Abstract
Medulloblastoma is the most common pediatric malignant brain tumor, arising from aberrant cerebellar precursors' development, a process mainly controlled by Hedgehog (Hh) signaling pathway. Histone deacetylase HDAC1 has been recently shown to modulate Hh signaling, deacetylating its effectors Gli1/2 and enhancing their transcriptional activity. Therefore, HDAC may represent a potential therapeutic target for Hh-dependent tumors, but still little information is available on the physiological mechanisms of HDAC regulation. The putative tumor suppressor RENKCTD11 acts through ubiquitination-dependent degradation of HDAC1, thereby affecting Hh activity and medulloblastoma growth. We identify and characterize here two RENKCTD11 homologues, defining a new family of proteins named KCASH, as “KCTD containing, Cullin3 adaptor, suppressor of Hedgehog.” Indeed, the novel genes (KCASH2KCTD21 and KCASH3KCTD6) share with RENKCTD11 a number of features, such as a BTB domain required for the formation of a Cullin3 ubiquitin ligase complex and HDAC1 ubiquitination and degradation capability, suppressing the acetylation-dependent Hh/Gli signaling. Expression of KCASH2 and -3 is observed in cerebellum, whereas epigenetic silencing and allelic deletion are observed in human medulloblastoma. Rescuing KCASHs expression reduces the Hedgehog-dependent medulloblastoma growth, suggesting that loss of members of this novel family of native HDAC inhibitors is crucial in sustaining Hh pathway-mediated tumorigenesis. Accordingly, they might represent a promising class of endogenous “agents” through which this pathway may be targeted.
Introduction
Medulloblastoma (MB) is the most common malignant brain tumor in children, which accounts for 10% to 20% of primary central nervous system (CNS) neoplasms, and requires aggressive surgery and combined radiation and chemotherapeutic treatments [1]. Although the prognosis has improved in the last few years, still a large number of MB patients undergo tumor relapse or serious neurologic and cognitive adverse effects. These facts highlight the need for an improved knowledge of the complex molecular biology of MB to develop new and more efficient therapeutic strategies [2–4]. MB arises from defects in control of cerebellar development and differentiation and, in particular, from cerebellar granule cell precursors that maintain their proliferating and undifferentiated state after the maturation age [5,6]. Granule cell precursor proliferation and differentiation is a process mainly controlled by the level of Hedgehog (Hh) pathway activity [7–9], and Hh dysregulation is now recognized as a leading cause of MB tumorigenesis [5,7,10–12]. As a consequence, the mechanism that regulates Hh pathway activity is now considered among the most important therapeutic targets for MB treatment [13–16].
Histone deacetylases (HDACs) play a pivotal role in developmental processes and tumorigenesis, regulating gene expression through the modulation of the acetylation status of histones and nonhistonic proteins [17]. HDACs have been recently involved in Hh modulation [18]. In particular, HDAC1 has been recently demonstrated to deacetylate the transcription factors Gli1 and Gli2 (the main Hh pathway effectors) and, through this mechanism, enhance their transcriptional activity [18].
Because an increased HDAC activity is a common finding in cancer cells, HDACs have been proposed as effective targets for cancer therapy [17]. Several chemical classes of HDAC inhibitors (HDACi) have been identified and are currently being tested for human cancer therapy (reviewed in Lane and Chabner [19]). Nevertheless, scant information is still available on the mechanism of the physiological regulation of HDAC proteins.
Indeed, although a few endogenous cell signals have been described to regulate HDAC function through posttranslational modifications (e.g., phosphorylation, sumoylation, acetylation, and ubiquitination), the role of these regulatory events in cancer is not yet defined [20–22] (reviewed in Di Marcotullio et al. [23]).
Growing evidence indicates the relevance of ubiquitination-dependent control of HDACs. Ubiquitination is a posttranslational modification that involves the action of different enzymes (E1, E2, E3) and ends with the transfer of ubiquitin to substrate proteins, which are then targeted to the proteasome and degraded [24]. The oncosuppressors RENKCTD11 and Chfr downregulate HDAC1 by inducing its ubiquitin-dependent degradation [18,25]. Importantly, the frequent deletion or silencing of RENKCTD11 and Chfr observed in medulloblastoma (MB) and other cancer types highlights the relevance of physiological mechanisms that, by repressing HDAC function, may prevent tumorigenesis.
We previously reported that RENKCTD11 plays an important role during cerebellar granule differentiation [26] and its loss, associated to chromosome 17p deletion, is linked to MB formation [27]. RENKCTD11 was recently identified as a part of Cullin3 (Cul3) E3 ubiquitin ligase complex, which inhibits HDAC1 function by inducing its ubiquitination and subsequent degradation [18]. HDAC1 suppression is in turn responsible for the inhibitory role of RENKCTD11 on Hh signaling [18].
While searching for additional Hh suppressors in human cancers, we have cloned and characterized two homologues of the RENKCTD11. This finding prompted us to hypothesize that RENKCTD11-mediated control of HDAC1 could represent a model shared by additional tumor suppressors, which may act in a concerted fashion. The two new genes in fact allowed us to define a new family of proteins that we named KCASH, as “KCTD containing, Cullin3 adaptor, suppressor of Hedgehog.” The two genes KCASH2KCTD21 and KCASH3KCTD6 share a number of features, such as high homology and a BTB domain responsible for oligomerization and interaction with Cul3, specific expression in brain and cerebellum, suppressor activity on acetylation-dependent Hh/Gli signaling through ubiquitination and degradation of HDAC1, genetic loss or silencing in human primary MBs, and growth-inhibitory activity on MB cells.
The results presented here indicate that the three KCASH genes belong to a subfamily of KCTD containing proteins. The discovery of such a novel family suggests that these negative regulators of Hh/Gli signaling act in concert as native HDAC inhibitors and that their genetic or epigenetic defects are crucial in sustaining Hh/Gli and HDAC-mediated tumorigenesis. Accordingly, they might represent a promising class of endogenous “agents” through which this oncogenic addiction pathway may be targeted.
Materials and Methods
Human and Mouse Tissue Samples
Human primary MB specimens were collected during surgical resection with the approval of institutional review board as previously described [28]. RNA of normal human cerebellum (nine adult samples from 22- to 82-year-old subjects and four fetal samples from 22 to 37 weeks) were purchased from Biocat (Heidelberg, Germany), Ambion (Applied Biosystems, Foster City, CA), and BD Biosciences (San Jose, CA). Mouse normal cerebella and tissues were obtained from C57Bl/6 mice (Charles River, Calco, Italy).
