(IL-10) cytokine responses was evaluated in vitro.
Results. Antibody to seasonal viruses was highest in school-age children. Cross-reactive HI/MN antibody to pH1N1 was low among participants <70 years of age (yoa; 6%/4% ≥ 40), but seroprevalence increased at 70–79 yoa (27%/6%), increased even more at 80–89 yoa (65%/47%), and was highest at ≥90 yoa (88%/76%). CMI to pH1N1 was evident in all 5 birth cohorts but was lower compared with seasonal strains. There was little differentiation by subtype priming, but the Th1:Th2 ratio for all viruses dropped significantly in the 2 oldest cohorts.
Conclusions. Preexisting antibody may have protected the very old from pH1N1 infection, while diminished CMI may have contributed to greater severity once infected. In the young, cross-reactive pH1N1 antibody was mostly absent, while more intact CMI may have protected against severe outcomes.
BACKGROUND
During the 20th century, there were 3 recognized pandemics of influenza A (1918, 1957, 1968). Each was caused by the emergence of a novel influenza A subtype, defined by new hemagglutinin (HA) and neuraminidase (NA) proteins on the virus’s surface. Each pandemic virus replaced previously circulating influenza viruses and then evolved as antigenic drift variants over subsequent years [1–2].
The most dramatic pandemic occurred in 1918 and was due to an H1N1 subtype. Descendants of this 1918 virus were replaced in 1957 by a reassortant H2N2 subtype that introduced novel HA, NA, and PB1 segments but retained 5 of the 1918 gene segments encoding internal proteins (PB2/PA/NP/M/NS) [2]. In 1968, the existing H2N2 subtype acquired novel HA and PB1 segments by reassortment and became the H3N2 subtype. This virus also retained the 5 RNA segments encoding internal proteins from 1918. In 1976, a swine-origin H1N1 virus caused a limited zoonotic outbreak at Fort Dix military camp but did not spread beyond that setting; however, in 1977, separate emergence of another H1N1 virus successfully propagated [1]. This 1977 virus was closely related to predecessor viruses of the 1950s and is believed to have emerged as a result of laboratory escape of a vaccine candidate strain. It caused a “pseudo-pandemic” mostly affecting people <25 years of age (yoa) [1]. Variant descendants of this 1977 H1N1 virus and the 1968 H3N2 subtype have since co-circulated in human populations globally. Of note, the internal components of both subtypes are derived from the 1918 virus. The 1918 virus has thus been called “the mother of all pandemics” since it is the likely ancestor of all currently recognized human and swine influenza lineages [3].
Swine influenza viruses have been recognized since 1933, but molecular studies suggest that they share common ancestry with the 1918 pandemic virus [4–6]. During the 1990s several reassortant events occurred in pigs, with sporadic human infections, but further transmission was limited [6–7]. In June 2009, the World Health Organization declared the first influenza pandemic of the 21st century. Unlike previous pandemics of the 20th century, this pandemic was not caused by a novel subtype but, rather, by an antigenically distant variant within an existing H1N1 subtype that originated in swine [8]. Sequence analysis revealed the 2009 pandemic H1N1 (pH1N1) virus to be a complex reassortant swine virus including HA, NP, and NS from classical North American swine H1N1; NA and M from Eurasian swine H1N1; PB2 and PA from North American avian virus; and PB1 from human H3N2 virus [5–6, 9].
The short lifespan of pigs has precluded the sort of selective immune pressure that drives evolutionary adaptation and antigenic variation in surface proteins of human influenza viruses [4]. Consequently, the HA sequence of the 2009 swine-origin pH1N1 virus most closely resembles viruses from 1918 or from the limited zoonotic outbreaks of 1976 or the 1990s but is antigenically very distant from seasonal H1N1 viruses circulating in the human population since 1977 (Appendix A) [5]. Conversely, considerable homology exists for the key internal proteins (notably M, NP, and polymerases) of pH1N1 and recent human viruses of both H1N1 and H3N2 subtypes (Appendix B) [10].
This chronology of 20th century pandemics and subsequent seasonal influenza epidemics has produced disparate, age-related patterns of antigenic exposure among the contemporary human population resulting in a complex immuno-epidemiologic patchwork [9]. The diversity in primary and preferentially recalled humoral and cell-mediated immunity (CMI) induced by this complex array of antigenic exposures is relevant to consider in relation to the epidemiologic profiles of pH1N1 disease burden. Surveillance reports have consistently shown that pH1N1 attack rates are highest in the young, whereas the risk of severe outcomes if infected is highest in the old [11–14]. This was seen also in laboratory, hospitalization, and mortality surveillance data assembled for the province of British Columbia, Canada [15].
In this report we attempt to reconcile divergent age-related surveillance trends through an immuno-epidemiologic hypothesis. Our main goal was to describe age-related patterns in cross-reactive pH1N1 humoral and cell-mediated immunity pre-pandemic that may correlate with protection against infection or severe outcomes. In an age-based design we assessed preexisting antibody to pH1N1 as well as recent seasonal H1N1 and H3N2 viruses in banked/residual sera collected across the life span. In a twin birth cohort design we assessed both antibody and T helper type 1 to type 2 (Th1:Th2) cytokine responses [16] in volunteers according to birth cohorts defined by original subtype priming experience in childhood.
