Abstract
Background. Poor tolerance and adverse drug reactions are main reasons for discontinuation of antiretroviral therapy (ART). Identifying predictors of ART discontinuation is a priority in HIV care.
Methods. A genetic association study in an observational cohort to evaluate the association of pharmacogenetic markers with time to treatment discontinuation during the first year of ART. Analysis included 577 treatment-naive individuals initiating tenofovir (n = 500) or abacavir (n = 77), with efavirenz (n = 272), lopinavir/ritonavir (n = 184), or atazanavir/ritonavir (n = 121). Genotyping included 23 genetic markers in 15 genes associated with toxicity or pharmacokinetics of the study medication. Rates of ART discontinuation between groups with and without genetic risk markers were assessed by survival analysis using Cox regression models.
Results. During the first year of ART, 190 individuals (33%) stopped 1 or more drugs. For efavirenz and atazanavir, individuals with genetic risk markers experienced higher discontinuation rates than individuals without (71.15% vs 28.10%, and 62.5% vs 14.6%, respectively). The efavirenz discontinuation hazard ratio (HR) was 3.14 (95% confidence interval (CI): 1.35–7.33, P = .008). The atazanavir discontinuation HR was 9.13 (95% CI: 3.38–24.69, P < .0001).
Conclusions. Several pharmacogenetic markers identify individuals at risk for early treatment discontinuation. These markers should be considered for validation in the clinical setting.
Pharmacogenetic research has focused on understanding the mechanisms of adverse drug reactions (ADR) and on finding biomarkers that identify people at risk, with the aims of decreasing the number of adverse drug reactions and increasing drug efficacy. In human immunodeficiency virus (HIV) therapeutics, pharmacogenetics has been pursued because of the prevalence of toxicity [1, 2], the long-term nature of treatment, and the complexity inherent in a multidrug therapy that could benefit from predictive tools to identify the drug combination most likely to be tolerated and effective. Treatment decisions at the initiation of a first-line antiretroviral therapy (ART) in treatment-naive individuals are mostly based on CD4 T cell counts, viremia, symptoms, comorbidities, and analysis of primary drug resistance. Unfortunately, up to 45% of individuals discontinue or change treatment during the first year of ART [3, 4], frequently due to poor treatment tolerance [4–6].
For abacavir, a randomized prospective trial confirmed the value of genetic assessment before prescription to avoid hypersensitivity drug reactions [7]. Similar information regarding the potential for a pharmacogenetic-driven intervention does not exist for other ART drugs. However, there is an increasing body of literature on genes and genetic markers that may relate to toxicity and or to pharmacokinetics of the drug. This includes extensive data on the pharmacokinetics/genetics of efavirenz, with some indication that drug levels may associate with central nervous system toxicity (CYP2B6, CYP2A6, and CYP3A4) [8–11]. Data from genome-wide association studies on dyslipidemia in the general population predict ritonavir-boosted protease inhibitor–associated dyslipidemia (APOA5, CET, DOCK7, GCKR, LPL, and TRIB1) [12, 13], and there are also genetic predictors of lopinavir drug levels [14, 15]. Atazanavir-associated hyperbilirubinemia is associated with alleles of UGT1A1 [16], and has been reported in association with alleles of ABCB1 (MDR1) [17] and NR1I2 (PXR) [18]. There is early information on the genetic basis of tenofovir tubulopathy (ABCC2, ABBC4) [19, 20].
Most of the above genetic markers are convincingly associated with an intermediate phenotype (ie, laboratory abnormality), which may or may not translate into symptoms and signs that lead to clinical actions such as treatment discontinuation. We hypothesized that if any of the markers presented above contribute to intolerance or toxicity, they would predict treatment modification or discontinuation. Therefore, this study aims to evaluate the association of recognized and proposed genetic predictors of toxicity or elevated drug levels with time to treatment discontinuation during the first year of first-line ART. As the study is retrospective, and the genotype was obtained a posteriori, it can be considered an unbiased assessment of genetic association with treatment discontinuation.
METHODS
Study Design and Population
In the Swiss HIV Cohort Study (SHCS, http://www.shcs.ch) database, clinical and laboratory information were collected prospectively on standardized questionnaires through a semiannual structured interview and a set of prespecified biological analyses (The Swiss HIV Cohort Study [21]). Eligible study participants were all ART naive, starting one of the study medications (n = 688) between January 2004 and December 2007, and had at least 2 regular follow-up visits within 18 months after treatment initiation. Study ART included tenofovir or abacavir, 3TC or FTC, associated with lopinavir or atazanavir boosted with ritonavir (r) or efavirenz. Participant selection process was performed with Stata (version 10, StataCorp LP). The SHCS Genetics Project was approved by the ethics committees of all participating centers, and participants gave written informed consent for genetic testing. As this laboratory has been active in the description of associations of genetic markers with efavirenz pharmacokinetics [9–11, 22], lopinavir/r lipid metabolism and pharmacokinetics [13, 23], and atazanavir hyperbilirubinemia [16], we identified patients that could have been included in such previous studies. Only 1 of 272 individuals receiving efavirenz, 20 of 184 individuals receiving lopinavir/r, and none of 121 individuals receiving atazanavir had contributed to previous studies in this laboratory.
We defined a discontinuation episode as the first event of stop of each individual drug, whether the event included discontinuation of 1 or more drugs in the combination ART. At the time of treatment discontinuation, SHCS physicians are requested to fill in limited information that codes the reason for discontinuation. Codes are available for the following categories: treatment failure (virological), drug-associated toxicity (which includes coding for dyslipidemia, hypersensitivity reaction, gastrointestinal, liver, central nervous system, and kidney toxicity, and other toxicities), patient decision, physician decision, and other. Efavirenz dose reduction and the introduction of lipid-lowering agents in patients taking lopinavir/r were also considered as study events, and were coded as drug-related treatment modifications.
Genotyping
On the basis of existing knowledge [24] and recent publications [13–15, 18, 20], 23 genetic markers in 15 genes were included in the study (Table 1). Markers were genotyped using the Veracode technology (Illumina) with the exception of the UGT1A1 promoter variants (number of TA repeats in the TATA box associated with hyperbilirubinemia), which were analyzed by sequencing.
Table 1.