Cell Cultures, Transfection, Treatments, and Luciferase Assay
HEK293T cells were cultured in Dulbecco modified Eagle medium (Gibco, Carlsbad, CA) plus 10% FBS, Daoy and D283 MB cells were cultured in minimum essential medium (Gibco, Carlsbad, CA), supplemented with 10 and 20% FBS, respectively, 1% sodium pyruvate, 1% nonessential amino acid solution, 1% l-glutamine, and penicillin/streptomycin. Transfections were performed with Lipofectamine 2000 or Plus (Invitrogen, Carlsbad, CA).
KCASH2 small interfering RNA (siRNA) was performed using SMART pool siRNA duplexes (30 nM; M-026714-01, Dharmacon) transfected with HiPerFect Reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. KCASH1 siRNA knockdown was performed with pools of siRNA duplexes (100 nM) (Ambion; cat. 4392420; IDs44957 and s44959) transfected with Lipofectamine 2000.
Retinoic acid (2.5 µM; all-trans-retinoic acid; Sigma-Aldrich, St Louis, MO), epidermal growth factor (Sigma-Aldrich; 20 ng/ml), and nerve growth factor (Upstate, Lake Placid, NY; 100 ng/ml) treatments were performed from 6 to 48 hours on D283 MB cells. Luciferase and Renilla activity were assayed with a dual-luciferase assay system (Promega, Madison, WI). Results are expressed as luciferase/Renilla ratios and represent the means ± SD of at least three experiments, each performed in triplicate.
Plasmids and Mutagenesis
The following plasmids were provided by other laboratories: 12x Gli-Luc (R. Tofgard, Karolinska Institutet, Sweden), pBJ5-HDAC1 (S. Shereiber, Harvard University, MA), pCMV-HDAC1 (P.L. Puri, The Burnham Institute, CA), pcDNACul1-HA, Cul2-myc, Cul3-myc (M. Pagano, New York University School of Medicine, NY). Polymerase chain reaction (PCR)-amplified complementary DNA (cDNA) of human Gli1, KCASH1, KCASH2, KCASH3, BTB-KCASH2, ΔBTBKCASH2, and mutated forms were tagged (HA, Flag, or Myc tag) and cloned in the pCXN mammalian expression vector. Single or multiple residues were mutated using the QuickChange site-directed or multisite mutagenesis kits (Stratagene, La Jolla, CA).
RNA Isolation and Quantitative Real-time PCR
Total RNA from tissue samples and cells was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions, and cDNA synthesis was performed using the SuperScript II First-Strand Synthesis kit (Invitrogen). Quantitative real-time PCR analysis of Gli1, KCTD11, KCASH2, KCASH3, β-actin, GAPDH, and HPRT messenger RNA (mRNA) was performed on cDNA using TaqMan gene expression assays according to the manufacturer's instructions (Applied Biosystems) and using the ABI Prism 7900HT (Applied Biosystems) as previously described [29]. The designed assays on demand were the following: human KCASH2 forward 5′-gcgagggcaggaggactact-3′, human KCASH2 reverse 5′-cgccagtgaggttgtatagagagctt-3′, human KCASH2 probe 6-FAM-caacccagtctcctgc-MGB; mouse KCASH2 forward 5′-gagcgagggcaggagtatttc-3′, mouse KCASH2 reverse 5′-cccccaacattcagtgtaatgg-3′, mouse KCASH2 probe 6-FAM-cacccagcctcctac-MGB. Each amplification reaction was performed in triplicate, and the average of the three threshold cycles was used to calculate the amount of transcripts in the sample (SDS software; ABI). mRNA quantification was expressed, in arbitrary units, as the ratio of the sample quantity to the calibrator or to the mean values of control samples. All values were normalized to three endogenous controls, GAPDH, β-actin, and HPRT.
Western Blot Analysis, Immunoprecipitation, and Ubiquitination Assay
Cells were lysed with buffer containing: 50 mM Tris-HCl (pH 7.6), deoxycholic acid sodium salt 1%, 150 mM NaCl, 1% NP40, 5 mM EDTA, 100 mM NaF, and protease inhibitors. Total protein extracts were then separated on a denaturing SDS-PAGE gel and evaluated by Western blot assay using the antibodies listed below.
For coimmunoprecipitation, lysates were incubated with agarose-conjugated Flag M2 beads (Sigma) for 2 hours at 4°C. Control sample antibody was saturated with peptide anti-Flag (Sigma). Beads were washed extensively with lysis buffer, and the complexes were evaluated by Western blot analysis. In vivo ubiquitination assays were performed as previously described [30]; briefly, cells were lysed with denaturing RIPA buffer to disrupt protein-protein interactions and then immunoprecipitated with the indicated antibodies.
Antibody sources were as follows: goat polyclonal antibody against actin (Santa Cruz Biotechnology, Santa Cruz, CA), secondary antibody anti-mouse/rabbit/goat IgG conjugated with HRP (Santa Cruz), and mouse monoclonal antibody against HA and against Myc (Santa Cruz) and against Flag (M2, Sigma). Anti-KCASH2 antibodies have been developed in-house or custom-made (Biosense, Milan, Italy).
Gene Copy Number Assay
KCASH2 and KCASH3 gene copy numbers were measured by quantitative real-time PCR. DNA from MB specimens and paired blood cells were isolated by overnight treatment with proteinase K at 50°C followed by phenol-chloroform extraction and precipitation with ethanol. PCRs were performed using TaqMan copy number assay according to the manufacturer's instructions (Applied Biosystems) on an ABI Prism 7900 Sequence Detection System (Applied Biosystems) according to the standard thermal profile (2 minutes at 50°C, 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C). For the relative quantification of allele copy number, two different methods have been used: the standard curve method and the comparative Ct method, in tumor DNA sample relative to the normal DNA from the same patient (calibrator) and relative to RNaseP as endogenous control.
Cell Proliferation and Colony Assays
Cell proliferation was evaluated by BrdU incorporation (4-hour pulse). Briefly, after the BrdU pulse, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100, and BrdU detection (Roche, Basel, Switzerland) was performed according to the manufacturer's instructions. Nuclei were counterstained with Hoechst reagent. At least 500 nuclei were counted in triplicate, and the number of BrdU-positive nuclei was recorded. For colony formation assays, 1 x 104 transfected Daoy MB cells were plated in 10-cm-diameter dishes, and after 2 weeks of neomycine selection, cell colonies were counted after staining in 20% methanol and crystal violet. In all experiments in which additive effects of KCASH1 and -2 or -3 were evaluated, the amount of total transfected plasmid was maintained the same, (i.e., 5 µg in the case of a single transfected plasmid, 2.5 µg of each plasmid, in the case of two transfected plasmids).