METHODS
Procedures described below for both age-based and birth cohort study components received the approval of the Research Ethics Review Committee of the University of British Columbia. Surveillance data indicate that pH1N1 activity in British Columbia, and the Lower Mainland area specifically, was low prior to the fall 2009 pandemic wave, which started gradually in late September/early October, began a steeper rise in mid-October, and peaked during the last week of October [17].
Age-Based Study of Antibody in Banked/Residual Sera
During the summer of 2009, the BC Centre for Disease Control (BCCDC) obtained sera collected between 1 June 2007 and 31 July 2009 from residents of the Lower Mainland area of British Columbia. The Lower Mainland is the most densely populated region of British Columbia and includes the Vancouver metropolitan area and municipalities of the Fraser Valley. Specimens were collected as a convenience sample from (1) banked sera submitted to the BCCDC (∼20%) and (2) residual sera from patients recently presenting to a community laboratory network (∼80%). Assuming seroprevalence of 10%, 100 sera were required for precision of +/−6% with 95% confidence interval (CI); if seroprevalence were instead 50%, corresponding precision would be +/−10%. Approximately 100 sera were assembled for each of the following age strata: 0–4, 5–9, 10–19, 20–29, 30–39, 40–49, 50–59, 60–69, 70–79, and ≥80 yoa. Accompanying information included collection date, age, sex, and city. All specimens were anonymized, and individual consent was not required.
Birth Cohort Study of Antibody and Cytokine Responses in Volunteers
In September–October 2009, whole blood was collected from a convenience sample of ∼50 community-dwelling adult volunteers from the Lower Mainland area per representative birth cohort defined by childhood subtype priming experience. Assuming most would have experienced a first influenza A exposure by 9 yoa, those born in 1919–1929 and in 1945–1949 were expected to have had childhood priming with H1N1 virus; those in 1958–1960, with H2N2 virus; those in 1969–1970, with H3N2; and those in 1978–1989, with H3N2 or H1N1. Given the specified birth cohorts, only adults ≥19 yoa were included, and those with potential immune-compromising conditions/medications were specifically excluded. Information collected included age, sex, comorbidity, city, occupation, influenza vaccination, and influenza-like-illness (ILI = fever plus cough or sore throat) during the 2008–2009 season and since 1 April 2009. All participants provided written informed consent.
Antibody Responses
Serology procedures were completed at the BCCDC provincial laboratory. Antibodies to A/California/7/09(pH1N1), A/Brisbane/59/07(H1N1)-like, and A/Brisbane/10/07(H3N2)-like viruses were measured. Although A/Brisbane/59/2007(H1N1) and A/Brisbane/10/2007(H3N2) strains were not introduced as seasonal vaccine components until the 2008–2009 season, isolates submitted by the British Columbia sentinel surveillance system indicated up to 16% and 100% of 2007–2008 isolates, respectively, were already closer to these antigenic variants, indicating that they were present in the population when most of the study sera were collected (from 1 October 2008). For the current study, antibody was measured by hemagglutination inhibition (HI) and microneutralization (MN) assays. Protocols, including sequence results and percent homology across viruses used, are described in detail in Appendix B.
Cytokine Responses
The Vancouver Coastal Health Research Institute VITALiTY laboratory conducted the cytokine assays. Peripheral blood mononuclear cells were prepared by Ficoll gradient purification and stimulated with live virus for 20 h, using the same virus strains as used for measuring antibody response for the birth cohort study, as described in Appendix B. Supernatants were harvested, and interferon γ (IFN- γ) and interleukin10 (IL-10) levels were evaluated using multiplex bead assays (Millipore; minimum level of detection [MLD] .4 pg/mL and .3 pg/mL, respectively) according to a validated protocol with sensitivity to detect 25% between-group difference in the IFN-γ:IL-10 ratio with 33 subjects per group [18].
Statistical Methods
HI and MN titers for each antigen were summarized as the geometric mean titer (GMT) of duplicate tests. By convention, seroprevalence or seroprotection was primarily defined at HI titers ≥40 but assessed also by MN [19–20]. Titers <10 were assigned a value of 5 (half MLD) for group GMT derivation. Group GMTs with 95% CIs and proportion with titers ≥40 were summarized per study overall and by age stratum or birth cohort. IFN-γ and IL-10 values (undetected = half MLD) were (natural) log-transformed, and means with 95% CIs were estimated by birth cohort, as were IFN-γ:IL-10 ratios as indications of the Th1:Th2 balance [21].
