Abstract
The cell wall of bacteria induces pro-inflammatory cytokines in monocytes and neutrophils in human blood. The nature of the stimulating component of bacterial cell walls is not well understood. We have previously shown polymeric peptidoglycan (PGN2) has this activity, and the cytokine response requires PGN internalization and trafficking to lysosomes. Here, we demonstrate that peptidoglycan monomers like muramyl dipeptide and soluble peptidoglycan fail to induce robust cytokine production in immune cells although they activate the NOD proteins in transfected cell models. We further show that lysosomal extracts from immune cells degrade intact peptidoglycan into simpler products and that the lysosomal digestion products activate the NOD proteins. We conclude that naïve innate immune cells recognize PGN in its polymeric form rather than monomers such as muramyl dipeptide and require PGN lysosomal hydrolysis to respond. These findings offer new opportunities in the treatment of sepsis, especially sepsis arising from Gram-positive organisms.
Introduction
Peptidoglycan (PGN), a component of the cell wall of all bacteria, is a polymer made of alternating N-acetyl glucosamine and N-acetyl muramic acid with a short stem peptide composed of alanine, glutamic acid/glutamine and diaminopimelic acid (DAP)/lysine covalently linked to the N-acetyl muramic acid. Two or more peptidoglycan strands are cross-linked by covalent linkages between the stem peptides of each strand (1). The monomeric components of PGN include muramyl dipeptide (MDP), which consists of muramic acid linked to Ala-Glu and the DAP- or Lys-containing stem peptides.
Studies on the biological activity of PGN have been confounded by bacterial cell wall impurities, including lipoteichoic acid and triacetylated lipopeptides that stimulate Toll-like receptor (TLR) 2 (2). PGN itself was thought to be recognized by TLR2 (3) but later studies revealed that the recognition was due to contaminants in the PGN preparations (4, 5). Subsequently, two members of the nucleotide oligomerization domain (NOD)-like receptor family were found to detect PGN monomers, as portions of the PGN stem peptide containing DAP (recognized by NOD1), or MDP (recognized by NOD2) (6–9). The notion of NOD recognition of PGN monomers is largely based on studies of transfected embryonic kidney cells over-expressing the NOD receptors (6–8). With the discovery of NOD function as PGN monomer sensors, it was thought that the PGN had no biological activity as a polymer. Indeed, polymeric PGN was shown to be inert towards innate immune cells (4, 5, 10) and intact intestinal epithelial cells (6), although there is evidence to the contrary (11–13).
The notion that immune cells recognize PGN monomers introduces a conundrum: how do water-soluble monomeric components enter immune cells? Accordingly, elaborate models have been invoked by which monomeric PGN fragments are delivered to immune cells: uptake of monomeric PGN fragments in clathrin-coated pits (14, 15), delivery of monomeric PGN fragments via bacterially-secreted vesicles (16), surface-expressed NOD proteins where monomeric fragments can be detected (17), or NOD ligands generated by bacterial hydrolytic enzymes and shed from growing bacteria [reviewed in (18). These models are based on transformed cell lines which may have different processes than naïve immune cells.
To resolve these controversies, we purified PGN from the Gram-positive sporulating bacteria Bacillus anthracis such that the material lacks detectable TLR2 contaminants, has insignificant levels of endotoxin, and minimal protein contamination (11, 12). We showed that the pure preparation of PGN induced production of pro-inflammatory cytokines in human innate immune cells (12) which required PGN internalization and lysosomal trafficking (11).
In the current report we compared intact polymeric PGN to monomeric MDP and soluble PGN and found the PGN polymer to be a much more potent inducer of pro-inflammatory cytokines in human innate immune cells. We demonstrate for the first time that lysosomal extracts obtained from human innate immune cells have the ability to degrade the PGN into soluble components, and that the lysosome-generated products of PGN hydrolysis activate the NOD proteins. Our findings support a new model of PGN recognition in which polymeric PGN is internalized, traffics to the lysosome and is degraded into simpler products that are efficiently recognized by the intracellular NOD receptors.