Genes, Genetic Variants, and Genetic Scores. Panel A: 23 genetic variants were selected on the basis of literature, and association with an intermediate pharmacokinetic or toxicity phenotype. Panel B: description of genetic risk scores. The numbers represent the count of variant alleles. Scores for efavirenz and lopinavir/r have been previously described. The atazanavir score has been explored post hoc
| A. | ||||||
| Drug | Gene | Genetic variant (rs number)a | Genetic variant change | Minor allele (nomenclature) | Expected minor allele effect | Reference |
| TDF | ABCC2 | rs2273697 | G>A | A | Risk | [19] |
| ABCC2 | rs717620 | C>T | T | Protective | [19, 20] | |
| ABCC4 | rs899494 | C>T | T | Risk | [19] | |
| EFV | CYP2A6 | rs28399433 | T>G | G (*9) | Risk | [10, 11] |
| CYP2B6 | rs3745274 | G>T | T (*6) | Risk | [8, 9, 22] | |
| rs35303484 | A>G | G (*11) | Risk | |||
| rs35979566 | T>A | A (*15) | Risk | |||
| rs28399499 | T>C | C (*18) | Risk | |||
| CYP3A4 | rs4646437 | C>T | T | Risk | [10, 11] | |
| LPV | APOA5 | rs3135506 | C>G | G | Risk | [12, 13] |
| rs662799 | T>C | C | Risk | |||
| CETP | rs708272 | G>A | A | Protective | ||
| DOCK7 | rs1748195 | C>G | G | Protective | ||
| GCKR | rs780094 | C>T | T | Risk | ||
| LPL | rs6586891 | A>C | C | Risk | ||
| rs328 | C>G | G | Protective | |||
| TRIB1 | rs17321515 | A>G | G | Protective | ||
| ABCC2 | rs717620 | C>T | T | Protective | [14] | |
| CYP3A | rs6945984 | T>C | C | Risk | [14] | |
| SLCO1B1 | rs11045819 | C>A | A (*4) | Protective | [14] | |
| rs4149056 | T>C | C (*5) | Risk | [14, 15] | ||
| ATV | ABCB1 | rs1045642 | C>T | T | Risk | [17] |
| NR1I2 | rs 2472677 | T>C | C | Protective | [18] | |
| UGT1A1 | rs8175347 | A(TA)6TAA > A(TA)5, 7 or 8TAA | (TA)7 (*28) | Risk | [16] | |
| (TA)5 (*36) | Protective | |||||
| (TA)8 (*37) | Risk | |||||
| B. | ||||||||||
| Efavirenz |
CYP2B6/2A6/3A4 six-group genetic score |
|||||||||
| Genes | Genetic variant | Score 1 | Score 2 | Score 3 | Score 4 | Score 5 | Score 6 | |||
| CYP2B6 | rs3745274 (*6) rs35303484 (*11) rs35979566 (*15) rs28399499 (*18) | 0 | 0 | 1 | 1 | 2 | 2 | |||
| CYP2A6 | rs28399433 (*9) | 0 | 1 to 4 | 0 | 1 to 4 | 0 | 1 to 4 | |||
| CYP3A4 | rs4646437 | |||||||||
| Lopinavir | Hypertriglyceridemia additive genetic scoreb,c | |||||||||
| Genes | Genetic variant | Score 1 | Score 2 | Score 3 | ||||||
| Risk variant | -4 to -2 | -1 to 0 | +1 to +4 | |||||||
| APOA5 | rs3135506 | |||||||||
| rs662799 | ||||||||||
| GCKR | rs780094 | |||||||||
| LPL | rs6586891 | |||||||||
| Protective variant | ||||||||||
| DOCK7 | rs1748195 | |||||||||
| TRIB1 | rs17321515 | |||||||||
| CETP | rs708272 | |||||||||
| LPL | rs328 | |||||||||
| Pharmacokinetic genetic scored | ||||||||||
| Score -2 | Score 0 | Score +2 | ||||||||
| SLCO1B1 | rs11045819 (*4) | 2 | 0 or 1 | 0 | ||||||
| rs4149056 (*5) | 0 | <2 | ≥2 | |||||||
| ABCC2 | rs717620, | |||||||||
| CYP3A | rs6945984 | |||||||||
| Atazanavir | UGT1A1/ABCB1 additive genetic score | |||||||||
| Genes | Genetic variant | Score 0 | Score 1 | Score 2 | Score 3 | Score 4 | ||||
| UGT1A1 | rs8175347 (*28, *37) | 0 | 0 | 1 | 1 | 0 | 2 | 1 | 2 | 2 |
| ABCB1 | rs1045642 | 0 | 1 | 0 | 1 | 2 | 0 | 2 | 1 | 2 |
NOTE. ABCB1: ATP-binding cassette, subfamily B (MDR/TAP), member 1; ABCC2 and 4: ATP-binding cassette subfamily C members 2 and 4; APOA5: apolipoprotein A5; ATV: atazanavir; CETP: cholesteryl ester transfer protein; CYP2A6, 2B6, and 3A4: Cytochrome P-450 2A6, 2B6, and 3A4; DOCK7: dedicator of cytokinesis 7; EFV: efavirenz; GCKR: glucokinase (hexokinase 4) regulator; HCP5: HLA complex P5; LPL: lipoprotein lipase; LPV: lopinavir/r; NR1I2: nuclear receptor subfamily 1, group I, member 2; SLCO1B1: solute carrier organic anion transporter family, member 1B1; TDF: tenofovir; TRIB1: tribbles homolog 1 (Drosophila); UGT1A1: UDP glucuronosyltransferase 1 family polypeptide A1.
a The rs numbers are the accession numbers in the National Center for Biotechnology Information single-nucleotide polymorphism database, dbSNP.
b Described previously by Arab-Alameddine et al. in the study of efavirenz plasma levels [11].
c Described previously by Rotger et al. The numbers represents the score values resulting from adding and subtracting the number of variant alleles of the risk and protective genetic variants, respectively [13].
d Described previously by Lubomirov et al. in the study of lopinavir plasma levels; patients with Score -2 have low plasma levels, and with Score +2 have high plasma levels compared to the Score 0 [14].
Genetic Risk and Scores
For each drug, we assessed the contribution of each independent allele (Table 1). When supported by the literature, we also assessed their joint contribution in a genetic score. Genetic scores were established a priori based on literature by using an additive model [25] (Table 1).