RESULTS
Age-Based Study
In the age-based design 993 banked/residual sera were included. Exact sample sizes, demographics, and dates of specimen collection are shown in Table 1. To enable finer age assessment in the elderly, the ≥80 yoa stratum was subdivided into those 80–89 yoa (n = 49) and 90–99 yoa (n = 51). All sera were collected before the second wave, and as shown in Table 1, virtually all were collected between 1 October 2008 and 31 July 2009, with slight variation by age group collected before/after 1 April 2009.
Table 1.
Sample Size, Sex, and Date of Specimen Collection for Age-Based Serosurvey of Banked/Residual Sera from Vancouver Metropolitan Area of British Columbia, Canada
| Age range (years) | Sample size | Median age (years) | % Female | Percent between 1 June 2007 and 1 October 2008 | Percent between 1 October 2008 and 1 April 2009 | Percent between 1 April 2009 and 31 July 2009 |
| All | 993 | 40 | 55 | .7 | 18.8 | 80.5 |
| 0–4 | 101 | 3 | 43 | 0 | 30.7 | 69.3 |
| 5–9 | 95 | 7 | 49 | 0 | 50.5 | 49.5 |
| 10–19 | 98 | 15 | 61 | 0 | 23.5 | 76.5 |
| 20–29 | 100 | 25 | 71 | 0 | 0 | 100 |
| 30–39 | 100 | 33 | 62 | 0 | 0 | 100 |
| 40–49 | 98 | 45 | 68 | 0 | 0 | 100 |
| 50–59 | 101 | 55 | 47 | 1.0 | 26.7 | 72.3 |
| 60–69 | 103 | 65 | 55 | 2.9 | 34.0 | 63.1 |
| 70–79 | 97 | 73 | 42 | 1.0 | 19.6 | 79.4 |
| 80–89 | 49 | 84 | 53 | 4.1 | 8.1 | 87.8 |
| 90–99 | 51 | 94 | 59 | 0 | 0 | 100 |
Antibody Responses by Age.
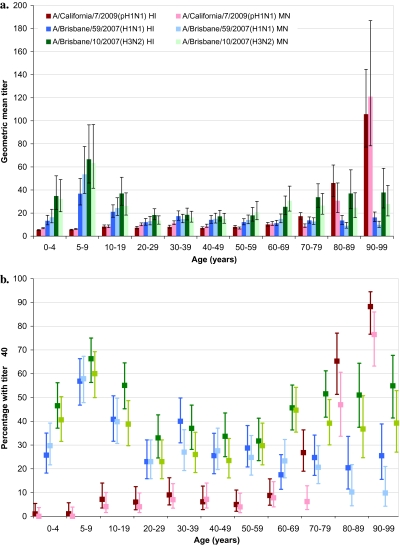
Antibody titers to pH1N1 and seasonal H1N1 and H3N2 strains are shown by age stratum in Figure 1A. The proportion with HI/MN titers ≥40 is shown in Figure 1B.
Figure 1.
Antibody titers to A/California/7/2009(pH1N1), seasonal A/Brisbane/59/2007(H1N1), and seasonal A/Brisbane/10/2007(H3N2) viruses, age-based serosurvey. (A) Geometric mean titers (95% confidence intervals). (B) Percentage with titers ≥40 and 95% confidence intervals. HI, hemagglutination inhibition assay; MN, microneutralization assa. NOTE. HI= hemagglutination inhibition assay; MN = microneutralization assay.
Seroprevalence of seasonal H1N1 and H3N2 viruses was highest in school-age children, especially those 5–9 yoa, among whom 57%/58% had HI/MN titers ≥40 to H1N1 and 66%/60% had HI/MN titers ≥40 to H3N2. Titers to both seasonal viruses appeared lower in adults, although H3N2 titers rose again in those ≥60 yoa, among whom 50%/41% showed HI/MN titers ≥40.
Overall, there was little or no preexisting pH1N1 antibody in those <70 yoa, among whom only 6%/4% had HI/MN titers ≥40. Among those <50 yoa overall, 5%/4% had HI/MN titers ≥40, including 3%/1% in children <20 yoa and 7%/6% in young adults 20–49 yoa. Similar proportions were found in older adults 50–69 yoa (7%/6%). Beginning at 70–79 yoa there was significant increase in the proportion with HI (27%) but not MN (6%) titers ≥40. Further substantial and significant increase in both HI/MN pH1N1 seroprevalence began at ≥80 yoa, with 77%/62% demonstrating HI/MN ≥40, including 65%/47% of those 80–89 yoa and 88%/76% of those 90–99 yoa. Further stratifying by age, seroprevalence among the small sample 80-84 yoa (n=25) was 52% (95%CI 34% - 70%) by HI and 24% (95%CI 12% - 43%) by MN; among those 85-89 yoa (n=24) it was 79% (95%CI 60% - 91%) and 71% (95%CI 51% - 85%), respectively.
Seroprevalence for the pH1N1 virus was significantly higher than for seasonal H1N1 by both HI and MN in those ≥80 yoa and also significantly exceeded the seroprevalence for seasonal H3N2 among those ≥90 yoa.