Results
Identification of KCASH Family Members
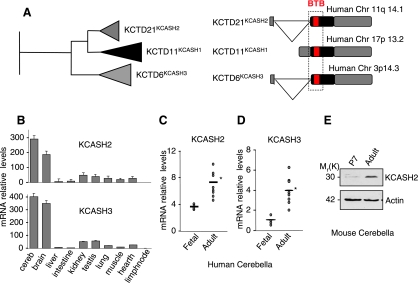
The search for protein homologues to RENKCTD11 on the public databases and subsequent analysis on the Ensemble genome browser [31] resulted in the identification of two ORF coding for potassium channel tetramerization domain containing 21 (KCTD21; NM_001029859.1) and potassium channel tetramerization domain containing 6 (KCTD6; NM_001128214.1) putative proteins (see Figures 1A and W1A). Both transcripts are encoded by biexonic genes mapping to human chromosomes 11q14.1 (KCTD21) and 3p14.3 (KCTD6), although the CDS are coded only by the second exon.
Figure 1.
Identification of KCASH family members. (A) Phylogenetic tree of KCTD11, KCTD21, and KCTD6 (left panel) and schematic representation of their human genetic locus (right panel). (B) KCASH2 and KCASH3 mRNA levels in adult mouse tissues evaluated by quantitative real-time PCR. Values shown are calculated relative to values in lymph nodes. KCASH2 (C) and KCASH3 (D) mRNA expression in fetal (n = 4) and adult (n = 9) human cerebella. Values obtained by quantitative real-time PCR. *P < .05 adult versus fetal cerebella. (E) Western blot analysis of endogenous levels of KCASH2 protein in immature (P7) and adult mouse cerebellum.
Similar to RENKCTD11, KCTD21 and KCTD6 contain a BTB domain sharing a high homology (58% [64/109 aa] and 54% [59/109 aa], respectively) versus the RENKCTD11 BTB motif, whereas the whole sequences present a 42% (111/260 aa) and 32% (77/237 aa) of homology, respectively (see Figure W1A).
These genes are conserved throughout most vertebrates (RENKCTD11 and KCTD6 in euteleostoma, KCTD21 in amniota) and have similarities to a gene coding for a KCTD-containing protein present in basal chordates such as the sea squirt Ciona intestinalis (ENSCSAVG00000010257, not shown). The phylogenetic tree seems to indicate that the KCTD6 gene branched earlier in the evolution than both RENKCTD11 and KCTD21 (Figure 1A).
The high homology within the three KCTD-containing proteins suggests that they belong to a gene family that shares common structural and functional features and may play similar biologic roles (see also below). For this reason and based on the functional features of RENKCTD11 [18], we have named this family KCASH (KCTD containing, Cullin3 adaptor, suppressor of Hedgehog). KCTD21 is therefore referred as KCASH2 and KCTD6 as KCASH3, whereas RENKCTD11 is renamed KCASH1.
Analysis of the expression of KCASH2 and KCASH3 in different mouse tissues indicated that both are highly expressed in cerebellum and brain, two organs in which also KCASH1 has been described to play a critical role in development and differentiation (Figure 1B and Argenti et al. [26]).
Accordingly, analysis of KCASH2 and KCASH3 mRNA expression levels in human cerebellar samples during development indicates an increase in the expression of the two genes in adult differentiated cerebellum compared with fetal cerebellum (Figure 1, C and D). To determine the expression of KCASH2- and KCASH3-encoded protein, we generated (and validate by siRNA) antibodies against KCASH2, which were able to recognize the endogenous 30-kDa protein (Figure W1B). Attempts to generate antibodies for KCASH3 have not been successful so far, probably because of the low immunogenicity of the protein. Analysis of KCASH2 protein levels in mouse adult and P7 cerebellar samples confirmed the modulation of the mRNA observed during development in human (Figure 1E).
KCASH Family Members Form Homo-oligomeric and Hetero-oligomeric Complexes
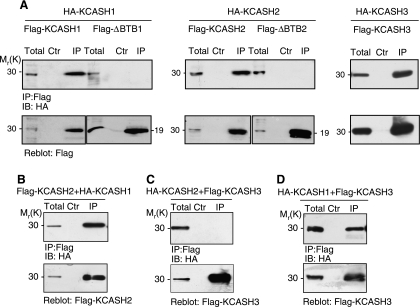
The BTB domain has been described to mediate the formation of oligomerization complexes [32]. Here we demonstrate that KCASH2 and KCASH3 are able to form homo-oligomers, as evaluated by coimmunoprecipitating KCASH2 or KCASH3 HA- and Flag-tagged proteins (Figure 2A). Similar results were observed for KCASH1REN/KCTD11 (Figure 2A and Correale et al. [33]). The oligomerization domain was confirmed to be located in the BTB motif because HA-KCASH1 and Flag-ΔBTBKCASH1 (deleted of the BTB motif, 4BTB1) or HAKCASH2 and Flag-ΔBTBKCASH2 (ΔBTB2) did not coimmunoprecipitate (Figure 2A). Interestingly, the three family members were able to form hetero-oligomers with each other, with the exception of KCASH2/KCASH3 (Figure 2, B–D). The lack of interaction between KCASH2 and KCASH3 confirms the increased divergence of these two family members (Figure 2C).
Figure 2.
KCASH family members form homodimeric and heterodimeric complexes. (A–D) Coimmunoprecipitation experiments performed in lysates from HEK293T cells transfected with expression vectors encoding for the indicated tagged proteins and immunoprecipitated (IP) with anti-Flag agarose beads. As a negative control (Ctr), anti-Flag agarose beads were preblocked with Flag peptide. Immunoprecipitated samples and a fraction of the total lysate (Total) were loaded and separated on SDS-PAGE gels. Blots were immunoblotted (IB) with anti-HA or anti-Flag antibody as indicated. ΔBTB1 and ΔBTB2 indicate BTB-deleted KCASH1 and KCASH2, respectively. Reblots with anti-Flag antibody were performed to verify immunoprecipitation.
Thus, the newly discovered KCASH family members share a number of features, such as high homology of both the BTB domain and whole sequence, specific expression in brain and cerebellum, and the ability to form hetero-oligomer and homo-oligomer complexes, suggesting that these proteins might also share some functional properties.