Materials and Methods
Cells
HEK293 cell lines stably-expressing vector only (HEK-Blue-Null1, HEK-Blue-Null2), or human Nod1 (HEK-Blue-hNOD1) or human Nod2 (HEK-Blue-hNOD2) genes and secreted alkaline phosphatase on an NF-κB promoter were purchased from Invivogen (San Diego, CA). The cells were cultured according to the manufacturer’s recommendations. Heparinized peripheral blood (PB) obtained by venipuncture was cultured as described (11, 12).
Preparation of B. anthracis peptidoglycan
Peptidoglycan was isolated from B. anthracis ΔSterne strain that lacks the capsule and toxins as described in (11, 12). FITC labeling of PGN was performed as described (11). The material lacks detectable endotoxin, lipopeptides or other TLR stimuli (11, 12).
Flow cytometry analysis
Detection of cytokines and chemokines produced by Brefeldin A (BFA)-treated blood cells was performed by intracellular cytokine staining as described (11). 200,000 cells per sample were analyzed.
Lysosome preparation
Cells were resuspended in 0.25 M sucrose, lysed and centrifuged 6,000 ×g. The supernatants were centrifuged 100,000 ×g for 60 minutes. The pellet was lysed in 50 mM sodium citrate buffer pH 5.5, 2 mM DTT, 0.5% Triton X-100. Preliminary experiments established that lysosomal enzymes were most active at pH 5.5. 20–50 μg of the lysosomal extract was incubated with 10 μg FITC-PGN for 24 hours at 37°C followed by centrifugation at 20,000 ×g for 10 minutes. The supernatants were analyzed using a luminescence spectrometer (Perkin Elmer, Waltham, MA). For experiments measuring NOD activation in lysosome treated PGN, the cells were sonicated to avoid detergent.
Liquid chromatography and mass spectroscopy analysis of lysosomal products
Samples were acid-hydrolyzed with 6N HCl for 3 hrs at 105°C. The acid-hydrolyzed sample was dried, re-dissolved in 10 mM ammonium acetate, 80% acetonitrile and injected on to a ZIC pHILC column (150 × 2.1mm, 5 um, 200 Å, The Nest Group, Inc., Southborough, MA). The material was eluted with an acetonitrile gradient and the eluted peaks were analyzed and quantitated by mass spectroscopy (Bruker Daltonics HCT Ultra Ion trap in positive mode). The elution time and mass spectra of standard amounts of purified muramic acid were determined in preliminary experiments.
NOD activation
HEK293 cell lines were treated with appropriate ligands for 24 hours. NF-κB activation was detected by measuring secreted embryonic alkaline phosphatase spectrophotometrically at 630 nm.
Statistical analysis
All statistical analyses were performed using GraphPad Prism. Statistical significance was determined by the non-parametric Mann-Whitney test and p values <0.05 were considered statistically significant.
Results
Polymeric PGN is a more potent stimulant of innate immune cells than MDP and/or soluble PGN
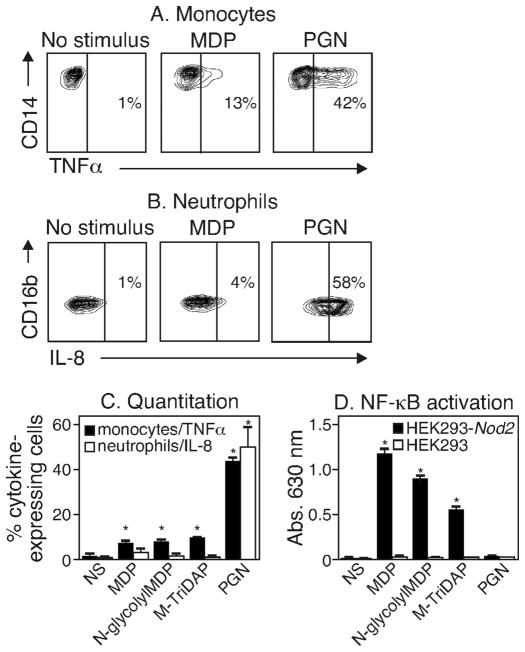