Birth Cohort Study
Blood was collected from 228 adult volunteers between 8 September and 8 October 2009: 50 participants each were in the 1919–1929 (79–90 yoa; median 82.5 yoa), 1945–1949 (60–64 yoa; median 63 yoa), and 1958–1960 (48–51 yoa; median 50 yoa) cohorts; and there were 51 participants in the 1978–1989 cohort (19–31 yoa; median 24 yoa). The 1969–1970 cohort specified a tighter age band (38–40 yoa; median 39 yoa) to capture those whose original exposure was to an H3N2 virus without possibility of childhood H1N1 priming after 1977. Consequently recruitment was more difficult, resulting in a smaller sample of only 27 participants, with implications for variability around estimates in that 1969–1970 cohort.
Seventy-five percent of participants were female, and 84% had ever received an influenza vaccine, ranging by proportion of youngest (1978–1989) to oldest (1919–1929) birth cohorts as 71%, 81%, 78%, 96%, and 94%. Sixty percent received 2008–2009 seasonal vaccine, increasing from the youngest to oldest cohort as 33%, 48%, 54%, 70%, and 88%, with a similar distribution for the 2007–2008 vaccine. The proportions reporting current work in a health care setting were 25%, 41%, 28%, 16%, and 0%, respectively. Just 2 of the 228 (1%) participants reported ILI since early April 2009 (one each from the oldest 2 cohorts). Eleven percent reported ILI between November 2008 and April 2009, decreasing from the youngest to oldest cohort as 22%, 15%, 10%, 8%, and 0%.
Antibody Responses by Birth Cohort.
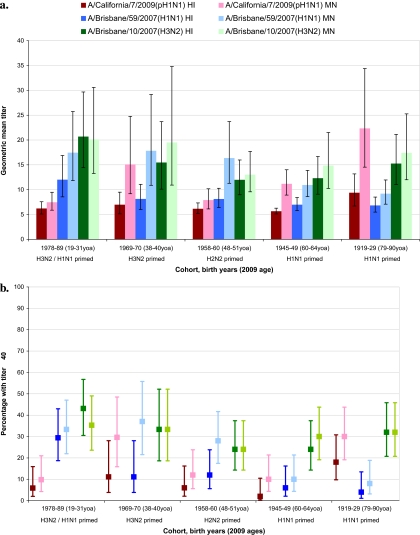
As shown in Figure 2, antibodies to seasonal H1N1 and H3N2 viruses were highest in the 2 youngest adult birth cohorts. Seasonal H1N1 titers decreased thereafter with increasing age, whether measured by HI or MN. Seasonal H3N2 titers showed the same general decline with age across the 3 youngest cohorts and, as in the age-based design, increased slightly in the 2 oldest cohorts (at ≥60 yoa).
Figure 2.
Antibody titers to A/California/7/2009(pH1N1), seasonal A/Brisbane/59/2007(H1N1), and seasonal A/Brisbane/10/2007(H3N2) viruses, birth cohort study. (A) Geometric mean titers and 95% confidence intervals. (B) Percentage with titers ≥40 and 95% confidence intervals. HI, hemagglutination inhibition assay; MN, microneutralization assay; yoa, years of age NOTE. HI= hemagglutination inhibition assay; MN = microneutralization assay; yoa= years of age.
Similar age-related trends in pH1N1 antibody were also observed for participants as in the age-based design—low until the oldest 1919–1929 cohort (79–90 yoa), in which 18%/30% had HI/MN titers ≥40. Antibody to pH1N1 was lower in this cohort compared with those ≥80 yoa in the age-based design, but this reflected a younger median age (82.5 yoa), with differences also in the pH1N1 virus used (Appendix B). Corresponding proportions with HI/MN titers ≥40 were 6%/10% for the 1978–1989 cohort, 6%/12% for 1958–1960, 12%/30% for 1969–1970, and 2%/10% for 1945–1949.
In general, antibody responses in the birth cohort design showed similar age-related patterns as in the age-based design without evidence of additional birth cohort effects based on childhood subtype priming.
Ctyokine Responses by Birth Cohort.
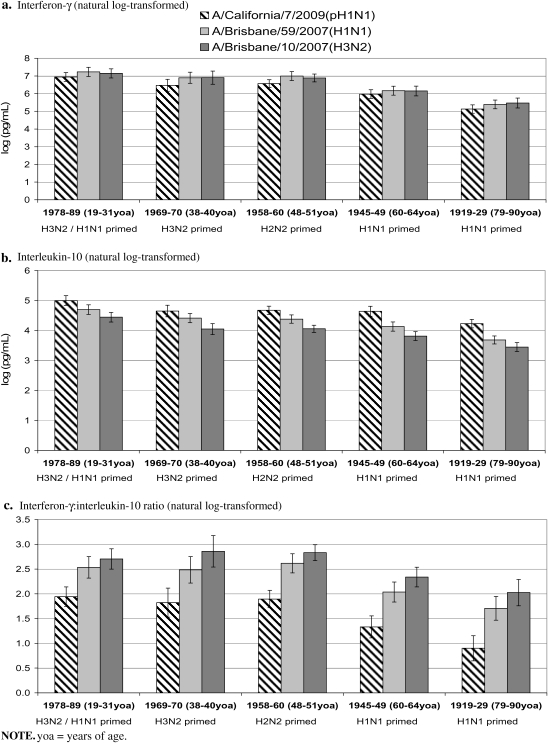
Within all adult birth cohorts assessed, IFN-γ responses were comparable between seasonal strains but slightly lower for pH1N1 (Figure 3A). For all viruses, IFN-γ responses were lowest for the 1919–1929 cohort and also lower for the 1945–1949 cohort relative to the 3 youngest cohorts(1978–1989, 1969–1970, 1958–1960). For seasonal viruses, IL-10 responses decreased with successive cohorts, a trend also evident but less consistently for pH1N1 (Figure 3B).