KCASH2 and KCASH3 Share with KCASH1 the Ability to Bind to Cullin3
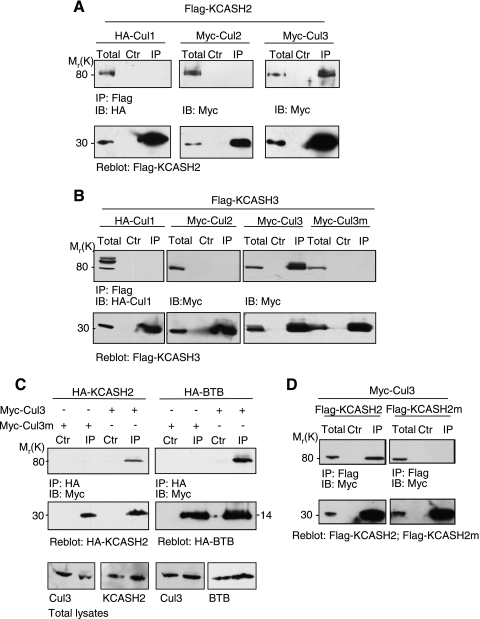
KCASH1REN/KCTD11 has been recently identified as a Cullin3 (Cul3) E3 ubiquitin ligase and has been shown to interact with Cul3 through the BTB domain and thus to form the active E3 ligase complex [18]. To verify whether KCASH2 and KCASH3 also share this property, we looked at the interaction of these proteins with Cullins. By coimmunoprecipitation experiments, we observed that both KCASH3 and KCASH2 were able to interact with Cul3, but not with Cul1 and Cul2 proteins (Figure 3, A and B), similar to what previously observed for KCASH1 [18]. We have also identified the amino acid residues of the Cul3 protein required for binding to KCASH2 and KCASH3. In fact, KCASH2 and KCASH3 coimmunoprecipitation with Cul3 was abolished by mutation of tyrosines Y58, Y62, and Y125 to lysines in the Cul3 protein (Cul3m; Figure 3, B and C). These results confirmed the relevance of these residues, previously demonstrated for the interaction with the other family member KCASH1REN/KCTD11 [18] and indicated a similar mechanism of interaction with Cul3 by all three KCASH family members. Similar to what previously described for KCASH1 [18], the KCASH2 protein domain critical for binding to Cul3 is the BTB-containing region, which is sufficient for coimmunoprecipitation with wild-type Cul3 (Figure 3C), whereas this interaction is abolished when Cul3 is mutated (Cul3m; Figure 3C). Accordingly, KCASH2 protein deleted of the BTB domain is not able to immunoprecipitate Cul3 (see Flag-ΔBTB; Figure 4A).
Figure 3.
KCASH2 and KCASH3 share with KCASH1 the ability to bind to Cullin3 E3 ligase through the same domains. (A–D) Coimmunoprecipitation experiments have been performed as in Figure 2, overexpressing KCASH members and HA or Myc-tagged Cul1-3 as indicated. Immunoprecipitation was carried out with anti-HA or anti-Flag antibodies, and immunocomplexes were revealed with the indicated antibodies. (A) KCASH2 form a complex with Cul3 but not Cul1 and Cul2. (B) KCASH3 does not bind Cul1 and Cul2 but forms a complex with Cul3, which is abolished bymutation of Cul3 in three critical residues (Y58/62/125K, indicated as Cul3m). (C) KCASH2 BTB domain (HA-BTB) is sufficient for Cul3 binding, and this interaction is lost bymutation of Cul3. Cells were transfected with myc-Cul3 or Myc-Cul3mut together with HA-KCASH2 or HA-BTB as indicated. (D) KCASH2 interaction with Cul3 is abolished by specific mutations in the BTB domain. Myc-Cul3 was transfected together with Flag-KCASH2 or Flag-KCASH2KKK mutant (KCASHm), and immunoprecipitations were performed as above.
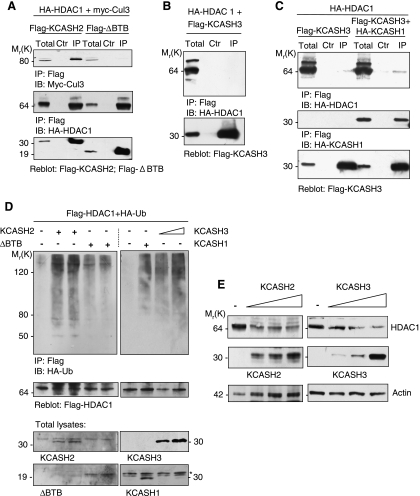
Figure 4.
KCASH2 and KCASH3 bind to and promote HDAC1 ubiquitination and degradation. (A) KCASH2 binds to HDAC1, through its C-terminal domain (ΔBTB domain), bringing together Cul3 and HDAC1. (B and C) KCASH3 alone does not bind efficiently to HDAC1 (B), but the interaction with HDAC1 is mediated by binding to KCASH1 (C). Lysates from HEK293T cells cotransfected with the indicated plasmids were immunoprecipitated and blotted as in Figure 3. (D) KCASH2 and KCASH3 expression induces HDAC1 ubiquitination. Expression of a KCASH2 protein devoid of the BTB domain (ΔBTB) indicates that this function is dependent on the presence of the BTB domain. KCASH1-dependent ubiquitination is also shown for a comparison. Cell lysates from MG132 (50 µM)-treated HEK293T cells, transfected with the indicated plasmids and HA-ubiquitin, were immunoprecipitated with anti-Flag agarose beads and immunoblotted with anti-HA to detect conjugated HA-Ub. The blot was reprobed with an anti-Flag antibody to monitor immunoprecipitation efficiency. Bottom, KCASH1-3 and KCASH2-ΔBTB (ΔBTB) total protein levels. The amount of each transfected plasmid is 1 µg except for KCASH3 (1 and 2 µg). (E) KCASH2 and KCASH3 expression reduces HDAC1 protein levels. HEK293T cells were transfected with Flag-HDAC1 in the presence or absence of increasing amounts (1, 2, and 4 µg) of Flag-KCASH2 or Flag-KCASH3 and cell lysates immunoblotted with anti-Flag and anti-Actin antibodies.
We were also able to map the Cul3 binding region identifying three critical residues on the BTB domain of KCASH2 (DFY; Figure W1A). Mutation of the DFY residues (to KKK) abolished Cul3 binding (Figure 3D), confirming results observed with KCASH1 protein [18]. Interestingly, these DFY residues are also conserved in the KCASH3 BTB domain (Figure W1A), supporting the critical role of this domain for Cul3 interaction.
KCASH2 and KCASH3 Promote the Ubiquitination of HDAC1
Binding to Cul3 and the functional properties of the KCASH1 homolog [18] suggest that KCASH2 and KCASH3 may also promote ubiquitination. Indeed, this property together with the functional role of the BTB domain was confirmed by the ability of KCASH2 to coimmunoprecipitate ubiquitinated proteins, whereas immunoprecipitation of KCASH2 devoid of the BTB domain failed to do so (Figure W2A).
The most likely candidate substrate for KCASH2 and 3/Cul3 complexes is HDAC1, based on the recently identified HDAC1 as a substrate of the KCASH1/Cul3 complex [18]. KCASH2 was found to bind to HDAC1 through the C-terminal region because KCASH2 protein truncated of the N-terminal BTB motif (ΔBTB) is still able to bind to HDAC1 (Figure 4A). Interestingly, the coimmunoprecipitated KCASH2/HDAC1 complex also contains Cul3 which instead is missing in the ΔBTB/HDAC1 complex (Figure 4A). These observations suggest that KCASH2, HDAC1, and Cul3 form a trimeric complex through the involvement of the Cul3 binding BTB motif.