We compared the ability of monomeric MDP vs. polymeric PGN to induce cytokine production in primary human blood cells and in Nod-transfected HEK293 cells. Human peripheral blood cells were treated with 10 μg/mL of MDP or 10 μg/mL intact PGN and cytokine production was measured as previously described (11, 12). As shown in Figure 1A, only about 13% of monocytes produced TNFα in response to monomeric MDP in contrast to 42% monocytes that produced TNFα in response to polymeric PGN. 18% of monocytes became TNFα+ with 12 hour MDP incubation, and a higher dose (50 μg/mL) did not improve the response at 6 or 12 hours. A similar trend was observed for IL-8 production in neutrophils: 4% of neutrophils produced IL-8 in response to MDP vs. nearly 60% in response to PGN (Figure 1B). The data from several experiments using polymeric PGN and a variety of commercial PGN monomeric forms including MTriDAP (N-acetyl-muramyl-L-Ala-γ-D-Glu-mDAP), MDP and N-glycolylMDP are shown in Figure 1C. In contrast, HEK293 cells stably expressing NOD2 and an NF-κB reporter gene responded robustly to the monomeric forms of PGN but failed to respond to polymeric PGN as measured by NF-κB activation (Figure 1D).
Figure 1. MDP is a poor stimulator of innate immune cells.
(A and B) Human peripheral blood cells treated with nothing (NS), 10 μg/mL MDP or 10 μg/mL PGN for 6 hours were identified using cell surface markers. TNFα production (A) in monocytes and IL-8 production (B) in neutrophils was determined by flow cytometry. (C) The percent of monocytes and neutrophils expressing TNFα and IL-8 respectively following treatment with MDP, N-glycolylMDP, M-TriDAP, PGN or no stimulus (NS) was determined by flow cytometry. Results are from three independent experiments using three different donors. (D) HEK293 cells untransfected (HEK293) or stably transfected with human Nod2 (HEK293-Nod2) were treated with 10 μg/mL PGN or 0.1 μg/mL MDP, N-glycolylMDP or M-TriDAP for 24 hours. NF-κB activation was determined spectrophotometrically at 630 nm. The results are typical of three identical experiments. * P values <0.05.
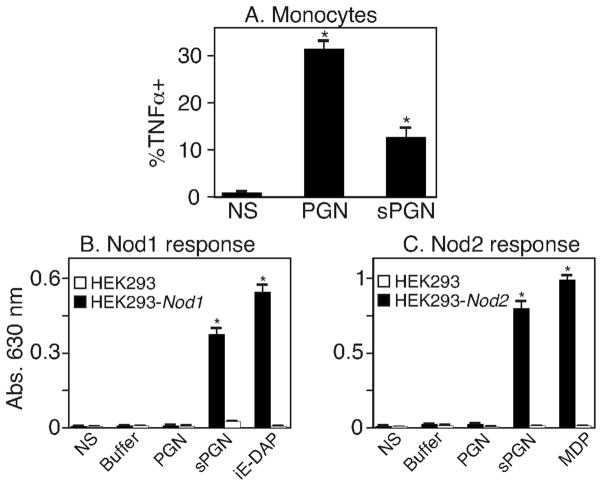
Since pure MDP may not truly represent naturally-occurring soluble PGN fragments, we digested intact PGN with mutanolysin (an enzyme isolated from Streptomyces having lysozyme activity) to generate soluble PGN (sPGN) degradation products. We then measured TNFα production in monocytes or NF-κB activation in the Nod-transfected HEK293 cells treated with digested or intact PGN. We found that intact PGN was a better stimulator of TNFα production than sPGN (Figure 2A). In contrast, sPGN was able to activate NF-κB in HEK293 cells over-expressing either NOD proteins (Figure 2B and C). Together, these data show that monomeric components of PGN are poor stimulants of innate immune cells but potent stimulants of artificial models where NOD receptors are over-expressed. In contrast, polymeric PGN is capable of efficiently stimulating immune cells.
Figure 2. Intact PGN is better at inducing TNFα production in peripheral blood monocytes.