Figure 3.
Log-transformed interferon-γ, interleukin-10, and their ratio for pH1N1, seasonal H1N1, and H3N2 viruses, birth cohort study. (a) Interferon-γ (natural log-transformed). (b) Interleukin-10 (natural log-transformed). (c) Interferon-γ:interleukin-10 ratio (natural log-transformed). yoa, years of age.
Within all cohorts, IFN-γ:IL-10 ratios were significantly lower for pH1N1 relative to seasonal strains (Figure 3C). For all viruses, there were no significant differences in IFN-γ:IL-10 ratios across the 3 youngest cohorts. Conversely, IFN-γ:IL-10 ratios dropped significantly for all viruses in the 1945–1949 cohort and were lowest in the 1919–1929 cohort.
In general, CMI responses were most consistent with age-related decline without evidence of additional birth cohort differences based on categories of childhood subtype priming.
DISCUSSION
This study assessed both humoral and cell-mediated immunity against influenza in order to better understand divergent age-related trends of pH1N1 infection versus severe outcomes in surveillance data. Findings show that in the oldest age groups, antibody to pH1N1 was highest while CMI was lowest; the reverse was seen in younger age groups. An immuno-epidemiologic hypothesis is proposed that reconciles these serologic and cytokine findings with surveillance observations and with historic variation in antigenic exposure among contemporary populations through the 20th century.
Antibody to surface proteins is the main determinant of protection from influenza infection [22]. Seroprotective levels for influenza are, by convention, defined at HI ≥ 40, although this has not been specifically validated for cross-reactive antibody to pH1N1 [23–24]. Our age-based serosurvey found that antibody to seasonal strains was highest in young school-aged children with good match between HI and MN responses. Similar to previous population serosurveys [25–29], however, little or no pH1N1 antibody (6%/4% with HI/MN titers ≥40) was found other than in the very old, suggesting broad pre-pandemic susceptibility to infection. A substantial increase in seroprotective antibody was found at 80–89 yoa (65%/47% with HI/MN ≥40), notably above 85 yoa and with most of those ≥90 yoa showing evidence of preexisting seroprotection against pH1N1 by both HI and MN assays (88%/76%). In the young, antibody and HI/MN alignment were higher for seasonal strains, whereas in the very old these were greatest for pH1N1. In the 70–79 yoa cohort (born in the 1930s), antibody for pH1N1 versus seasonal strains was intermediate between younger and older age groups, with some increase in HI antibody to pH1N1 that was not evident by MN assay (27%/6% ≥40).
The 2009 swine-origin pH1N1 virus bears close antigenic relatedness to the 1918 pandemic virus (Appendices A–B) [5]. The doctrine of original antigenic sin (OAS) dictates that antibody to dominant epitopes of first-infecting influenza viruses is preferentially recalled through subsequent infection with new and antigenically distinct virus, sometimes at the expense of response to the new virus [30]. OAS with immunologic imprinting to closely related viruses of early childhood, further amplified through accumulated lifetime boosting, may explain the high proportion of the very old we found with cross-reactive pH1N1 antibodies. The observation that antibody to pH1N1 exceeded that of seasonal H1N1 and H3N2 strains in these older individuals is also consistent with OAS. Sequence analysis underscores the further significance of the 1930–1940 decade as a cut point for pH1N1 cross-reactivity (Appendix A) [31]. Reichert has described the first appearance of several new sites for glycosylation on the globular head of historic H1N1 viruses beginning in the 1930s and 1940s, with implications for antibody binding that became more prominent from 1949 to 1950 [32]. Viruses circulating thereafter, including the reemerged 1977 H1N1 virus and its descendants, bear only distant antigenic relatedness to pH1N1 (Appendix A). These sequencing findings, in addition to surveillance data, are mirrored in the current serosurvey results.