Surprisingly, KCASH3 was instead unable to bind HDAC1 protein in coimmunoprecipitation experiments (Figure 4B). Because KCASH3 and KCASH1 are able to heterodimerize (Figure 2D), we hypothesized that KCASH1 may tether the HDAC1 protein into a complex with KCASH3, in which both KCASH3 and KCASH1 contribute to Cul3 recruitment. Indeed, coimmunoprecipitation experiments performed by overexpression of both KCASH3 and KCASH1 indicated that KCASH3 was able to immunoprecipitate HDAC1 in the presence of KCASH1 (Figure 4C).
Formation of the KCASH2-Cul3-HDAC1 complex resulted in the ubiquitination of HDAC1, whereas ΔBTB-KCASH2 was unable to promote HDAC1 ubiquitination (Figure 4D, left panel). Consistently, KCASH3 also promoted the HDAC1 ubiquitination (Figure 4D, right panel). As a consequence of such ubiquitination, both KCASH2 and KCASH3 expression induced a decrease in the levels of HDAC1 protein, indicating that ubiquitination promotes HDAC1 proteolytic degradation (Figure 4E).
KCASH2 and KCASH3 Downregulate Gli Transcriptional Activity in an Acetylation-Dependent Way
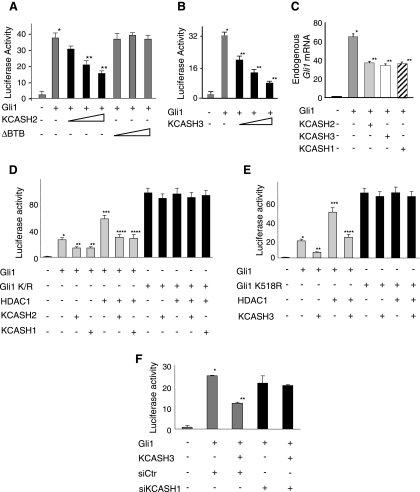
One of the main pathways involved in cerebellar development and differentiation is the Hh pathway. This pathway has been previously shown to be regulated by KCASH1 that promotes the HDAC1 degradation, thus increasing Gli acetylation and suppressing its transcriptional function [18,26,34]. On the basis of the features shared by KCASH2 and KCASH3 with KCASH1, we investigated whether the two proteins were able to control Hh pathway through a similar mechanism. Here, we demonstrated that overexpression of KCASH2 or KCASH3 significantly decreased the transcriptional activity of ectopic Gli1 on a Gli-responsive luciferase reporter (Figure 5, A and B) and the levels of endogenous Gli1 mRNA, as a read out of Hh signaling (Figure 5C; for a comparison, levels of Gli mRNA reduction by KCASH1 expression are also shown). As expected, the KCASH2 protein devoid of BTB domain was not able to reduce the luciferase activity (Figure 5A). To investigate whether KCASH2 and KCASH3 activity was due to the acetylation of Gli1, we tested the activity of the ectopic proteins by using an unacetylatable Gli1 mutant (Gli1K518R) [18]. K518R displayed an increased ability to activate the luciferase activity, confirming the critical role of lysine 518 deacetylation in enhancing Gli1 function. As expected, KCASH2 and KCASH3 (and KCASH1, as also previously reported in Canettieri et al. [18]) were unable to antagonize Gli1K518R activity (Figure 5, D and E), thus suggesting that lysine 518 acetylation is required for their action. However, because KCASH3 was unable to directly bind HDAC1, we hypothesized that its HDAC1-dependent Gli antagonismcould require the cooperation of endogenous KCASH1, which is able to recruit HDAC1 to the complex with Cul3 and KCASH3 as shown in Figure 4C. Indeed, siRNA-mediated depletion of endogenous KCASH1 abrogated the inhibitory effect of KCASH3 on Gli1-induced luciferase reporter activity (Figure 5F).
Figure 5.
KCASH2 and KCASH3 downregulate Gli transcriptional function in an acetylation-dependent way. (A and B) KCASH2 and KCASH3 downregulate transcriptional Gli activity. Relative luciferase activity in HEK293T cells transfected with 12x Gli-Luc and pRL-TK Renilla (as a normalizer) alone (ctrl) or with increasing amounts (100, 200, and 400 ng) of KCASH2 or KCASH2-ΔBTB (ΔBTB) (A) or KCASH3 (B). Data are indicated as mean ratios with respect to pRL-TK Renilla luciferase signal. *P < .05 Gli1 versus control, **P < .05 KCASH2 or KCASH3 + Gli1 versus Gli1. (C) Endogenous Gli1 mRNA levels evaluated by quantitative real-time PCR in HEK293T cells cotransfected with Gli1, KCASH2, KCASH3 or KCASH1 (shown for comparison). *P < .05 Gli1 versus control, **P < .05 KCASH2 or KCASH3 + Gli1 versus Gli1. (D, E) KCASH2 and KCASH3 downregulatory activity depends on acetylation of HDAC1. Relative luciferase activity in HEK293T cells cotransfected with 12x Gli-Luc and pRL-TK Renilla plus Gli1, Gli1K518R, HDAC1, KCASH2, and KCASH3 expression vectors as indicated. KCASH1 was also transfected for comparison. *P < .05, Gli1 versus control. **P < .05 KCASH2 or KCASH3 + Gli1 versus Gli1. ***P < .05 HDAC1 + Gli1 versus Gli1. ****P < .05 KCASH2 or KCASH3 + HDAC1 + Gli1 versus HDAC1 + Gli1. (F) KCASH3 function on Gli1 transcriptional activity requires the presence of endogenous KCASH1. Relative luciferase activity in HEK293T cells cotransfected with 12x Gli-Luc and pRL-TK Renilla plus Gli1, KCASH3 in the presence of siKCASH1 or siCTR. *P < .05, Gli1 + siCTR versus control. **P < .05 KCASH3 + Gli1 + siCTR versus Gli1 + siCTR.