Data Analyses
Rates of ART discontinuation were compared between individuals with and without genetic risk markers using Cox proportional hazards regression models. The nongenetic predictors included: age (stratified in 3 categories: <40, 40–49, and >50 years), body weight (stratified in 4 categories: <60, 60–69, 70–79, and >80 kg), sex, ethnicity (Caucasian, African, Asian, Hispanic, and other), CD4 T cell count (in 100-cell strata) and viral RNA concentration (log10 transformed) at the time of starting ART (baseline), transmission risk group (men who have sex with men [MSM], heterosexual, blood, injection drug user [IDU], other, and unknown), year of starting ART (2004, 2005, 2006, and 2007), ART regimen, center, and pregnancy status. Markers with potential prognostic value after univariable analyses were included in a multivariable Cox model for association with the ART treatment discontinuation during the 1-year follow-up period. The proportional hazards assumptions were assessed for all models using Schoenfeld residuals. We used Stata software, version 11 (StataCorp LP) for analyses.
RESULTS
Demographic and Genetic Characteristics of Participants
Of a total of 668 patients starting ART with the study medication (ie, tenofovir, efavirenz, lopinavir/r, or atazanavir), 577 individuals were successfully genotyped and included in the analysis. Reasons for exclusion included an incomplete genetic consent (n = 5), unavailable biological material (n = 7), unsuccessful genotyping (n = 2), and samples that were not included because the standard size of the genotyping array was reached (n = 77). The 91 individuals not included in the analysis were comparable in age, sex, and ethnicity to those included, but more likely to receive efavirenz (86% vs 47%) because such samples were preferentially excluded once the array capacity was reached. Most of the participants were Caucasian (80%) and men (73%). ART regimen included tenofovir/emtricitabine or lamivudine (n = 500) or abacavir/emtricitabine or lamivudine (n = 77), and efavirenz (n = 272) or lopinavir/r (n = 184) or atazanavir boosted with ritonavir (n = 121). Age was median 44 years (interquartile range [IQR] 37–50 years). The presumed mode of HIV transmission was men who had sex with men (44.4%), heterosexual (40.6%), injection drug users (11.4%), and unknown/other (3.6%). Baseline (ART start) CD4+ T cell count was median 209 cells/μL (IQR: 125–282 cells/μL), and HIV viral load was median 4.9 log copies/mL (IQR: 4.4–5.4 log copies/mL). The exact time span of the study was from 5 January 2004 (earliest ART initiation) to 4 November 2008 (last registered follow-up). The median follow-up time (interval between ART initiation date and last registered follow-up date) was 618 days (IQR: 463–914 days). A total of 53 (9%) participants were lost for follow-up during the first year after ART initiation, and were censored. Distribution of alleles and genotypes were comparable for the populations receiving or not receiving the drug concerned by each specific genetic variant, with the exception of CYP2B6 rs28399499, a genetic variant only present in Africans. A total of 190 (33%) individuals discontinued ART; in 117 instances, this concerned a single drug; in 62 instances, two or more components of the regimen; and 11 individuals stopped 2 or more drugs at different times.
Genetic Risk and Discontinuation of Tenofovir
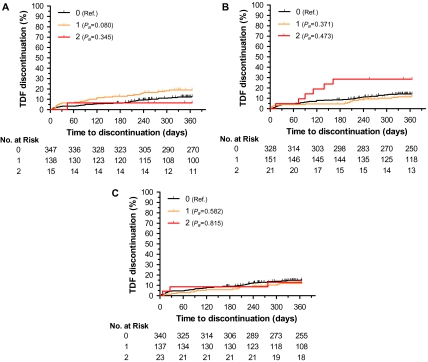
Among 500 patients receiving tenofovir, 70 (14%) discontinued treatment in the first year. The various genetic variants evaluated did not present a statistically significant association with rates of treatment discontinuation (Table 2). In the first year, individuals carrying 1 (CT) or 2 (TT) risk alleles of ABCC4 rs899494 discontinued tenofovir with cumulative rates of 19% and 7%, respectively, compared with 13% among individuals without the genetic risk allele (CC): adjusted hazard ratio (HRa) (CT) 1.60 (95% CI: 0.95–2.70), Pa = .080; and HRa (TT) 0.38 (95% CI: 0.05–2.85), Pa = .345, respectively (Figure 1A). Individuals carrying 1 (GA) or 2 (AA) risk alleles of ABCC2 rs2273697 variant discontinued tenofovir with cumulative rates of 12.26% and 28.60%, respectively, compared with 14.23% among individuals without the genetic risk allele (GG): HRa (GA) 0.77 (95% CI: 0.44–1.36), Pa = .371; and HRa (AA) 1.39, (95% CI: 0.58–3.40), Pa = .473, respectively (Figure 1B). We did not observe differences in discontinuation rates associated with ABCC2 rs717620 genotypes (Figure 1C). The final Cox analysis identified body weight as significant covariable. Individuals with body weight lower than 60 kg or between 60 and 69 kg had higher risk of discontinuation (HRa = 2.71, 95% CI: 1.14–6.44, Pa = .024; and HRa = 2.43, 95% CI: 1.13–5.26, Pa = .024, respectively) compared with the reference group (70–79 kg).
Table 2.