While protection against influenza infection is primarily mediated through antibody, protection against severe outcomes is instead mediated through cellular immune responses effecting viral clearance [22, 33]. In addition, unlike antibody that is induced by rapidly mutating surface proteins, CMI to influenza is primarily induced by the major internal virus proteins that are generally more conserved across subtypes, allowing for greater heterologous cross-reactivity [22, 34]. Indeed, sequencing results show that internal components are more highly conserved across twentieth-century subtypes, in humans and swine and finally, in pH1N1 (Appendix B) [5, 10]. To assess CMI, we defined a birth cohort approach based on original subtype exposure in childhood and explored the balance of Th1 versus Th2 cytokine responses to pH1N1 and seasonal strains in vitro [16]. This allowed simultaneous assessment of age-related effects while accounting for major differences in childhood subtype priming based on viral surface proteins—an approach also recently recommended by Morens et al [30]. We showed cross-reactive CMI responses to the pH1N1 virus with Th1:Th2 ratios significantly lower compared with seasonal strains in all adult birth cohorts assessed. The Th1:Th2 ratio for all viruses was significantly lower in the 2 older cohorts and notably lower in the oldest category. CMI responses to pH1N1 were not significantly different across the 3 youngest cohorts despite major differences in subtype priming, whereas the 2 oldest cohorts showed a decrease despite common H1N1 subtype priming. We interpret CMI findings as diminished cross-reactive CMI for pH1N1 relative to seasonal strains and further diminished with advanced age as a consequence of immunosenescence [35]—that is, a direct age-related effect rather than a birth cohort phenomenon.
There are several caveats to the interpretation of these results. First, surveillance data are useful for identifying age-related trends, but their limitations for precise quantification of disease burden or for addressing other concomitant influences (such as comorbidity, propensity for care seeking, or laboratory testing) should be recognized. For the age-based serosurvey convenience sampling drew from banked/residual sera submitted from patients who presented for other clinical testing; sera were anonymized and without additional detail provided related to comorbidity (such as immunosuppression) or vaccination history. The low rates of seroprotection we measured in the young are not unexpected given the novelty of the pH1N1 virus, and methodological influences are unlikely to explain the high rates of seroprotection we found in the very old. There was some variation by age in the date of original specimen collection (before/after 1 April 2009), and we cannot rule out contribution from pH1N1 infection during the first pandemic wave. However, surveillance data in British Columbia suggest that this was unlikely [17].
For the birth cohort design we included only adults ≥19 yoa and specifically excluded participants with potential immune-compromising conditions/medications. Recruitment to this study occurred during the late summer/early fall, and per above we cannot rule out recent pH1N1 infection. A high proportion, especially among the 1969–1970 cohort, worked in health care, and this may have increased opportunities for influenza exposure. Younger cohorts may also have had more recent infections with seasonal strains reinforcing cross-reactive CMI responses, whereas a greater proportion of the older cohorts were vaccinated. Although seasonal vaccine has been associated with only low levels of cross-reactivity to pH1N1 [26], repeat immunization with effective vaccine may block opportunities for more robust heterotypic cross-immunity induced by heterologous infection, particularly relevant to CMI responses [36–38].
We have focused on immuno-epidemiologic correlates of disease burden, but other factors such as social mixing patterns or underlying health status are also known to influence attack rates and severe outcomes by age. For both studies we recruited participants from an urban area to assess the “best-case scenario” for preexisting cross-reactive antibodies to pH1N1 [39]. We found little preexisting antibody in this study population, but levels could be even lower in remote or rural areas. Considerable laboratory variation can occur with the assays employed, and identical strains were not used in both study arms. Nevertheless, we found the same age-related trends and have provided detailed laboratory protocols and virus sequencing to aid in the interpretation of results (Appendix B). Finally, we assessed IFN-γ:IL-10 ratios as markers of Th1:Th2 balance and interpreted lower ratios as indicative of less effective viral clearance and increased risk of severe outcomes [16, 32, 40–41]. However, specific cytokine correlates or CMI thresholds for protection against severe outcomes have not been established, and other markers may also be relevant. We correlate cytokine findings with surveillance data, but case-control evaluation to directly assess the association between in vitro CMI responses and clinical outcomes of pH1N1, with adjustment for other covariates, is still warranted.
In summary, we present divergent age-related patterns of humoral and cell-mediated immunity to pH1N1 that plausibly explain age-related differences in the risks of pH1N1 evident in surveillance data. In the very old, high levels of preexisting antibody may have protected against pH1N1 infection, while immunosenescence and diminished CMI responses may have contributed to greater severity if infected. Conversely, preexisting antibody to pH1N1 was mostly absent in people <80 yoa, suggesting broad pre-pandemic susceptibility to infection, while more intact cross-reactive CMI responses may have protected the young against severe outcomes once infected. This immuno-epidemiologic hypothesis offers a unifying theory that may reconcile sequencing, surveillance, serologic, and cytokine data. It has implications for the targeting of prevention and treatment measures for subsequent influenza seasons or future pandemics and warrants further direct evaluation in other settings.
Funding
This work was supported by the Michael Smith Foundation for Health Research (grant # OT-GIA-00012091). The funding organization did not have a role in study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
Acknowledgments
We gratefully acknowledge the contributions of Leslie Love, as well as the TASC Research team for birth cohort study coordination and field work and Hochan Kim of the BC Centre for Disease Control for assistance with virus sequencing.