KCASH1, KCASH2, and KCASH3 Are Downregulated in Human Primary MB
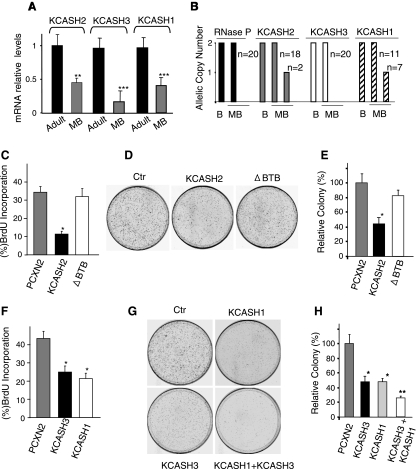
Given the role of KCASH2 and KCASH3 in regulation of Hh activity, and the role of Hh in cerebellar tumorigenesis, we analyzed the levels of KCASHs expression in a set of human sporadic MB samples. Indeed, both KCASH2 and KCASH3 showed a significant reduction of their expression levels in most tumors analyzed (Figures 6A and W3A), suggesting that loss of both genes may contribute to MB. KCASH1 expression is also downregulated in the same MB samples (Figure 6A), confirming the previously reported silencing on this gene [34,35]. In fact, downregulated expression of KCASH1 has been reported to be due both to promoter methylation [34,35] and to genetic alteration because deletion of the 17p 13.1 chromosome region (where this gene maps) occurs very frequently in human MB [34]. Interestingly, human KCASH2 is located on chromosome 11q14.1, with chromosome 11q loss being involved in several tumors including MB [36,37]. The localization of KCASH3 to the 3p14.3 chromosomal region is also intriguing. Deletions and other rearrangements of this region have been reported in a variety of tumor types, including malignant mesotheliomas and carcinomas of the lung, kidney, breast, and ovary [38]. The existence of recurrent losses of chromosome 3p implicates the involvement of a tumor suppressor gene in this region.
Figure 6.
Concerted KCASH1, KCASH2, and KCASH3 downregulated expression in human primary MBs and growth inhibitory role of the KCASH family members. (A) Quantitative real-time PCR analysis of KCASH2, KCASH3 and KCASH1 expression in samples from adult human cerebella (n = 9) and human primary medulloblastoma (n = 32). Data are means ± SD. **P = .0016, ***P = .0001 medulloblastoma versus adult human tissue. (B) Quantitative real-time PCR evaluation of KCASH1, KCASH2, and KCASH3 allelic copy number (mean ± SD) in primary human MB (n, number of cases tested; B, blood). RNaseP was used as an endogenous control. (C, F) BrdU proliferation assays: Daoy MB cells were transfected with indicated vectors, and percent of BrdU incorporating cells were measured to monitor cell proliferation. *P <.05 versus empty vector (PCXN2). (D, G) Colony formation assay: Daoy cells were transfected with the indicated vectors and colony formation assay was performed to monitor cell proliferation. Representative images from experiments are shown. (E, H) Counts from the colony formation assays are represented. *P < .05 KCASH2, KCASH3, and KCASH1 versus control vector. **P < .05 KCASH1 + KCASH3 versus KCASH3 or KCASH1. Unless otherwise indicated, all experiments were performed in triplicate, and mean ± SD is shown.
Therefore, we measured allelic loss of KCASH1, KCASH2, and KCASH3 in genomic DNA from our MB samples. Although we did not observe any loss of KCASH3 (hinting for a different epigenetic silencing mechanism), we observed deletion of KCASH2 in 10% of samples (Figure 6B), thus suggesting that genetic loss of this gene is a relevant mechanism in MB formation. As expected, deletion of KCASH1 was also present in 35%of samples (Figure 6B). Interestingly, none of the MB analyzed presented deletion of both KCASH1 and KCASH2 (not shown).
The observations presented here suggest that a reduced expression of all three members of KCASH family may occur in MB, indicating a contribution of the KCASH family in regulating proper development and preventing Hh dysregulation leading to MB.
KCASH2 and KCASH3 Downregulate the Growth of MB Cells
Hh signaling is critical for promoting the growth of MB cells [10]. The inhibitory role of KCASH2 and KCASH3 on Hh signaling and their reduced expression in primary MB suggested that these proteins could suppress tumor cell growth, similar to KCASH1 that displays an antitumor activity [18,34]. Accordingly, KCASH2 and KCASH3 overexpression significantly reduced the growth of MB cell lines as demonstrated by BrdU incorporation and colony formation assays (Figure 6, C–H). In contrast, KCASH2 protein deleted of the Cul3 interacting BTB domain (ΔBTB) did not suppress cell growth (Figure 6, C–E). Of note, we observed a cooperation between KCASH family members in MB cell growth suppression as a general trait of the family. In particular, the cooperative role of KCASH3 and KCASH1 was indicated by the higher suppression of cell growth induced by combined KCASH3 and KCASH1 overexpression compared with single agents (Figure 6, F–H).
Such a cooperative role was also confirmed as a more general trait of the family by the observation of the higher suppression of tumor cell growth induced by combined KCASH2 and KCASH1 (Figure W3, B–C). These results are in agreement with the inhibitory activity on Gli1-induced Gli-reporter luciferase activity of coexpressed KCASH1 and -2 or KCASH1 and -3 proteins compared with overexpressed individual proteins (Figure W3, D and E).
Overall, our findings suggest that the above described Cul3-dependent mechanism is involved in KCASH2 and KCASH3 tumor growth suppressor function.
Discussion
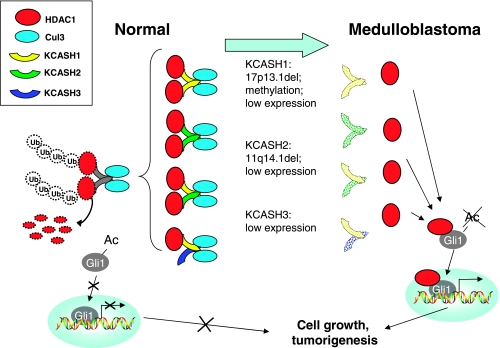
We describe here two novel regulators of HDAC/Hh signaling and a model by which the newly identified KCASH family of Cul3 ubiquitin E3 ligase adaptors acts in concert to suppress HDAC1 and Hh/Gli signaling (Figure 7). KCASH2 and KCASH3 share with KCASH1 (previously termed RENKCTD11) the ability to bind Cul3. By recruiting Cul3 into a complex with HDAC1, KCASH2 promotes the ubiquitination and degradation of HDAC1, thereby inhibiting the previously described deacetylation-mediated transcriptional activation of the Hh effectors Gli1 and Gli2 [18]. In contrast, KCASH3, which is unable to bind directly HDAC1, by exploiting its ability to heterodimerize with KCASH1, recruits this deacetylase into a complex with Cul3, leading to HDAC1 ubiquitination and degradation as well as suppression of Gli activity. Like KCASH1 [34], KCASH2 and KCASH3 expression was also significantly reduced in human MB. In addition to still unidentified epigenetic silencing events, KCASH2 allelic deletion was also observed to contribute to the reduced expression of this gene. Finally, rescuing the low expression of KCASH2 and KCASH3 reduces the growth of Hh-dependent MB cells. This is of particular interest because Hh/Gli signaling represents the master regulator of cerebellar granule cell progenitor development (reviewed in Ruiz i Altaba [39]) as well as an oncogenic addiction pathway in MB [10,40]. These observations suggest that the increased expression of KCASH family members in adult compared with fetal cerebellum and their genetic and epigenetic downregulated expression in human MB may have an in vivo relevance.