Detailed Results From the Cox Regression Models for Each Drug
| Drug | SNP/score | Genotype/score group | Adjusted hazard ratio (95% confidence interval) | Adjusted P value | Cumulative discontinuation rate (%) |
| Tenofovir | ABCC4 rs899494 | CC (0) | 1 (Reference) | — | 12.66 |
| CT (1) | 1.60 (0.95–2.70) | .080 | 19.00 | ||
| TT (2) | 0.38 (0.05–2.85) | .345 | 6.70 | ||
| ABCC2 rs2273697 | GG (0) | 1 (Reference) | — | 14.23 | |
| GA (1) | 0.77 (0.44–1.36) | .371 | 12.26 | ||
| AA (2) | 1.39 (0.58–3.40) | .473 | 28.60 | ||
| ABCC2 rs717620 | CC (0) | 1 (Reference) | — | 15.30 | |
| CT (1) | 0.85 (0.48–1.52) | .582 | 11.80 | ||
| TT (2) | 1.15 (0.34–3.87) | .815 | 13.04 | ||
| Efavirenz | CYP2B6/2A6/3A4 score | Score 1 | 1 (Reference) | — | 32.30 |
| Score 2 | 0.85 (0.43–1.70) | .648 | 28.90 | ||
| Score 3 | 0.57 (0.28–1.17) | .125 | 22.00 | ||
| Score 4 | 0.61 (0.27–1.38) | .234 | 29.90 | ||
| Score 5 | 0.86 (0.24–3.10) | .820 | 23.00 | ||
| Score 6 | 2.10 (0.70–6.01) | .185 | 71.15 | ||
| CYP2B6/2A6/3A4 score (dichotomized) | Score 1–5 | 1 (Reference) | — | 28.10 | |
| Score 6 | 3.14 (1.35–7.33) | .008 | 71.15 | ||
| Lopinavir | Hypertriglyceridemia additive score | Score 1 | 1 (Reference) | — | 30.00 |
| Score 2 | 0.99 (0.52–1.88) | .967 | 35.90 | ||
| Score 3 | 1.27 (0.61–2.61) | .523 | 36.40 | ||
| Pharmacokinetic score | Score -2 | 1 (Reference) | — | 20.00 | |
| Score 0 | 2.03 (0.26–15.87) | .499 | 34.00 | ||
| Score +2 | 2.15 (0.27–17.27) | .473 | 36.00 | ||
| Atazanavir | UGT1A1 rs8175347 | *1/*1 | 1 (Reference) | — | 14.60 |
| *1/*28, *37 | 1.97 (0.77–5.03) | .158 | 23.80 | ||
| *28/*28, *37 | 9.13 (3.38–24.69) | <.0001 | 62.50 | ||
| NR1I2 rs2472677 | TT (0) | 1 (Reference) | — | 21.80 | |
| TC (1) | 1.42 (0.61–3.26) | .413 | 28.90 | ||
| CC (2) | 1.1 (0.32–3.67) | .905 | 22.30 | ||
| ABCB1 rs1045642 | CC (0) | 1 (Reference) | — | 21.30 | |
| CT (1) | 1.54 (0.55–4.32) | .416 | 22.50 | ||
| TT (2) | 2.33 (0.84–6.50) | .105 | 34.80 | ||
| UGT1A1/ABCB1 additive score | Score 0+1 | 1 (Reference) | — | 14.61 | |
| Score 2 | 2.53 (0.95–6.70) | .062 | 29.16 | ||
| Score 3 | 5.44 (1.86–15.90) | .002 | 39.40 | ||
| Score 4 | 7.22 (1.90–27.50) | .004 | 57.14 |
NOTE. SNP: single-nucleotide polymorphism.
The nongenetic covariables evaluated and included when appropriate included body weight, sex, ethnicity, CD4 T cell count and viral RNA concentration, transmission category, year of starting ART, treatment regimen, clinical center, and pregnancy status.
Figure 1.
Genetic risk of tenofovir discontinuation at 1 year. Cumulative rates of discontinuation for 500 participants stratified by ABCC4 and ABCC2 genetic variants: (A) by ABCC4 rs899494 genotypes CC (0), CT (1), and TT (2); (B) by ABCC2 rs2273697 genotypes GG (0), GA (1), and AA (2); and (C) by ABCC2 rs717620 genotypes CC (0), CT (1), and TT (2). Pa, covariate adjusted P value, estimated from Cox regression models. NOTE. CC: individuals without the genetic risk allele ABCC4 rs899494; CT: individuals carrying 1 risk allele ABCC4 rs899494; TT: individuals carrying 2 risk alleles ABCC4 rs899494; GG: individuals without genetic risk allele ABCC2 rs2273697; GA: individuals carrying 1 genetic risk allele ABCC2 rs2273697; AA: individuals carrying 2 genetic risk alleles ABCC2 rs2273697.
Genetic Risk and Discontinuation of Efavirenz
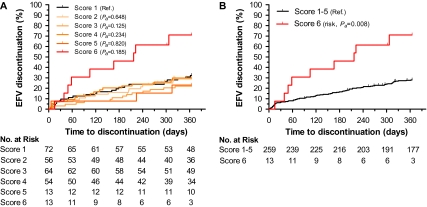
Among 272 patients receiving efavirenz, 81 discontinued treatment in the first year with a cumulative rate of 30%. For 3 individuals, the event was a dose reduction rather than full discontinuation; 2 of the 3 had neuropsychological toxicity as the stated reason for dose reduction. The various genetic variants and scores were associated with different rates of treatment discontinuation (Table 2). Consistent with the known pharmacokinetic associations, the data indicated that loss of CYP2B6 function (homozygocity, decrease/loss of function alleles) with a concomitant decrease of function in accessory metabolic pathways (decrease/loss of function alleles of CYP2A6 and/or CYP3A4) was associated with a higher risk of discontinuation (Figure 2). Individuals with the highest genetic risk score (score 6) discontinued efavirenz more frequently than individuals with lower genetic risk scores, with cumulative rates of 71.15% vs 28.10% (HRa = 3.14, 95% CI: 1.35–7.33, Pa = .008) (Table 2). Among covariates, sex showed an independent statistically significant effect on efavirenz discontinuation, with women showing a higher risk (HRa = 3.08, 95% CI: 1.62–5.85, Pa = .001), consistent with published estimates [26–28].
Figure 2.
Genetic risk of efavirenz discontinuation at 1 year. Cumulative rates of discontinuation for 272 participants stratified by CYP2B6, CYP2A6, and CYP3A4 genetic variants by six-group genetic score (A) and by dichotomized genetic score. Pa, covariate adjusted P value (B).
Genetic Risk and Discontinuation of Lopinavir
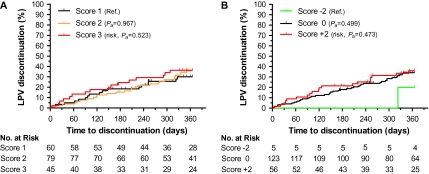
Among 184 patients receiving lopinavir, 60 discontinued treatment in the first year, with a cumulative rate of 34%. Eight genetic variants associated with hypertriglyceridemia [12, 13] were evaluated individually and in unweighted additive scores. Only CETP rs708272 G>A genotypes displayed when assessed in isolation, assuming a dominant genetic model (GG vs GA + AA), a marginally significant protection from early treatment discontinuation (HRa = 0.60, 95% CI: 0.35–1.01, Pa = .056). The unweighted 3-level additive score (Table 1) did not show an association of genetic variants with early treatment discontinuation (Figure 3A and Table 2).