The seed viruses used to prepare the reagents for a number of the antibody assays were kindly provided by Dr. Yan Li from the National Microbiology Laboratory in Winnipeg, Manitoba, Canada.
References
- 1.Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis. 2006;12(1):9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belshe RB. The origins of pandemic influenza—lessons from the 1918 virus. N Engl J Med. 2005;353(21):2209–11. doi: 10.1056/NEJMp058281. [DOI] [PubMed] [Google Scholar]
- 3.Taubenberger JK, Morens DM. 1918 influenza: The mother of all pandemics. Emerg Infect Dis. 2006;12(1):15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reid AH, Fanning TG, Hultin JV, Taubenberger JK. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc Natl Acad Sci U S A. 1999;96(4):1651–6. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garten RJ, Davis CT, Russell CA, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325(5937):197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmer SM, Burke DS. Historical perspective—emergence of influenza A (H1N1) viruses. N Engl J Med. 2009;361(3):279–85. doi: 10.1056/NEJMra0904322. [DOI] [PubMed] [Google Scholar]
- 7.Myers KP, Olsen CW, Gray GC. Cases of swine influenza in humans: a review of the literature. Clin Infect Dis. 2007;44(8):1084–8. doi: 10.1086/512813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zarocostas J. World Health Organization declares A (H1N1) influenza pandemic. BMJ. 2009;338:b2425. doi: 10.1136/bmj.b2425. [DOI] [PubMed] [Google Scholar]
- 9.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459(7249):931–9. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skowronski DM, De Serres G, Crowcroft NS, et al. Association between the 2008–09 seasonal influenza vaccine pandemic H1N1 illness during the spring–summer 2009: four observational studies from Canada. PLoS Med. 2010;7(4):e1000258. doi: 10.1371/journal.pmed.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker MG, Wilson N, Huang QS, et al. Pandemic influenza A(H1N1)v in New Zealand: the experience from April to August 2009. Euro Surveill. 2009;14(34):pii 19319. doi: 10.2807/ese.14.34.19319-en. [DOI] [PubMed] [Google Scholar]
- 12.Donaldson LJ, Rutter PD, Ellis BM, et al. Mortality from pandemic A/H1N1 2009 influenza in England: public health surveillance study. BMJ. 2009;339:b5213. doi: 10.1136/bmj.b5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Louie JK, Acosta M, Winter K, et al. Factors associated with death or hospitalization due to pandemic 2009 influenza A(H1N1) infection in California. JAMA. 2009;302(17):1896–1902. doi: 10.1001/jama.2009.1583. [DOI] [PubMed] [Google Scholar]
- 14.Vaillant L, La Ruche G, Tarantola A, Barboza P Epidemic Intelligence Team at InVS. Epidemiology of fatal cases associated with pandemic H1N1 influenza 2009. Euro Surveill. 2009;14(33):pii 19309. doi: 10.2807/ese.14.33.19309-en. [DOI] [PubMed] [Google Scholar]
- 15.British Columbia Office of the Provincial Health Officer. B.C.’s response to the H1N1 pandemic: a summary report June 2010. Available at: http://www.health.gov.bc.ca/pho/pdf/PHO_Report_BC_Response_to_the_H1N1_Pandemic_June2010.pdf. Accessed 1 December 2010. [Google Scholar]
- 16.McElhaney JE, Xie D, Hager D, et al. T cell responses are better correlates of vaccine protection in the elderly. J Immunol. 2006;176:6333–9. doi: 10.4049/jimmunol.176.10.6333. [DOI] [PubMed] [Google Scholar]
- 17.BC Centre for Disease Control. BC influenza surveillance bulletins: 2009–2010. Available at: http://www.bccdc.ca/dis-cond/DiseaseStatsReports/influSurveillanceReports.htm. Accessed 1 December 2010. [Google Scholar]
- 18.Gijzen K, Liu WM, Visontai I, et al. Standardization validation of assays determining cellular immune responses against influenza. Vaccine. 2010;28:3416–22. doi: 10.1016/j.vaccine.2010.02.076. [DOI] [PubMed] [Google Scholar]
- 19.Committee for Proprietary Medicinal Products. Note for guidance on harmonisation of requirements for influenza vaccines. London: European Agency for the Evaluation of Medicinal Products; 1997. CPMP/BWP/214/96 (circular no. 96-0666):1–22. Available at: http://www.emea.europa.eu/pdfs/human/bwp/021496en.pdf. Accessed 1 December 2010. [Google Scholar]
- 20.Potter CW, Oxford JS. Determinants of immunity to influenza infection in man. Br Med Bull. 1979;35(1):69–75. doi: 10.1093/oxfordjournals.bmb.a071545. [DOI] [PubMed] [Google Scholar]
- 21.Spellberg B, Edwards JE. Type 1/type 2 immunity in infectious diseases. Clin Infect Dis. 2001;32(1):76–102. doi: 10.1086/317537. [DOI] [PubMed] [Google Scholar]