Figure 7.
The KCASH family acts in concert to suppress Hh/Gli signaling and tumorigenesis. We propose a model in which all members of the KCASH family operate in concert, either as homo-oligomers or as hetero-oligomers to bind, ubiquitinate, and promote degradation of HDAC1, thereby preserving Gli1 acetylation and suppressing Hh/Gli signaling (left side of the panel). Events that reduce the expression of KCASH members (genetically or epigenetically) allow the accumulation in the cell of HDAC protein and thus the deacetylation of Gli1, which becomes fully transcriptionally active, driving the cells toward uncontrolled proliferation and eventually tumorigenesis (right side of the panel).
KCASH Family of HDAC1 Inhibitors Determine Concerted Suppression of Hh Signaling
All three members of the KCASH family of Cul3 adaptors mediate the suppression of HDAC1. However, the role of the three proteins is not equal because KCASH3, while keeping the ability to bind Cul3, does not bind directly HDAC1. Although KCASH3 may also target other significant substrates to be still identified, it does indeed control HDAC1 by exploiting the ability of KCASH members to homodimerize and heterodimerize. In particular, KCASH3 recruits HDAC1 into an active complex with Cul3 by binding KCASH1, which tether HDAC1. Therefore, whereas the KCASH3/KCASH3 homodimer does not have access to HDAC1 and is thus inactive, the KCASH3/KCASH1 heterodimer is fully active on HDAC1 ubiquitination. These findings suggest the presence of two functionally distinct classes of KCASH family members: i) KCASH1 and 2, which are fully active on HDAC1 also as homodimers; and ii) KCASH3, which requires heterodimerization with the fully active KCASH1. Our observations also suggest that KCASH family members act in cooperation to ubiquitinate HDAC1 because their combination is functionally active and, in certain conditions (KCASH3), is required. The different features of these two classes of KCASH family members (inability of KCASH3 to interact with KCASH2 or HDAC1 and requirement for KCASH3 to bind KCASH1 to recruit HDAC1 into the active Cul3 E3 ligase complex) may reflect the divergence of these proteins during phylogenesis, where KCASH3 earlier separates from both KCASH1 and 2, which display more conserved functions.
Redundancy of KCASH-Mediated Regulation of HDAC1 and Hh in MB
The identification of the KCASH family adds new information to the emerging mechanism of regulation of HDAC by inducing its ubiquitin-dependent degradation (which is exploited also by Chfr [25]). Whereas the targets regulated by the HDAC1/Chfr pathway have not been specifically identified, the most interesting aspect of our observations is that the KCASH family control of HDAC1 results in inhibition of the Hh signaling through the regulation of deacetylation-dependent transcriptional activation of Gli1 (this study and that of Canettieri et al. [18]).
The role of HDAC pharmacologic inhibition in the control of tumor growth, the previously reported HDAC overexpression in cancer [17–19], and the deletion or silencing of the HDAC-suppressor genes observed in human tumors (reviewed in Di Marcotullio et al. [23]), suggest their relevance as physiological mechanisms that, by repressing HDAC function, prevent tumorigenesis.
Whereas HDACs comprise a heterogenous family with multiple targets, suggesting redundancy of HDAC-dependent control of cell functions involved in tumorigenesis, cancers are usually sustained by specific oncogene addiction events representing the master tumorigenic pathways [40]. Our findings suggest a redundant regulatory mechanism occurring within the HDAC-dependent control of the unique oncogenic addiction Hh/Gli pathway. Such a redundancy including the contribution of the three members of the KCASH family implies that they act in concert to optimally inhibit HDAC1 function and the Hh pathway. This has been directly shown in this study by the requirement of KCASH1 in the KCASH3-dependent regulation of HDAC1/Hh pathway and by the cooperative activity of KCASH1 and KCASH2.
Furthermore, all KCASH family members are downregulated in most human MB. Deletion of KCASH1 and 2 by 17p13.1 and 11q14.1 loss, respectively, contributes to their downregulated expression (this study and that of Di Marcotullio et al. [34]). Promoter methylation of KCASH1 has also been reported to be responsible for its downregulated expression in human MB [35]. Whether methylation is also contributing to the downregulated expression of KCASH2 and 3 remains to be elucidated. The combined downregulated expression of KCASH1, 2, and 3 in human MB suggests the requirement of the misregulation of the mechanisms affecting such a redundant KCASH-mediated control of HDAC1/Hh-Gli pathway in this tumor (Figure 7).
In conclusion, the discovery of the above described KCASH family members suggests that these negative regulators of Hh/Gli signaling may act in concert as native HDAC inhibitors during development and that their combined genetic or epigenetic defects are crucial in sustaining Hh/Gli and HDAC-mediated tumorigenesis, proposing their role as a promising class of endogenous “agents” by which this oncogenic addiction pathways may be targeted.
Supplementary Material
Abbreviations
- Hh
Hedgehog
- KCASH
KCTD containing
- Cullin3 adaptor
suppressor of Hedgehog
- HDAC
histone deacetylase
- Cul
Cullin
Footnotes
This work was supported, in part, by Associazione Italiana Ricerca Cancro, Telethon grant GGP07118, MIUR FIRB and PRIN projects, Ministry of Health, Fondazione Roma, Fondazione Mariani, EU Healing grant, Italian Institute of Technology, and Agenzia Spaziale Italiana. M.P. and M.M. are supported by fellowships from the Pasteur Institute, Cenci Bolognetti Foundation. The authors declare that they have no conflict of interest.
