Abstract
Understanding how human genetic variation impacts individual response to immunogens is fundamental for rational vaccine development. To explore host mechanisms involved in cellular immune responses to the MRKAd5 human immunodeficiency virus type 1 (HIV-1) gag/pol/nef vaccine tested in the Step trial, we performed a genome-wide association study of determinants of HIV-specific T cell responses, measured by interferon γ enzyme-linked immunospot assays. No human genetic variant reached genome-wide significance, but polymorphisms located in the major histocompatibility complex (MHC) region showed the strongest association with response to the HIV-1 Gag protein: HLA-B alleles known to be associated with differences in HIV-1 control were responsible for these associations. The implication of the same HLA alleles in vaccine-induced cellular immunity and in natural immune control is of relevance for vaccine design. Furthermore, our results demonstrate the importance of considering the host immunogenetic background in the analysis of immune responses to T cell vaccines.
The Step trial was a phase 2b proof-of-concept study conducted by the HIV Vaccine Trials Network (HVTN) and Merck that was designed to assess the safety and efficacy of a 3-dose regimen of the Merck adenovirus serotype 5 (MRKAd5) human immunodeficiency virus type 1 (HIV-1) gag/pol/nef vaccine, a replication-defective adenovirus serotype 5 (Ad5) vaccine expressing Gag, Pol, and Nef proteins from an HIV-1 clade B strain, which aimed to elicit cellular immune responses. It was a multicenter, double-blind, randomized, placebo-controlled study performed in adults at high risk of HIV-1 infection [1]. Vaccinations in the trial were terminated following a planned interim analysis in September 2007 because of a lack of efficacy of the tested vaccine. Vaccination was not associated with a lower viral load in participants who became infected, and the rate of HIV-1 acquisition was actually higher among vaccine recipients than among placebo recipients, particularly in the subgroup of uncircumcised male participants with high pre-existing Ad5 neutralizing antibody titers [1, 2]. Understanding this disappointing and paradoxical outcome is a high priority for the field.
Recombinant Ad5 vaccine candidates had previously been shown to elicit strong cellular immune responses against HIV-1 epitopes [3, 4]. Accordingly, the MRKAd5 HIV-1 gag/pol/nef vaccine was highly immunogenic for inducing HIV-specific T cell responses: at least three-quarters of vaccine recipients generated a response to 1 or more of the HIV-1 proteins included in the vaccine [2]. Still, those responses were quantitatively highly variable: higher response rates were observed in participants with low pre-existing Ad5 neutralizing antibody titers [2], but a significant fraction of variability in response intensity remains unexplained and could be at least partially attributable to human genetic variation.
Thanks to recent advances in genomics, technology, and bioinformatics, it is now possible to comprehensively assess associations between common human genetic variants and biological phenotypes [5]. Here we combine whole-genome genotyping results and HLA class I typing data to explore the host mechanisms associated with variability in T cell responses to the HIV-1 proteins present in the MRKAd5 HIV-1 gag/pol/nef vaccine tested in the Step trial.
METHODS
Participants
Study participant enrollment inclusion and exclusion criteria for the Step study have been described elsewhere [1]. Each participant provided informed consent for genetic testing. All male Step trial participants who received at least 2 doses of the MRKAd5 HIV-1 gag/pol/nef vaccine and were HIV-1 seronegative at week 8 (ie, 4 weeks after the second vaccination) were eligible for our study. This investigation was initiated after the interim analysis when all but 1 of the case individuals identified were male [1]. Most immunogenicity and covariate analyses subsequently performed were confined to male participants, and accordingly this study was restricted to male vaccine recipients.