- 22.Thomas PG, Keating R, Hulse-Post DJ, Doherty PC. Cell-mediated protection in influenza infection. Emerg Infect Dis. 2006;12(1):48–54. doi: 10.3201/eid1201.051237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montomoli E. Correlates of protection against influenza. In: Rappuoli R, Del Guidice G, editors. Influenza vaccines for the future. Basel, Switzerland: Birkhäuser Verlag; 2008. pp. 139–59. [Google Scholar]
- 24.de Jong JC, Palache AM, Beyer WEP, Rimmelzwaan GF, Boon ACM, Osterhaus ADME. Haemagglutination-inhibiting antibody to influenza virus. In: Brown F, Haaheim LR, Schild GC, editors. Laboratory correlates of immunity to influenza—a reassessment. Devl Biol. Vol 115. Basel, Switzerland: Karger; 2003. pp. 63–73. [PubMed] [Google Scholar]
- 25.World Health Organization. Seroepidemiological studies of pandemic influenza A (H1N1) 2009 virus. kly Epidemiol Rec. 2010;85:229–36. [PubMed] [Google Scholar]
- 26.Hancock K, Veguilla V, Xiuhu L, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361(20):1945–52. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 27.Itoh Y, Shinya K, Kiso M, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460(7258):1021–7. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller E, Hoschler K, Hardelid P, Stanford E, Andrews N, Zambon M. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross-sectional serological survey. Lancet. 2010;375(9720):1100–8. doi: 10.1016/S0140-6736(09)62126-7. [DOI] [PubMed] [Google Scholar]
- 29.Rizzo C, Rota MC, Bella A, et al. Cross-reactive antibody responses to the 2009 A/H1N1v influenza virus in the Italian population in the pre-pandemic period. Vaccine. 2010;28(20):3558–62. doi: 10.1016/j.vaccine.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Morens DM, Burke DS, Halstead SB. The wages of original antigenic sin. Emerg Infect Dis. 2010;16(6):1023–4. doi: 10.3201/eid1606.100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Igarashi M, Ito K, Yoshida R, Tomabechi D, Kida H, Takada A. Predicting the antigenic structure of the pandemic (H1N1) 2009 influenza virus hemagglutinin. PLoS One. 2010;5(1):e8553. doi: 10.1371/journal.pone.0008553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reichert T, Chowell G, Nishiura H, Christensen RA, McCullers JA. Does glycosylation as a modifier of original antigenic sin explain the case age distribution and unusual toxicity in pandemic novel H1N1 influenza? BMC Infect Dis. 2010;10:5. doi: 10.1186/1471-2334-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stambas J, Guillonneau C, Kedzierska K, Mintern JD, Doherty PC, La Gruta NL. Killer T cells in influenza. Pharmacol Ther. 2008;120(2):186–96. doi: 10.1016/j.pharmthera.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Tu W, Mao H, Zheng J, et al. Cytotoxic T lymphocytes established by seasonal human influenza cross-react against 2009 pandemic H1N1 influenza virus. J Virol. 2010;84:6527–35. doi: 10.1128/JVI.00519-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McElhaney JE, Effros RB. Immunosenescence: what does it mean to health outcomes in older adults? Curr Opin Immunol. 2009;21:418–24. doi: 10.1016/j.coi.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bodewes R, Kreijtz JHCM, Baas C, et al. Vaccination against human influenza A/H3N2 virus prevents the induction of heterosubtypic immunity against lethal infection with avian influenza A/H5N1 virus. PLoS One. 2009;4(5):e5538. doi: 10.1371/journal.pone.0005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bodewes R, Kreijtz JHCM, Rimmelzwaan GF. Yearly influenza vaccinations: a double-edged sword? Lancet Infect Dis. 2009;9(12):784–8. doi: 10.1016/S1473-3099(09)70263-4. [DOI] [PubMed] [Google Scholar]
- 38.Laurie KL, Carolan LA, Middleton D, Lowther S, Kelso A, Barr IG. Multiple infections with seasonal influenza A virus induce cross-protective immunity against A(H1N1) pandemic influenza virus in a ferret model. J Infect Dis. 2010;202 doi: 10.1086/656188. 1011–20. [DOI] [PubMed] [Google Scholar]
- 39.Mathews JD, Chesson JM, McCaw JM, McVernon J. Understanding influenza transmission, immunity and pandemic threats. Influenza Other Respi Viruses. 2009;3(4):143–9. doi: 10.1111/j.1750-2659.2009.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graham MB, Braciale VL, Braciale TJ. Influenza virus-specific CD4+ T helper type 2 T lymphocytes do not promote recovery from experimental virus infection. J Exp Med. 1994;180:1273–82. doi: 10.1084/jem.180.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaji M, Kobayashi M, Pollard RB, Suzuki F. Influence of type 2 T cell responses on the severity of encephalitis associated with influenza virus infection. J Leukoc Biol. 2000;68:180–6. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.