(A) PB cells were treated for 6 hours with 10 μg/mL intact PGN (PGN) or 10 μg/mL soluble PGN (sPGN). TNFα production in monocytes was measured by intracellular staining and flow cytometry. Results shown are from three independent experiments performed with three different donors. (B and C) HEK293 cells untransfected (HEK293) or stably transfected with human Nod1 (B) or Nod2 (C) were untreated (NS) or treated with buffer used for digesting PGN (Buffer), 10 μg/mL PGN, 10 μg/mL digested PGN (sPGN), iE-DAP or MDP for 24 hours. NF-κB activation was determined spectrophotometrically at 630 nm. Similar results were obtained in three additional experiments. * P values <0.05.
PGN is digested by lysosomal extracts
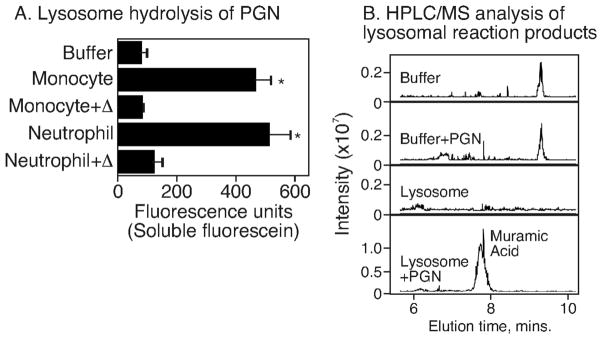
We have previously shown that lysosomotropic agents block cytokine production in innate immune cells responding to PGN. The observation suggests that lysosomal activity is required for cytokine production in innate immune cells exposed to intact polymeric PGN. To determine if lysosomes from innate immune cells had the capacity to degrade intact PGN, we labeled PGN with FITC. The FITC covalently-bound to PGN was completely recoverable by low-speed centrifugation. We reasoned that digestion of the PGN disaccharide backbone would render a fraction of the FITC to become water-soluble and thus present in the supernatant of a lysosomal digest. For this experiment, the FITC-labeled PGN was incubated with lysosomal extracts from monocytes and neutrophils and PGN hydrolysis was determined by the appearance of water-soluble fluorescein in the supernatant. We found (Figure 3A) that the lysosomal fraction of both monocytes and neutrophils was able to solubilize FITC-PGN and that the activity was sensitive to heat inactivation. We verified that the PGN disaccharide backbone was degraded by assaying for muramic acid in the lysosomal reactions. Here, the lysosome-generated PGN hydrolytic products were acid-hydrolyzed and monitored by HPLC and mass spectroscopy. We found that muramic acid was detected only in the supernatants of lysosomal reactions incubated with PGN and not in the controls (Figure 3B). These data establish that PGN is hydrolyzed to water-soluble products by the lysosomal enzymes of innate immune cells.
Figure 3. PGN is degraded by lysosomal enzymes of monocytes and neutrophils.
(A) Lysosomal extracts prepared from monocytes or neutrophils were incubated with 10 μg of FITC-PGN for 24 hours and FITC in the supernatant was measured. Results shown are from three independent experiments performed with lysosomal extracts prepared from three different donors. (B) Unlabeled PGN was incubated with lysosomal extracts obtained from neutrophils. The supernatants of these reactions were hydrolyzed with 6N HCl for 3 hours at 105°C followed by HPLC-MS analysis. Results from one representative experiment are shown. * P values <0.05.
Neutrophil lysosomal extracts degrade PGN to generate NOD ligands
Since lysosomes can degrade PGN into soluble moieties, we asked whether PGN fragments generated by the lysosome fraction contained a ligand for NOD1 or NOD2. HEK293 cells stably expressing Nod1 or Nod2 were treated with the supernatants of lysosome-digested PGN or with PGN incubated with buffer alone or the lysosomal fractions alone as controls. We found that PGN digested by the lysosomal fraction of neutrophils was able to stimulate both NOD1- and NOD2-expressing HEK293 cells, indicated by NF-κB activation. Untransfected HEK293 cells failed to respond (not shown). In accordance with the above data, no NOD activation was observed when we used heat inactivated lysosomal extracts to degrade PGN.