Figure 3.
Genetic risk of lopinavir discontinuation at 1 year. Cumulative rates of discontinuation for 184 participants stratified by hypertriglyceridemia (A) and pharmacokinetic (B) genetic scores. Pa, covariate adjusted P value.
Separately, we assessed the contribution of 4 genetic variants in 3 genes that are associated with lopinavir/r pharmacokinetics [14]. None of the 4 single-nucleotide polymorphisms (SNPs) were individually associated with treatment discontinuation. A 3-level genetic score (Table 1) did not identify a significant statistical association with treatment discontinuation, although we observed low rates of treatment discontinuation among the rare individuals homozygous for SLCO1B1*4 (rs11045819), a gain-of-function variant associated with low lopinavir/r levels (Figure 3B). No other covariates were associated with lopinavir/r discontinuation at 1 year.
Genetic Risk and Discontinuation of Atazanavir
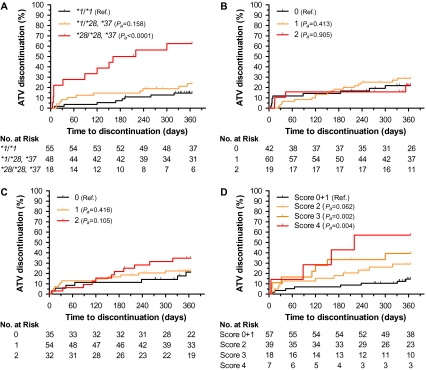
Among 121 patients receiving atazanavir boosted with ritonavir, 30 discontinued treatment in the first year with a cumulative rate of 25%. Homozygocity of decreased function UGT1A1 alleles (*28/*28 or *28/*37) was associated with risk of treatment discontinuation (HRa = 9.13, 95% CI: 3.38–24.69, Pa < .0001) (Figure 4A and Table 2). There was a statistically nonsignificant increase in risk associated with the carrier state (HRa = 1.97, 95% CI: 0.77–5.03, Pa =.158). First-year cumulative rates of treatment discontinuation were 62.5% for homozygous, 23.8% for heterozygous, and 14.6% for noncarrier individuals. No other covariates were associated with atazanavir discontinuation at 1 year. We explored whether differences in rates of discontinuation across centers (varying from 0% to 47%) could reflect center-specific policy and threshold of response to the specific toxicity. For the 4 centers prescribing atazanavir to more than 10 individuals, there was a statistically significant correlation between discontinuation rates and frequency of UGT1A1*28 homozygocity (r2 = 0.858, P =.048).
Figure 4.
Genetic risk of atazanavir discontinuation at 1 year. Cumulative rates of discontinuation for 121 participants stratified by UGT1A1, NR1I2, and ABCB1 genetic variants: (A) by UGT1A1 rs8175347 genotypes *1/*1, *1/*28 or *37, and *28/*28 or *37; (B) by NR1I2 rs2472677 genotypes TT (0), CT (1), and CC (2); (C) by ABCB1 rs1045642 genotypes CC (0), CT (1), and TT (2); (D) by UGT1A1/ABCB1 additive genetic score. Pa, covariate adjusted P value. NOTE. TT: individuals carrying 2 risk alleles ABCC4 rs899494; CT: individuals carrying 1 risk allele ABCC4 rs899494; CC: individuals without the genetic risk allele ABCC4 rs899494.
Two other studies have described the association of variants of NR1I2 and ABCB1 genes to atazanavir pharmacokinetics and/or hyperbilirubinemia [17, 18]. We did not observe an association of NR1I2 variant and treatment discontinuation rates (Figure 4B). Variant rs1045642 G>A of ABCB1 was associated with different rates of treatment discontinuation (Figure 4C). Homozygous individuals had an HRa of 2.33 (95% CI: 0.84–6.50), although the results did not reach statistical significance (Pa =.105). The inclusion of ABCB1 variant in an additive UGT1A1/ABCB1 genetic score (Table 1) stratified the risk of discontinuation conferred by UGT1A1 variants (Figure 4D). In the joint analysis, the cumulative discontinuation rate ranged from 29% to 57% across genetic risk scores. However, these UGT1A1/ABCB1 genetic score–containing Cox regression model did not reduce the Akaike information criterion compared with the model containing only UGT1A1 (262.93 vs 257.40, respectively).
Early Discontinuation
Eleven (2%) of the participants discontinued treatment within the first week—the earliest discontinuation was on day 2. Six patients discontinued tenofovir, none were homozygous for the risk allele of ABCC2 rs2273697. Four patients discontinued efavirenz; none had a genetic risk score of 6. Three patients discontinued lopinavir/r; 1 had a pharmacokinetic genetic risk score of +2, and 2 had a dyslipidemia genetic risk score of 3. Two patients discontinued atazanavir; both were homozygous for the risk allele of UGT1A1 rs8175347.
Evaluation of Coded Reasons for Discontinuation
During routine care, the SHCS physician indicates a coded reason for discontinuation at the time of treatment stop or modification. Retrospective assessment of the codes for the overall study identified the following categories as main reasons: drug-associated toxicity (15%), physician decision (8%), and patient decision (6%), which are in accordance with previously reported reasons for treatment change [4]. The reported reasons for treatment discontinuation are presented for each drug in Table 3. Based on the data presented in the previous sections, we used a score of 6 to define efavirenz genetic risk. For atazanavir, we defined risk by the simpler UGT1A1-based risk (promoter allele *28/28 or *28/*37) based on the Akaike information criterion. There is no defined genetic risk for treatment discontinuation for tenofovir or lopinavir/r. For tenofovir, we explored whether the trend observed for ABCC2 rs2273697 was further supported in the clinical analysis. For lopinavir/r, we considered the genetic risk for dyslipidemia as surrogate for the genetic risk of treatment discontinuation. The code “drug-associated toxicity” was invoked in 62% of instances when efavirenz was discontinued among individuals with a genetic risk versus 12% in those without (P < .0001). The code “drug-associated toxicity” was invoked in 33% of instances when atazanavir was discontinued among individuals with a genetic risk versus 7% in those without (P = .004).
Table 3.