Immunological Assays
Validated interferon γ (IFN-γ) enzyme-linked immunospot (ELISPOT) assays were performed on previously cryopreserved peripheral blood mononuclear cells (PBMCs) that were obtained at week 8; the Merck laboratory (hereafter Merck) tested one-quarter of the samples, which were randomly selected and stratified by treatment assignment and study site. The HVTN laboratory at Fred Hutchinson Cancer Research Center laboratory (Seattle, WA; hereafter HVTN) tested the remaining samples. Cells were stimulated ex vivo with pools of peptides that were 15 amino acids in length and overlapping in sequence by 11 amino acids. The peptide sequences matched the HIV-1 proteins encoded by the vaccine, and a total of 4 nonoverlapping pools of peptides were tested: 1 Gag pool, 2 Pol pools, and 1 Nef pool. If any of the protein-level responses was positive, then the overall response was considered positive. Responses were reported as the number of spot-forming cells (SFCs) per 106 PBMCs. A sample was considered positive if the background-adjusted mean of the experimental wells was ≥4 times the mean of the negative controls and ≥55 SFCs per 106 PBMCs for the assays performed at Merck, whereas positivity was assessed using a bootstrapping method specifically developed for in-house ELISPOT data for the samples tested at HVTN [6, 7]. We also compared the HIV-1-specific IFN-γ ELISPOT results to CD4+ and CD8+ T cell responses measured on a subset of samples by IFN-γ and interleukin 2 (IL-2) intracellular cytokine staining assays [2].
Genotyping
DNA samples were genotyped using the Human1M-Duo Infinium HD BeadChip (Illumina), which features >1 million single-nucleotide polymorphisms (SNPs) and an additional 52,167 markers designed to specifically target copy number variant regions. We also inferred larger copy number variants from the genotyping data using PennCNV software (2009Aug27version) [8]. A series of data cleaning and quality control procedures were performed: SNPs were filtered on the basis of missingness (dropped if called in <99% of participants), minor allele frequency (dropped if the value was <.005), and deviation from Hardy-Weinberg equilibrium. Study participants were filtered on the basis of genotyping quality, a sex check (heterozygosity testing), and cryptic relatedness (the sharing of genetic information was first predicted by estimating identity by descent [IBD], and then 1 sample in each pair of DNA samples showing >12.5% of IBD was excluded). High-resolution HLA class I typing (4 digits; HLA-A, HLA-B, and HLA-C) was obtained using sequence-based methods.
Association Analysis
SNPs, copy number variants, and HLA class I alleles were tested for association with IFN-γ ELISPOT responses to Gag, Pol, and Nef in regression models that also included age, prevaccination Ad5 serology titers, and the laboratory in which the ELISPOT assay was performed (HVTN vs Merck) as covariates. In addition, to correct for population stratification, we applied a modified Eigenstrat method [9], which is a principal component analysis of the genotyping data, to define ethnic groups and correct for residual population ancestry within each group. The analyses were performed separately in 3 ethnic groups by means of Plink software (version 1.06) [10]. Combined P values for all 3 populations were obtained using the Stouffer weighted Z method [11]. We used linear regression to test for association between genetic variants and natural log-transformed quantitative ELISPOT responses, and we used logistic regression for qualitative assessment of the ELISPOT responses (case-control comparisons between individuals with and individuals without detectable responses to the HIV-1 peptide pools). Significance was assessed with a straight Bonferroni correction (threshold for genome-wide significance, P = 5 × 10−8). Power calculations were performed using GWASpower/QT software (available at http://humangenome.duke.edu/software).
RESULTS
Participants and Samples
A total of 831 participants were eligible for the study. They all were male vaccine recipients and HIV-1 negative at week 8. However, 36 of them became infected later in the study. Participants were enrolled in 9 countries in the Americas and Australia, and self-reported ethnicities were very diverse, with a majority of the individuals being white and multiracial individuals (Table 1). All DNA samples were tested on Human1M-Duo chips: 22 samples were excluded because of insufficient genotyping quality; 3 individuals were excluded because of cryptic relatedness (1 in each of 2 pairs of 100% identical genotypes and 1 in a pair of 50% identical genotypes); and 2 participants were excluded because of a sex discrepancy between genotype and the phenotypic database (female participants misclassified as male).
Table 1.