Discussion
Although PGN is a well characterized cell wall component of bacteria, whether or not immune cells recognize and respond to PGN and how they respond is controversial. It has been well documented that transformed cell lines expressing NOD receptors respond to the water-soluble monomeric moieties that make up PGN. In this report we show that MDP and other bona fide ligands weakly induce the production of TNFα in primary naïve monocytes and IL-8 in primary naïve neutrophils while the intact, polymeric PGN is a more potent inducer of these pro-inflammatory molecules. Since PGN but not monomeric versions of PGN or mutanolysin-solubilized PGN fragments were able to induce cytokine production in monocytes, we conclude that innate immune cells efficiently recognize and respond to PGN only in its polymeric form, but the cells must process it intracellularly.
These data are in contrast to current models of monomeric PGN recognition by innate immune cells (14, 15) and indeed to rather abundant literature showing that murine innate immune cells respond vigorously to various monomeric fragments of PGN. We note that many such studies showing responses to PGN monomers manipulate the responder mouse macrophages in some way prior to stimulation: pretreatment of macrophages with LPS [e.g., (19)] or cytochalasin D [e.g., (20)], direct transfection of MDP into macrophages [e.g., (21)], or the studies rely exclusively on Nod-transfected HEK293, HL-60, HT-29, Caco-2 or HEK293T cell lines [e.g., (22)]. We found that a 12 hour pretreatment of bone marrow-derived mouse macrophages or a 3 hour pretreatment of human blood with LPS (1 ng/ml) caused the cells to be essentially equally responsive to monomeric MDP and polymeric PGN. LPS pretreatment might cause the responding cells to express a transporter of MDP or DAP-containing stem peptides or alter a signaling cascade (23) to enable a response to these monomeric components. However, our data here establish that primary unmanipulated innate immune cells in blood do not respond well to the PGN monomers like MDP and sPGN while they respond vigorously to the PGN polymer.
Our earlier report (11) showed that polymeric PGN bound some blood cells (B cells, neutrophils and monocytes) but not others (T cells). The observation suggests that some immune cells express an extracellular receptor capable of binding PGN. We found (not shown) that only 7% of HEK293 cells bind polymeric PGN, suggesting that they fail to respond in part because they lack the PGN receptor.
The nature of the NOD ligand(s) derived from lysosomal digestion of PGN are not known. The monomeric components MDP (6) and DAP-containing peptides (24) have been isolated from mutanolysin-digested PGN derived from N. meningitidis and B. subtilis. Clearly, lysosomes of innate immune cells express lysozyme (25), suggesting the monomers previously identified might be natural products of innate immune cell lysosomal digestion of PGN. We speculate that the need for PGN endocytosis and/or lysozyme digestion and/or NOD expression levels might be mechanism(s) by which intestinal epithelial cells ignore the copious quantities of PGN to which they are exposed (26).
We conclude that the polymeric nature of PGN is required for naïve immune cells to efficiently phagocytose it and traffic it to the lysosome. We provide direct evidence that lysosomes derived from innate immune cells are able to degrade intact PGN, and show that lysosomal degradation generates a NOD ligand in the process. Our model where intact PGN rather than PGN monomers is phagocytosed and digested in the lysosome is also supported by a recent study performed in murine macrophages using PGN from Staphylococcus aureus (13). These findings provide a novel mechanism on how human monocytes and neutrophils in peripheral blood recognize and respond to PGN.
Figure 4. Degradation of PGN by lysosomes generate NOD ligands.