Reasons for Drug Discontinuation According to the Coding Provided by the Treating Physician.
| Tenofovira |
Efavirenzb |
Lopinavir/rc |
Atazanavird |
|||||||||
| Reason for drug discontinuation | Population with genetic risk, n = 21Discontinuation events (%) | Population without genetic risk, n = 479Discontinuation events (%) | Pe | Population with genetic risk, n = 13Discontinuation events (%) | Population without genetic risk, n = 259Discontinuation events (%) | Pe | Population with HTG genetic risk, n = 45Discontinuation events (%) | Population without HTG genetic risk, n = 139Discontinuation events (%) | Pe | Population with genetic risk, n = 18Discontinuation events (%) | Population without genetic risk, n = 103Discontinuation events (%) | Pe |
| Virological treatment failure | 0 (0.00) | 6 (1.25) | 1 | 0 (0.00) | 15 (5.79) | 1 | 0 (0.00) | 0 (0.00) | 1 | 0 (0.00) | 0 (0.00) | 1 |
| Drug associated toxicity | 3 (14.29) | 22 (4.59) | .081 | 8 (61.54)f | 32 (12.36) | <.0001 | 6 (13.33) | 12 (8.63) | .389 | 6 (33.33) | 7 (6.80) | .004 |
| Patient decision | 1 (4.76) | 22 (4.59) | 1 | 1 (7.69) | 8 (3.09) | .361 | 5 (11.11) | 11 (7.91) | .545 | 2 (11.11) | 6 (5.83) | .339 |
| Physician decision | 0 (0.00) | 12 (2.51) | 1 | 0 (0.00) | 12 (4.63) | 1 | 5 (11.11) | 20 (14.39) | .803 | 2 (11.11) | 5 (4.85) | .279 |
| Other | 2 (9.52) | 2 (0.42) | .009 | 0 (0.00) | 5 (1.93) | 1 | 0 (0.00) | 1 (0.72) | 1 | 1 (5.56) | 1 (0.97) | .274 |
NOTE. Based on the data presented in the text, we used score 6 to define efavirenz genetic risk. For atazanavir, we defined risk by the simpler UGT1A1-based risk (promoter allele *28/28 or *28/*37) based on the Akaike information criterion. Because there is no defined genetic risk for treatment discontinuation of tenofovir, homozygocity ABCC2 rs2273697 was further explored for the analysis of reasons for treatment discontinuation. For lopinavir/r, we considered the genetic risk for dyslipidemia as surrogate for the analysis of reasons for treatment discontinuation.
Population with (AA or GA) and without (GG) genetic risk of tenofovir discontinuation according to the ABCC2 rs2273697 variant genotypes.
Population with (score 6) and without (scores 1 to 5) genetic risk of efavirenz discontinuation according to the CYP2B6/2A6/3A genetic score.
Population with (score 3) and without (scores 1 and 2) genetic risk of lopinavir/r discontinuation according to the risk of hypertriglyceridemia (HTG) additive genetic score.
Population with (*28/*28 or *28/*37) and without (*1/*1, *1/*28, or *1/*36) genetic risk of atazanavir discontinuation according to the UGT1A1 rs8175347 variant genotypes.
Fisher exact test P value.
Includes 3 instances of treatment dose modification.
DISCUSSION
This study tested the hypothesis that biological consequences of genetic variants will, if significant or severe, lead to treatment discontinuation, a well defined clinical outcome. Among 23 genetic variants included in the study, those in CYP2B6, 2A6, and 3A4 previously associated with plasma levels of efavirenz, and those in UGT1A1 previously associated with atazanavir-induced hyperbilirubinemia were also associated with early treatment discontinuation. The retrospective study was not designed to assess other clinical endpoints such as specific toxicities; however, assessment of the codes used in routine care to indicate the reasons for treatment discontinuation convincingly associated genetic risk to toxicity for those 2 drugs.
Thus, the study suggests that assessment of the genetic markers could lead to improved prescription of atazanavir and efavirenz. Unconjugated hyperbilirubinemia, the main adverse effect of atazanavir, is widely considered as irrelevant in clinical care [29]. However, despite the reversibility and the absence of clinical consequences of hyperbilirubinemia, 2 of 3 individuals homozygous for the UGT1A1 promoter variant discontinued atazanavir in the present study. Differences in rates of discontinuation across centers reflected the frequency of homozygous individuals receiving the drug. Overall, 10% of Caucasians carry this genetic risk, but its frequency varies across ethnic groups [30].
The basis of treatment discontinuation of efavirenz is more complex. While the genetic determinants of efavirenz blood levels are by now well understood [10, 11], there is no consensus as to whether high drug levels are associated with neuropsychotoxicity, or on the usefulness of therapeutic drug monitoring and dose adjustment [31]. These considerations notwithstanding, the study suggests that treatment discontinuation correlates with decreased metabolism of efavirenz, and that the loss of the primary metabolic pathway, CYP2B6, is necessary but not sufficient [32] for an increased rate in efavirenz discontinuation. Extremely high plasma drug levels of efavirenz results from loss of both primary (CYP2B6) and accessory (CYP3A4 and/or CYP2A6) metabolic pathways [10]. Consistent with such pharmacokinetic data, individuals that have loss of accessory metabolisms in the context of homozygous loss of function of CYP2B6 present the highest risk of treatment discontinuation. As pointed out for atazanavir, these data question the interest to initiate efavirenz in individuals with the genetic risk if two-thirds of them will subsequently discontinue the drug.
The study included tenofovir despite the paucity of genetic data associated with toxicity for this drug. We analyzed the genetic variants as proposed by Izzedine et al. [19] in a study of 13 individuals with tubular dysfunction on tenofovir, and by Rodriguez-Novoa et al. [20] in a study of 115 HIV-infected patients of whom 19 had tubular dysfunction. The current study suggests a possible association of rs2273697 G>A variant in ABCC2 with higher rates of drug discontinuation. The SHCS database did not allow for detailed analysis of the reasons for discontinuation in these individuals; in particular, for renal function (see below). Thus, the data on rs2273697 G>A should be considered preliminary; additional discovery efforts on the genetics of tenofovir are needed.
The case of lopinavir is a pertinent example of how a well-documented association of genetic variants and a laboratory phenotype (dyslipidemia) with potential long-term consequences—cardiovascular disease—may not translate into short-term clinical decisions. We did not document an association between the proposed risk score and treatment discontinuation or introduction of a lipid-lowering agent. However, the most appropriate clinical endpoint here would have been the actual laboratory data (reaching clinically relevant cut-offs of dyslipidemia) [12]. The patient carrying the genetic risk could be prescribed an alternative drug on that basis. Prediction of dyslipidemia will improve with the increasing availability of genetic markers [13, 33].