Characteristics of Study Participants
| Characteristic |
Eligible participants | Participants in association analyses | |
| Male sex, no. (%) | 831 (100) | 768 (100) | |
| Vaccine recipient, no. (%) | 831 (100) | 768 (100) | |
| Age in years, mean (SD) | 30.4 (7.7) | 30.4 (7.7) | |
| Circumcision, no. (%)a | 458 (55.8)) | 429 (55.9) | |
| HIV infection during trial, no. (%) | 36 (4.3) | 34 (4.4) | |
| Ethnicity, no. (%) | White | 410 (49.3) | 375 (48.8) |
| Black | 84 (10.1) | 76 (9.9) | |
| Hispanic American | 72 (8.7) | 69 (9.0) | |
| Asian | 13 (1.6) | 13 (1.7) | |
| Native American | 8 (1.0) | 7 (.9) | |
| Polynesian | 1 (.1) | 1 (.1) | |
| Multiracial | 242 (29.1) | 226 (29.4) | |
| Country, no. (%) | United States | 484 (58.2) | 449 (58.5) |
| Peru | 217 (26.1) | 202 (26.3) | |
| Brazil | 50 (6.0) | 47 (6.1) | |
| Canada | 27 (3.3) | 24 (3.1) | |
| Haiti | 18 (2.2) | 15 (2.0) | |
| Dominican Republic | 13 (1.6) | 11 (1.4) | |
| Australia | 10 (1.2) | 9 (1.2) | |
| Puerto Rico | 10 (1.2) | 10 (1.3) | |
| Jamaica | 2 (.2) | 1 (.1) | |
NOTE.
Information was missing for 10 participants.
Immunological Assays
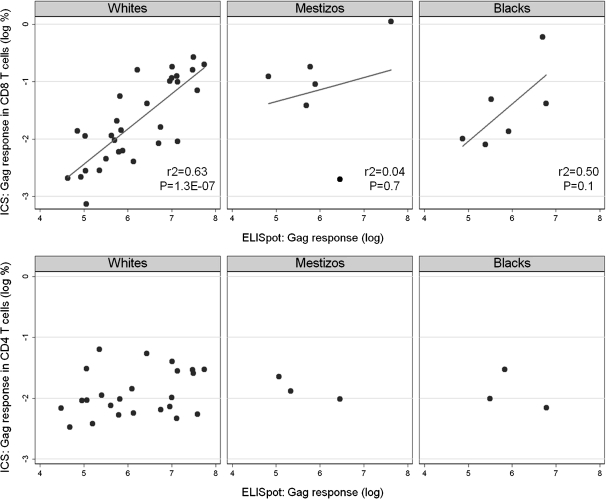
Of the 831 samples, 792 had sufficient cryopreserved PBMCs to be tested in IFN-γ ELISPOT assays: 30.6% were analyzed by Merck (N = 242) and 69.4% by HVTN (N = 550). IFN-γ ELISPOT responses were detected in 675 (85.2%) of 792 vaccine recipients. The percentages of participants with positive responses were 75.6% for Gag, 69.3% for Nef, and 63.6% and 55.3% for the 2 Pol peptide pools. Gag-specific expression of IFN-γ and/or IL-2 measured by intracellular cytokine staining assays gated on CD4+ and CD8+ T cell populations, available for 150 samples [2], were compared to Gag-specific responses measured by IFN-γ ELISPOT assays (Figure 1). We observed a strong correlation between ELISPOT results and CD8+ T cell responses (r2 = .4; P < .001), but no correlation with CD4+ T cell responses (r2 = .05; P = .1).
Figure 1.
Comparison between Gag-specific enzyme-linked immunospot (ELISPOT) and intracellular cytokine staining (ICS) results: interferon γ (IFN-γ) ELISPOT and CD8+ T cell IFN-γ and interleukin 2 (IL-2) ICS results strongly correlate (top), whereas there is no correlation between IFN-γ ELISPOT and CD4+ T cell IFN-γ and IL-2 ICS results (bottom).
Population Structure
Altogether, 768 participants had complete genotype and phenotype results (Table 1). To avoid spurious associations due to population structure, we ran separate analyses in 3 ethnic groups corresponding to the major clusters identified in the principal component analysis of the genotyping data. After removal of individuals who could not be classified into a homogeneous group, a total of 318 white, 87 black, and 115 Mestizo participants were available for regression analyses (Table 2). The study had 80% power to detect genetic variants explaining at least 6.6% of the variability in quantitative HIV-1-specific responses in the combined population.
Table 2.