This article refers to supplementary materials, which are designated by Figures W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Crawford JR, MacDonald TJ, Packer RJ. Medulloblastoma in childhood: new biological advances. Lancet Neurol. 2007;6:1073–1085. doi: 10.1016/S1474-4422(07)70289-2. [DOI] [PubMed] [Google Scholar]
- 2.Saran A. Medulloblastoma: role of developmental pathways, DNA repair signaling, and other players. Curr Mol Med. 2009;9:1046–1057. doi: 10.2174/156652409789839080. [DOI] [PubMed] [Google Scholar]
- 3.Gulino A, Arcella A, Giangaspero F. Pathological and molecular heterogeneity of medulloblastoma. Curr Opin Oncol. 2008;20:668–675. doi: 10.1097/CCO.0b013e32831369f4. [DOI] [PubMed] [Google Scholar]
- 4.Guessous F, Li Y, Abounader R. Signaling pathways inmedulloblastoma. J Cell Physiol. 2008;217:577–583. doi: 10.1002/jcp.21542. [DOI] [PubMed] [Google Scholar]
- 5.Marino S. Medulloblastoma: developmental mechanisms out of control. Trends Mol Med. 2005;11:17–22. doi: 10.1016/j.molmed.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Behesti H, Marino S. Cerebellar granule cells: insights into proliferation, differentiation, and role in medulloblastoma pathogenesis. Int J Biochem Cell Biol. 2009;41:435–445. doi: 10.1016/j.biocel.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 7.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 8.Ruizi Altaba A, Palma V, Dahmane N. Hedgehog-Gli signalling and the growth of the brain. Nat Rev Neurosci. 2002;3:24–33. doi: 10.1038/nrn704. [DOI] [PubMed] [Google Scholar]
- 9.Fuccillo M, Joyner AL, Fishell G. Morphogen to mitogen: the multiple roles of Hedgehog signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:772–783. doi: 10.1038/nrn1990. [DOI] [PubMed] [Google Scholar]
- 10.Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, et al. Medulloblastoma growth inhibition by Hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- 11.Gulino A, Di Marcotullio L, Ferretti E, De Smaele E, Screpanti I. Hedgehog signaling pathway in neural development and disease. Psychoneuroendocrinology. 2007;32(suppl 1):S52–S56. doi: 10.1016/j.psyneuen.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Jacob L, Lum L. Deconstructing the Hedgehog pathway in development and disease. Science. 2007;318:66–68. doi: 10.1126/science.1147314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyman JM, Firestone AJ, Heine VM, Zhao Y, Ocasio CA, Han K, Sun M, Rack PG, Sinha S, Wu JJ, et al. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc Natl Acad Sci USA. 2009;106:14132–14137. doi: 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 15.Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, et al. Treatment of medulloblastoma with Hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Smaele E, Ferretti E, Gulino A. Vismodegib, a small-molecule inhibitor of the Hedgehog pathway for the treatment of advanced cancers. Curr Opin Investig Drugs. 2010;11:707–718. [PubMed] [Google Scholar]
- 17.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 18.Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, Pietrosanti L, De Smaele E, Ferretti E, Miele E, et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol. 2010;12:132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 19.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 20.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu Y, Zhao Y, Becker M, John S, Parekh BS, Huang S, Hendarwanto A, Martinez ED, Chen Y, Lu H, et al. HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol Cell. 2006;22:669–679. doi: 10.1016/j.molcel.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, Bhalla K, Bai W. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Marcotullio L, Canettieri G, Infante P, Greco A, Gulino A. Protected from the inside: endogenous histone deacetylase inhibitors and the road to cancer. Biochim Biophys Acta. doi: 10.1016/j.bbcan.2011.01.002. (in press) [DOI] [PubMed] [Google Scholar]
- 24.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 25.Oh YM, Kwon YE, Kim JM, Bae SJ, Lee BK, Yoo SJ, Chung CH, Deshaies RJ, Seol JH. Chfr is linked to tumour metastasis through the downregulation of HDAC1. Nat Cell Biol. 2009;11:295–302. doi: 10.1038/ncb1837. [DOI] [PubMed] [Google Scholar]
- 26.Argenti B, Gallo R, Di Marcotullio L, Ferretti E, Napolitano M, Canterini S, De Smaele E, Greco A, Fiorenza MT, Maroder M, et al. Hedgehog antagonist REN(KCTD11) regulates proliferation and apoptosis of developing granule cell progenitors. J Neurosci. 2005;25:8338–8346. doi: 10.1523/JNEUROSCI.2438-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Smaele E, Di Marcotullio L, Ferretti E, Screpanti I, Alesse E, Gulino A. Chromosome 17p deletion in human medulloblastoma: a missing checkpoint in the Hedgehog pathway. Cell Cycle. 2004;3:1263–1266. doi: 10.4161/cc.3.10.1200. [DOI] [PubMed] [Google Scholar]
- 28.Ferretti E, Di Marcotullio L, Gessi M, Mattei T, Greco A, Po A, De Smaele E, Giangaspero F, Riccardi R, Di Rocco C, et al. Alternative splicing of the ErbB-4 cytoplasmic domain and its regulation by Hedgehog signaling identify distinct medulloblastoma subsets. Oncogene. 2006;25:7267–7273. doi: 10.1038/sj.onc.1209716. [DOI] [PubMed] [Google Scholar]
- 29.De Smaele E, Fragomeli C, Ferretti E, Pelloni M, Po A, Canettieri G, Coni S, Di Marcotullio L, Greco A, Moretti M, et al. An integrated approach identifies Nhlh1 and Insm1 as sonic Hedgehog-regulated genes in developing cerebellum and medulloblastoma. Neoplasia. 2008;10:89–98. doi: 10.1593/neo.07891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiMarcotullio L, Greco A, Mazza D, Canettieri G, Pietrosanti L, Infante P, Coni S, Moretti M, De Smaele E, Ferretti E, et al. Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene. 2011;30:65–76. doi: 10.1038/onc.2010.394. [DOI] [PubMed] [Google Scholar]
- 31.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 33.Correale S, Pirone L, Di Marcotullio L, De Smaele E, Greco A, Mazzà D, Moretti M, Alterio V, Vitagliano L, Di Gaetano S, et al. Molecular organization of the cullin E3 ligase adaptor KCTD11. Biochimie. doi: 10.1016/j.biochi.2010.12.014. (in press) [DOI] [PubMed] [Google Scholar]
- 34.Di Marcotullio L, Ferretti E, De Smaele E, Argenti B, Mincione C, Zazzeroni F, Gallo R, Masuelli L, Napolitano M, Maroder M, et al. REN(KCTD11) is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc Natl Acad Sci USA. 2004;101:10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mancarelli MM, Zazzeroni F, Ciccocioppo L, Capece D, Po A, Murgo S, Di Camillo R, Rinaldi C, Ferretti E, Gulino A, et al. The tumor suppressor gene KCTD11 (REN) is regulated by Sp1 and methylation and its expression is reduced in tumors. Mol Cancer. 2010;9:172. doi: 10.1186/1476-4598-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reardon DA, Jenkins JJ, Sublett JE, Burger PC, Kun LK. Multiple genomic alterations including N-myc amplification in a primary large cell medulloblastoma. Pediatr Neurosurg. 2000;32:187–191. doi: 10.1159/000028932. [DOI] [PubMed] [Google Scholar]
- 37.Lescop S, Lellouch-Tubiana A, Vassal G, Besnard-Guerin C. Molecular genetic studies of chromosome 11 and chromosome 22q DNA sequences in pediatric medulloblastomas. J Neurooncol. 1999;44:119–127. doi: 10.1023/a:1006387518100. [DOI] [PubMed] [Google Scholar]
- 38.Kok K, Naylor SL, Buys CH. Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv Cancer Res. 1997;71:27–92. doi: 10.1016/s0065-230x(08)60096-2. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz I, Altaba A, editors. Hedgehog-Gli Signaling in Human Disease. Georgetown: Plenum Publisher; 2006. [Google Scholar]
- 40.Weinstein IB, Joe AK. Mechanisms of disease: oncogene addiction—a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3:448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.