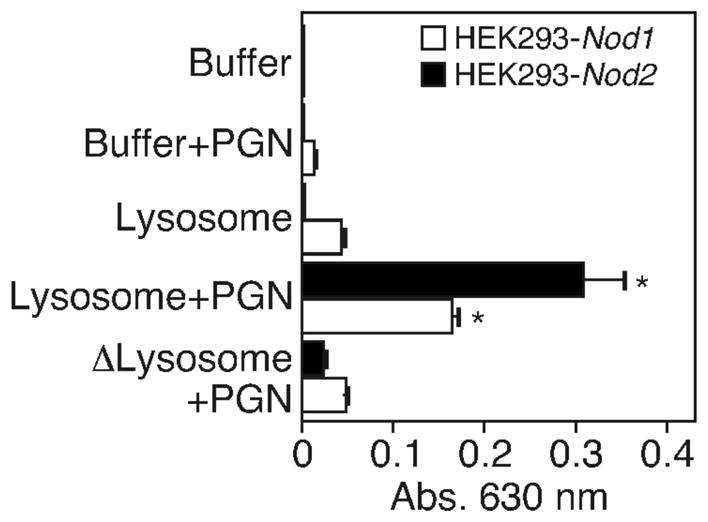
20 μg PGN was incubated with lysosomal extracts obtained from neutrophils or buffer alone. HEK293 cells stably expressing human NOD1 (HEK293-Nod1) or NOD2 (HEK293-Nod2) were treated with the supernatants of these reactions for 24 hours at 37°C. NF-κB activation was determined spectrophotometrically at 630 nm. Similar results were obtained in three additional experiments. * P values <0.05.
Acknowledgments
We thank the Laboratory for Molecular Biology and Cytometry Research at the University of Oklahoma Health Sciences Center for the LC/MS studies. We are grateful to Erich and Heike Gulbins (University of Duisburg-Essen, Essen Germany), Don Capra and Paul Kincade (Oklahoma Medical Research Foundation) for reviewing the manuscript.
Footnotes
Supported by NIH grant 2 U19 AI062629.
List of abbreviation: BFA, Brefeldin A; DAP, diaminopimelic acid; FITC, fluorescein isothiocyanate; iE-DAP, isoGlutamine-diaminopimelic acid dipeptide; M-TriDAP, Nacetyl-muramyl-L-Ala-γ-D-Glu-mDAP; MDP, muramyl dipeptide; NOD, nucleotide-binding oligomerization domain; PB, peripheral blood; PGN, peptidoglycan; sPGN, soluble peptidoglycan; TLR, Toll-like receptor.
References
- 1.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22:240–273. doi: 10.1128/CMR.00046-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J Biol Chem. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 4.Travassos LH, Girardin SE, Philpott DJ, Blanot D, Nahori MA, Werts C, Boneca IG. Toll-like receptor 2-dependent bacterial sensing does not occur via peptidoglycan recognition. EMBO Rep. 2004;5:1000–1006. doi: 10.1038/sj.embor.7400248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volz T, Nega M, Buschmann J, Kaesler S, Guenova E, Peschel A, Rocken M, Gotz F, Biedermann T. Natural Staphylococcus aureus-derived peptidoglycan fragments activate NOD2 and act as potent costimulators of the innate immune system exclusively in the presence of TLR signals. FASEB J. 2010;24:4089–4102. doi: 10.1096/fj.09-151001. [DOI] [PubMed] [Google Scholar]
- 6.Girardin SE, I, Boneca G, Carneiro LA, Antignac A, Jehanno M, Viala J, Tedin K, Taha MK, Labigne A, Zahringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ, Philpott DJ. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 7.Girardin SE, I, Boneca G, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 8.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nunez G, Inohara N. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 9.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nunez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 10.Rockel C, Hartung T, Hermann C. Different Staphylococcus aureus whole bacteria mutated in putative pro-inflammatory membrane components have similar cytokine inducing activity. Immunobiology. 2010 doi: 10.1016/j.imbio.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Iyer JK, Khurana T, Langer M, West CM, Ballard JD, Metcalf JP, Merkel TJ, Coggeshall KM. Inflammatory cytokine response to Bacillus anthracis peptidoglycan requires phagocytosis and lysosomal trafficking. Infect Immun. 2010;78:2418–2428. doi: 10.1128/IAI.00170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langer M, Malykhin A, Maeda K, Chakrabarty K, Williamson KS, Feasley CL, West CM, Metcalf JP, Coggeshall KM. Bacillus anthracis peptidoglycan stimulates an inflammatory response in monocytes through the p38 mitogen-activated protein kinase pathway. PLoS ONE. 2008;3:e3706. doi: 10.1371/journal.pone.0003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimada T, Park BG, Wolf AJ, Brikos C, Goodridge HS, Becker CA, Reyes CN, Miao EA, Aderem A, Gotz F, Liu GY, Underhill DM. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe. 2010;7:38–49. doi: 10.1016/j.chom.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marina-Garcia N, Franchi L, Kim YG, Hu Y, Smith DE, Boons GJ, Nunez G. Clathrin- and dynamin-dependent endocytic pathway regulates muramyl dipeptide internalization and NOD2 activation. J Immunol. 2009;182:4321–4327. doi: 10.4049/jimmunol.0802197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Tattoli I, Wojtal KA, Vavricka SR, Philpott DJ, Girardin SE. pH-dependent internalization of muramyl peptides from early endosomes enables Nod1 and Nod2 signaling. J Biol Chem. 2009;284:23818–23829. doi: 10.1074/jbc.M109.033670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaparakis M, Turnbull L, Carneiro L, Firth S, Coleman HA, Parkington HC, Le Bourhis L, Karrar A, Viala J, Mak J, Hutton ML, Davies JK, Crack PJ, Hertzog PJ, Philpott DJ, Girardin SE, Whitchurch CB, Ferrero RL. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol. 2010;12:372–385. doi: 10.1111/j.1462-5822.2009.01404.x. [DOI] [PubMed] [Google Scholar]
- 17.Kufer TA, Kremmer E, Adam AC, Philpott DJ, Sansonetti PJ. The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell Microbiol. 2008;10:477–486. doi: 10.1111/j.1462-5822.2007.01062.x. [DOI] [PubMed] [Google Scholar]
- 18.Humann J, Lenz LL. Bacterial peptidoglycan degrading enzymes and their impact on host muropeptide detection. J Innate Immun. 2009;1:88–97. doi: 10.1159/000181181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coulombe F, Divangahi M, Veyrier F, de Leseleuc L, Gleason JL, Yang Y, Kelliher MA, Pandey AK, Sassetti CM, Reed MB, Behr MA. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J Exp Med. 2009;206:1709–1716. doi: 10.1084/jem.20081779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, Lee BL, Shiffin N, Advincula R, Malynn BA, Werts C, Ma A. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertrand MJ, Doiron K, Labbe K, Korneluk RG, Barker PA, Saleh M. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30:789–801. doi: 10.1016/j.immuni.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Clark NM, Marinis JM, Cobb BA, Abbott DW. MEKK4 sequesters RIP2 to dictate NOD2 signal specificity. Curr Biol. 2008;18:1402–1408. doi: 10.1016/j.cub.2008.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfert MA, Murray TF, Boons GJ, Moore JN. The origin of the synergistic effect of muramyl dipeptide with endotoxin and peptidoglycan. J Biol Chem. 2002;277:39179–39186. doi: 10.1074/jbc.M204885200. [DOI] [PubMed] [Google Scholar]
- 24.Girardin SE, Travassos LH, Herve M, Blanot D, Boneca IG, Philpott DJ, Sansonetti PJ, Mengin-Lecreulx D. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003;278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- 25.Pryzwansky KB, Martin LE, Spitznagel JK. Immunocytochemical localization of myeloperoxidase, lactoferrin, lysozyme and neutral proteases in human monocytes and neutrophilic granulocytes. J Reticuloendothel Soc. 1978;24:295–310. [PubMed] [Google Scholar]
- 26.Melmed G, Thomas LS, Lee N, Tesfay SY, Lukasek K, Michelsen KS, Zhou Y, Hu B, Arditi M, Abreu MT. Human intestinal epithelial cells are broadly unresponsive to Toll-like receptor 2-dependent bacterial ligands: implications for host-microbial interactions in the gut. J Immunol. 2003;170:1406–1415. doi: 10.4049/jimmunol.170.3.1406. [DOI] [PubMed] [Google Scholar]