Participants Included in Linear Regression Analyses, by Ethnic Group
| Ethnic group | Total no. of participants | No. (%) of participants with positive responses to any HIV-1peptide pool | No. (%) of participants with positive Gag-specific responses |
| White | 318 | 283 (89.0) | 265 (83.3) |
| Black | 87 | 72 (87.8) | 59 (73.0) |
| Mestizoa | 115 | 101 (82.8) | 84 (67.8) |
NOTE. a Mestizo (“mixed race”) is used by Peruvians to describe their ethnic group.
Association Analysis
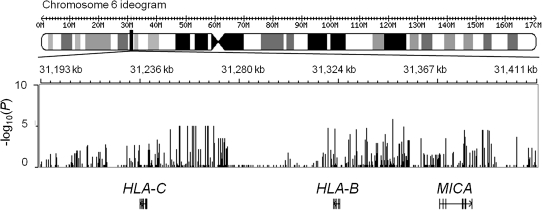
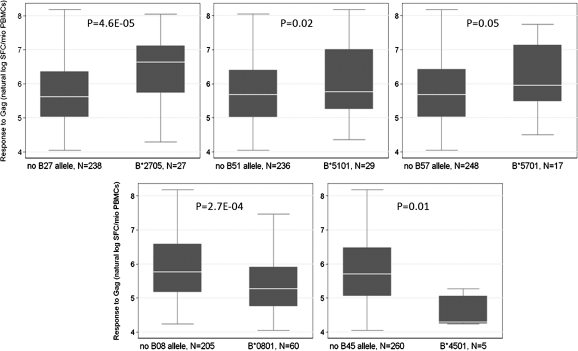
The genome-wide analyses did not reveal any significant association between IFN-γ ELISPOT responses to Gag, Pol, or Nef and SNPs or copy number variants after correction for multiple testing. However, in the analysis of association with quantitative responses to Gag, the SNPs with the lowest P values clustered in the MHC region, around the HLA-B and HLA-C genes (Figure 2); no such clustering was observed for Pol or Nef associations. Therefore, we tested all 4-digit HLA class I alleles present in the study population for association with the same phenotype. In white participants with positive Gag-specific responses (N = 265), 3 HLA-B alleles were associated with higher responses: HLA-B*2705 (N = 27; P = 4.6 × 10−5), B*5101 (N = 29; P = .02), and B*5701 (N = 17; P = .05); whereas 2 alleles were associated with lower responses: HLA-B*0801 (N = 60; P = 2.7 × 10−4) and B*4501 (N = 5; P = .01) (Figure 3). Together, these HLA-B alleles accounted for 13.6% of the variability in Gag responses. No significant association was found in the smaller groups of black and Mestizo individuals, although a trend toward higher Gag responses was observed for HLA-B*5703 (P = .1). We did not observe any significant association between HLA-C alleles and quantitative responses to Gag. We then tested whether the identified HLA-B alleles were responsible for the association signals detected in the MHC region in the genome-wide scan, using nested linear regression. When the SNPs that showed the strongest associations in the genome-wide association study (GWAS) were added to regression models that already incorporated HLA-B*2705, B*5101, B*5701, B*0801, and B*4501 as covariates, these SNPs did not result in a significant increase in the explained variation, which confirms that the GWAS signals are indeed explained by the combined effect of functional HLA alleles (Table 3).
Figure 2.
Genomic overview of the major histocompatibility complex (MHC) region that includes the most associated variants. Indicated are the P values for association with Gag-specific responses of all genotyped single-nucleotide polymorphisms in the region and the structures of the surrounding genes. A P value of <5 × 10−8 was required to declare significance at the genome-wide level. HLA-B, major histocompatibility complex, class I, B; HLA-C, major histocompatibility complex, class I, C; MICA, MHC class I polypeptide-related sequence A.
Figure 3.
HLA-B alleles associated with quantitative T cell responses to Gag in the interferon γ enzyme-linked immunospot assay. Only white participants are included. PBMC, peripheral blood mononuclear cell; SFC, spot-forming cell.
Table 3.