This study did not include abacavir hypersensitivity because clinical testing for HLA*B5701 was introduced during the study period. We did, however, notice that the allelic frequency for the risk marker was lower in patients that received abacavir than among those that did not (data not shown)—confirming that the patients were increasingly screened before treatment initiation. Another relevant observation of the present study is that although genetic predisposition to toxicity might lead, through poor adherence, to virological treatment failure, there were few (n = 15) instances coded as virological treatment failure, and none among participants with the genetic risk marker. The current analysis did not consider drug adherence; its inclusion would have been informative.
This study should be considered as a pilot analysis that can provide data for power calculation, and support further discovery efforts toward identification of additional genetic markers. Thereafter, a prospective clinical trial should ideally formalize the analysis, and provide the basis for measuring the cost effectiveness of this approach [34].
Funding
This work has been partially financed within the framework of the SHCS funded by the Swiss National science foundation (grant number 33CSC0-108787 and 324730-12493), by the SHCS research foundation, and by the SHCS project number 548.
Acknowledgments
We thank the patients for participating in the Swiss HIV Cohort study, all the physicians and study nurses of the centers for excellent patient care, and the data center for data management.
The members of the Swiss HIV Cohort Study (SHCS) are M. Battegay, E. Bernasconi, J. Böni, H.C. Bucher, P. Bürgisser, A. Calmy, S. Cattacin, M. Cavassini, R. Dubs, M. Egger, L. Elzi, M. Fischer, M. Flepp, A. Fontana, P. Francioli (President of the SHCS), H. Furrer (Chairman of the Clinical and Laboratory Committee), C.A. Fux, M. Gorgievski, H.F. Günthard (Chairman of the Scientific Board), H.H. Hirsch, B. Hirschel, I. Hösli, C. Kahlert, L. Kaiser, U. Karrer, C. Kind, T. Klimkait, B. Ledergerber, G. Martinetti, N. Müller, D. Nadal, F. Paccaud, G. Pantaleo, A. Rauch, S. Regenass, M. Rickenbach (Head of Data Center), C. Rudin (Chairman of the Mother & Child Substudy), P. Schmid, D. Schultze, J. Schüpbach, R. Speck, B.M. de Tejada, P. Taffé, A. Telenti, A. Trkola, P. Vernazza, R. Weber, and S. Yerly.
Author Contributions: ICMJE criteria for authorship read and met: R.L., S.C., and A.T. Agree with the manuscript’s results and conclusions: R.L., S.C., J.d.I., R.M., B.L., M.C., B.H., E.B., L.E., P.V., H.F., H.F.G., and A.T. Designed the experiments/the study: R.L., S.C., B.L., and A.T. Analyzed the data: R.L., B.L., and S.C. Collected data/did experiments for the study: S.C., J.d.I., and R.M. Contributed clinical data: M.C., B.H., E.B., L.E., P.V., H.F., and H.F.G. Wrote the first draft of the paper: R.L., S.C., and A.T. Contributed to the writing of the paper: all authors.
References
- 1.Fellay J, Boubaker K, Ledergerber B, et al. Prevalence of adverse events associated with potent antiretroviral treatment: Swiss HIV Cohort Study. Lancet. 2001;358:1322–7. doi: 10.1016/s0140-6736(01)06413-3. [DOI] [PubMed] [Google Scholar]
- 2.Keiser O, Fellay J, Opravil M, et al. Adverse events to antiretrovirals in the Swiss HIV Cohort Study: effect on mortality and treatment modification. Antivir Ther. 2007;12:1157–64. [PubMed] [Google Scholar]
- 3.Vo TT, Ledergerber B, Keiser O, et al. Durability and outcome of initial antiretroviral treatments received during 2000–2005 by patients in the Swiss HIV Cohort Study. J Infect Dis. 2008;197:1685–94. doi: 10.1086/588141. [DOI] [PubMed] [Google Scholar]
- 4.Elzi L, Marzolini C, Furrer H, et al. Treatment modification in human immunodeficiency virus-infected individuals starting combination antiretroviral therapy between 2005 and 2008. Arch Intern Med. 2010;170:57–65. doi: 10.1001/archinternmed.2009.432. [DOI] [PubMed] [Google Scholar]
- 5.d'Arminio Monforte A, Lepri AC, Rezza G, et al. Insights into the reasons for discontinuation of the first highly active antiretroviral therapy (HAART) regimen in a cohort of antiretroviral naive patients. I.CO.N.A. Study Group. Italian Cohort of Antiretroviral-Naive Patients. AIDS. 2000;14:499–507. doi: 10.1097/00002030-200003310-00005. [DOI] [PubMed] [Google Scholar]
- 6.Sabin CA, Smith CJ, Delpech V, et al. The associations between age and the development of laboratory abnormalities and treatment discontinuation for reasons other than virological failure in the first year of highly active antiretroviral therapy. HIV Med. 2009;10:35–43. doi: 10.1111/j.1468-1293.2008.00654.x. [DOI] [PubMed] [Google Scholar]
- 7.Mallal S, Phillips E, Carosi G, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358:568–79. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 8.Haas DW, Ribaudo HJ, Kim RB, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS. 2004;18:2391–400. [PubMed] [Google Scholar]
- 9.Rotger M, Colombo S, Furrer H, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics. 2005;15:1–5. doi: 10.1097/01213011-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 10.di Iulio J, Fayet A, Arab-Alameddine M, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genomics. 2009;19:300–9. doi: 10.1097/FPC.0b013e328328d577. [DOI] [PubMed] [Google Scholar]
- 11.Arab-Alameddine M, di Iulio J, Buclin T, et al. Pharmacogenetics-based population pharmacokinetic analysis of efavirenz in HIV-1-infected individuals. Clin Pharmacol Ther. 2009;85:485–94. doi: 10.1038/clpt.2008.271. [DOI] [PubMed] [Google Scholar]