P Values for Single-nucleotide Polymorphism (SNP) Associations for Top 5 SNPs in Genome-wide Association Study (GWAS)
| Top 5 SNPs in GWAS |
P values for SNP associations |
|
| GWAS P value | Adjusted P valuea | |
| rs4713462 | 1.9 × 10−6 | .02 |
| rs4713460 | 2.4 × 10−6 | .03 |
| rs2247056 | 1.4 × 10−5 | .07 |
| rs2853935 | 1.4 × 10−5 | .06 |
| rs2523535 | 2.2 × 10−5 | .11 |
NOTE. The HLA-B alleles HLA-B*2705, *5101, *5701, *0801, and *4501 together explain most of the genome-wide association signals detected in the region. Results are shown for the 5 most associated SNPs in the analysis of Gag-specific responses in white participants. All linear regression models include age, adenovirus serotype 5 titers, laboratory (HIV Vaccine Trials Network vs Merck), and EIGENSTRAT values as covariates.
The adjusted P value is obtained for each SNP in a model in which the 5 HLA-B alleles are incorporated.
DISCUSSION
In an attempt to better delineate the host mechanisms involved in the immune response to T cell vaccines, we performed a GWAS to search for human genetic determinants of the interindividual variability observed in cellular immune responses to the MRKAd5 HIV-1 gag/pol/nef vaccine. HIV-1-specific T cell responses were measured by IFN-γ ELISPOT assays in a large cohort of vaccine recipients. We observed a good correlation between the magnitudes of T cell responses detected by IFN-γ ELISPOT and those detected by CD8+ intracellular cytokine staining, which is not surprising since the vaccine preferentially induced CD8+ T cells, and the CD4+ T cells that were elicited primarily secreted IL-2 and tumor necrosis factor α [2]. Although no SNP reached genome-wide significance, of all tested polymorphisms, those located in the MHC region showed the strongest association with responses to Gag, leading us to look for possible associations between HLA class I alleles and that immunological outcome.
Several HLA-B alleles were found to be responsible for the SNP-associated signal: HLA-B*2705, B*5101, and B*5701 were associated with higher Gag responses, whereas HLA-B*0801 and B*4501 were associated with lower responses. Strikingly, all 5 alleles are also known to be associated with differences in HIV-1 control [12]. HLA-B*57 is the human genetic factor that has been most consistently associated with potent control of HIV-1 [13–17], with HLAB*5701 observed almost exclusively in white individuals and HLA-B*5703 mostly found in individuals of African ancestry. There is clear epidemiological and functional evidence for effective restriction of HIV-1 by HLA-B*27 [18, 19]. HLA-B*5101 was also known to be associated with slower progression to AIDS [20], and its protective effect was nicely confirmed in a recent study that showed that the allele drives HIV-1 adaptation at a population level [21]. Conversely, HLA-B*0801 and HLA-B*4501 have both been associated with higher viral load and more rapid disease progression [20, 22, 23]. The identification of the same HLA alleles, acting in comparable directions, strongly suggests that overlapping mechanisms are implicated in the development of vaccine-induced cellular immunity to HIV-1 and of natural immune control of HIV-1. Of course, the ELISPOT results used in this study only provide information about the immunogenicity of the vaccine and do not represent a correlate of protection for the tested vaccine. It is possible that the vaccine elicited appropriate CD8+ T cell immune responses to enhance viral control, as suggested by recent data [24], but that these are either too weak in function or magnitude, occur in too small a subset of the population, or both to confirm meaningfully enhanced control at a population level.
Comparable results had been reported previously in an immunogenetic study of an HIV-1 vaccine. Responses to HIV-1 proteins were measured in samples from ALVAC HIV recombinant canarypox vaccine trials by means of a lytic cytotoxic T lymphocyte assay, and the proportions of samples responding to Gag or Env were found to be significantly higher among those carrying HLA-B*27 or B*57 [25]. The canarypox T cell vaccine that was used was poorly immunogenic for eliciting CD8+ T cells, and the study population was smaller; still, the concordance between our observations and those earlier results strengthens the case for an important role of protective HLA-B alleles in differential cellular immune responses to HIV-1 vaccines.
Gag-specific CD8+ T cell responses have repeatedly been found to be related to improved immune control of HIV-1 infection [12, 26, 27], which is explained largely because Gag is both highly immunogenic and highly conserved in sequence, and because mutational escapes often result in a decrease in viral fitness. The results presented here show that Gag-specific immune responses to T cell vaccines are modulated by the host immunogenetic background. HLA class I typing data should thus be discussed at the design stage and included in the analysis of future studies of candidate vaccines eliciting robust CD8+ T cells.
Finally, it is noteworthy that most of the interindividual variation in cellular responses to the MRKAd5 HIV-1 gag/pol/nef vaccine remains unexplained. To help understand the rest of the variation, it seems appropriate to perform additional studies, for example, by applying resequencing technology to uncover rare causal genetic variants, since we here confirmed the existence of a connection between natural viral control and vaccine response.
Funding
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) Center for HIV/AIDS Vaccine Immunology (grant number U19 AI067854-05); and the NIAID HIV Vaccine Trials Network.
References
- 1.Buchbinder SP, Mehrotra DV, Duerr A, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–93. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McElrath MJ, De Rosa SC, Moodie Z, et al. HIV-1 vaccine-induced immunity in the test-of-concept Step study: a case-cohort analysis. Lancet. 2008;372:1894–905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catanzaro AT, Koup RA, Roederer M, et al. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector. J Infect Dis. 2006;194:1638–49. doi: 10.1086/509258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Priddy FH, Brown D, Kublin J, et al. Safety and immunogenicity of a replication-incompetent adenovirus type 5 HIV-1 clade B gag/pol/nef vaccine in healthy adults. Clin Infect Dis. 2008;46:1769–81. doi: 10.1086/587993. [DOI] [PubMed] [Google Scholar]
- 5.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 6.Moodie Z, Huang Y, Gu L, Hural J, Self SG. Statistical positivity criteria for the analysis of ELISpot assay data in HIV-1 vaccine trials. J Immunol Methods. 2006;315:121–32. doi: 10.1016/j.jim.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 7.Moodie Z, Price L, Gouttefangeas C, et al. Response definition criteria for ELISPOT assays revisited. Cancer Immunol Immunother. 2010;59:1489–501. doi: 10.1007/s00262-010-0875-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 10.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J Evol Biol. 2005;18:1368–73. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 12.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–30. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Migueles SA, Sabbaghian MS, Shupert WL, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97:2709–14. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altfeld M, Addo MM, Rosenberg ES, et al. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. IDS. 2003;17:2581–91. doi: 10.1097/00002030-200312050-00005. [DOI] [PubMed] [Google Scholar]
- 15.Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–7. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fellay J, Ge D, Shianna KV, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pelak K, Goldstein DB, Walley NM, et al. Host determinants of HIV-1 control in African Americans. J Infect Dis. 2010;201:1141–9. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goulder PJ, Phillips RE, Colbert RA, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–7. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 19.Schneidewind A, Brockman MA, Yang R, et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J virol. 2007;81:12382–93. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaslow RA, Carrington M, Apple R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–11. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 21.Kawashima Y, Pfafferott K, Frater J, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–5. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steel CM, Ludlam CA, Beatson D, et al. HLA haplotype A1 B8 DR3 as a risk factor for HIV-related disease. Lancet. 1988;1:1185–8. doi: 10.1016/s0140-6736(88)92009-0. [DOI] [PubMed] [Google Scholar]
- 23.Kiepiela P, Leslie AJ, Honeyborne I, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–75. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 24.Frahm N, Janes H, Friedrich DP, et al. Beneficial effects of protective HLA class I allele expression and breadth of epitope recognition after vaccination on HIV viral load post-infection. AIDS Vaccine. 2010;P17.20. Atlanta, Georgia, USA [Google Scholar]
- 25.Kaslow RA, Rivers C, Tang J, et al. Polymorphisms in HLA class I genes associated with both favorable prognosis of human immunodeficiency virus (HIV) type 1 infection and positive cytotoxic T-lymphocyte responses to ALVAC-HIV recombinant canarypox vaccines. J Virol. 2001;75:8681–9. doi: 10.1128/JVI.75.18.8681-8689.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riviere Y, McChesney MB, Porrot F, et al. Gag-specific cytotoxic responses to HIV type 1 are associated with a decreased risk of progression to AIDS-related complex or AIDS. AIDS Res Hum Retroviruses. 1995;11:903–7. doi: 10.1089/aid.1995.11.903. [DOI] [PubMed] [Google Scholar]
- 27.Kiepiela P, Ngumbela K, Thobakgale C, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]