- 12.Arnedo M, Taffe P, Sahli R, et al. Contribution of 20 single-nucleotide polymorphisms of 13 genes to dyslipidemia associated with antiretroviral therapy. Pharmacogenet Genomics. 2007;17:755–64. doi: 10.1097/FPC.0b013e32814db8b7. [DOI] [PubMed] [Google Scholar]
- 13.Rotger M, Bayard C, Taffe P, et al. Contribution of genome-wide significant single-nucleotide polymorphisms and antiretroviral therapy to dyslipidemia in HIV-infected individuals: a longitudinal study. Circ Cardiovasc Genet. 2009;2:621–8. doi: 10.1161/CIRCGENETICS.109.874412. [DOI] [PubMed] [Google Scholar]
- 14.Lubomirov R, di Iulio J, Fayet A, et al. ADME pharmacogenetics: investigation of the pharmacokinetics of the antiretroviral agent lopinavir coformulated with ritonavir. Pharmacogenet Genomics. 2010;20:217–30. doi: 10.1097/FPC.0b013e328336eee4. [DOI] [PubMed] [Google Scholar]
- 15.Hartkoorn RC, Kwan WS, Shallcross V, et al. HIV protease inhibitors are substrates for OATP1A2, OATP1B1 and OATP1B3 and lopinavir plasma concentrations are influenced by SLCO1B1 polymorphisms. Pharmacogenet Genomics. 2010;20:112–20. doi: 10.1097/FPC.0b013e328335b02d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotger M, Taffe P, Bleiber G, et al. Gilbert syndrome and the development of antiretroviral therapy–associated hyperbilirubinemia. J Infect Dis. 2005;192:1381–6. doi: 10.1086/466531. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Novoa S, Martin-Carbonero L, Barreiro P, et al. Genetic factors influencing atazanavir plasma concentrations and the risk of severe hyperbilirubinemia. AIDS. 2007;21:41–6. doi: 10.1097/QAD.0b013e328011d7c1. [DOI] [PubMed] [Google Scholar]
- 18.Siccardi M, D'Avolio A, Baietto L, et al. Association of a single-nucleotide polymorphism in the pregnane X receptor (PXR63396 C–>T) with reduced concentrations of unboosted atazanavir. Clin Infect Dis. 2008;47:1222–5. doi: 10.1086/592304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izzedine H, Hulot JS, Villard E, et al. Association between ABCC2 gene haplotypes tenofovir-induced proximal tubulopathy. J Infect Dis. 2006;194:1481–91. doi: 10.1086/508546. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez-Novoa S, Labarga P, Soriano V, et al. Predictors of kidney tubular dysfunction in HIV-infected patients treated with tenofovir: a pharmacogenetic study. Clin Infect Dis. 2009;48:e108–16. doi: 10.1086/598507. [DOI] [PubMed] [Google Scholar]
- 21.The Swiss HIV Cohort Study. Int J Epidemiol. 39. 2010. Cohort profile: the Swiss HIV cohort study; pp. 1179–89. [DOI] [PubMed] [Google Scholar]
- 22.Rotger M, Tegude H, Colombo S, et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther. 2007;81:557–66. doi: 10.1038/sj.clpt.6100072. [DOI] [PubMed] [Google Scholar]
- 23.Tarr PE, Taffe P, Bleiber G, et al. Modeling the influence of APOC3, APOE, TNF polymorphisms on the risk of antiretroviral therapy–associated lipid disorders. J Infect Dis. 2005;191:1419–26. doi: 10.1086/429295. [DOI] [PubMed] [Google Scholar]
- 24.Telenti A, Zanger UM. Pharmacogenetics of anti-HIV drugs. Annu Rev Pharmacol Toxicol. 2008;48:227–56. doi: 10.1146/annurev.pharmtox.48.113006.094753. [DOI] [PubMed] [Google Scholar]
- 25.Kathiresan S, Melander O, Anevski D, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–9. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 26.Spire B, Carrieri P, Garzot MA, L’henaff M, Obadia Y. Factors associated with efavirenz discontinuation in a large community-based sample of patients. AIDS Care. 2004;16:558–64. doi: 10.1080/09540120410001716342. [DOI] [PubMed] [Google Scholar]
- 27.Domingo P, Suarez-Lozano I, Torres F, et al. First-line antiretroviral therapy with efavirenz or lopinavir/ritonavir plus two nucleoside analogues: The SUSKA study, a nonrandomized comparison from the VACH cohort. J Antimicrob Chemother. 2008;61:1348–58. doi: 10.1093/jac/dkn121. [DOI] [PubMed] [Google Scholar]
- 28.Robison LS, Westfall AO, Mugavero MJ, et al. Short-term discontinuation of HAART regimens more common in vulnerable patient populations. AIDS Res Hum Retroviruses. 2008;24:1347–55. doi: 10.1089/aid.2008.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nettles RE, Child MJ, Bertz RJ, Schnittman S. Gilbert syndrome and the development of antiretroviral therapy–associated hyperbilirubinemia: genetic screening is unnecessary. J Infect Dis. 2006;193:1611–2. doi: 10.1086/503814. [DOI] [PubMed] [Google Scholar]
- 30.Innocenti F, Grimsley C, Das S, et al. Haplotype structure of the UDP-glucuronosyltransferase 1A1 promoter in different ethnic groups. Pharmacogenetics. 2002;12:725–33. doi: 10.1097/00008571-200212000-00006. [DOI] [PubMed] [Google Scholar]
- 31.Rotger M, Telenti A. Optimizing efavirenz treatment: CYP2B6 genotyping or therapeutic drug monitoring? Eur J Clin Pharmacol. 2008;64:335–6. doi: 10.1007/s00228-007-0440-z. [DOI] [PubMed] [Google Scholar]
- 32.Powers V, Ward J, Gompels M. CYP2B6 G516T genotyping in a UK cohort of HIV-positive patients: polymorphism frequency and influence on efavirenz discontinuation. HIV Med. 2009;10:520–3. doi: 10.1111/j.1468-1293.2009.00718.x. [DOI] [PubMed] [Google Scholar]
- 33.Manolio TA. Cohort studies and the genetics of complex disease. Nat Genet. 2009;41:5–6. doi: 10.1038/ng0109-5. [DOI] [PubMed] [Google Scholar]
- 34.Telenti A. Time (again) for a randomized trial of pharmacogenetics of antiretroviral therapy. Pharmacogenomics. 2009;10:515–6. doi: 10.2217/pgs.09.3. [DOI] [PubMed] [Google Scholar]