Abstract
O-class forkhead box (FOXO) transcription factors are critical regulators of diverse cellular processes, including apoptosis, cell-cycle arrest, DNA damage repair and oxidative stress resistance. Here, we show that FOXO1 and FOXO3a have an essential function in promoting cell detachment-induced anoikis, resistance to which is implicated in cancer development and metastasis. In contrast, the oncoprotein cyclin D1 inhibits anoikis. We further show that cyclin D1 interacts with FOXO proteins and impedes their transcriptional regulatory and anoikis-promoting functions. This effect of cyclin D1 requires its transcription repression domain but is independent of cyclin-dependent kinases CDK4 and CDK6. Moreover, we show that cancer-derived mutants of cyclin D1 are much more stable than wild-type cyclin D1 under anchorage-independent conditions and possess a greater antagonistic effect on FOXO-regulated anoikis and anchorage-independent growth of cancer cells. These data suggest that cyclin D1 may have a critical function in tumorigenesis and cancer metastasis by inhibiting the anoikis-promoting function of FOXO proteins.
Keywords: FOXO1, FOXO3a, cyclin D1, cyclin-dependent kinase, anoikis, prostate cancer
Adhesion to an appropriate extracellular matrix (ECM) is important for normal cells to survive, and detachment from such supportive matrices usually triggers a specific type of apoptosis termed anoikis.1 However, most cancer cells are commonly resistant to anoikis and can survive in the presence of an inappropriate ECM or in the absence of anchorage to the ECM.2 By binding to the ECM, members of the integrin family become activated, setting off a myriad of downstream/intracellular apoptosis-inhibitory pathways, thereby protecting cells from anoikis.3 In contrast, loss of integrin engagement with the ECM has been shown to promote anoikis in breast epithelial cells,4 and this process is likely mediated by increased expression of the pro-apoptotic gene, Bim, because silencing of the Bim gene prevents integrin inactivation-induced anoikis.4 Bim transcription is also upregulated during anoikis in many other types of cells,5–7 suggesting that elevated Bim gene expression is a common event that precedes anoikis. However, the transcription factors responsible for the increased expression of Bim during anoikis are unknown.
FOXO proteins, which include FOXO1 (FKHR), FOXO3a (FKHRL1), FOXO4 (AFX), and FOXO6 in humans, belong to the superfamily of forkhead transcription factors characterized by a highly conserved DNA-binding domain (also called the forkhead domain).8 Activation of FOXO proteins induces cell-cycle arrest at G1 by upregulating the cyclin-dependent kinase (CDK) inhibitors p27KIP1 and p21WAF1. Moreover, a number of pro-apoptotic genes, including Bim, Fas ligand (FasL), and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) are regulated by members of the FOXO family.9–11 Indeed, expression of FOXO proteins induces apoptosis in cells of various tissue types.12 These findings suggest that FOXO proteins possess tumor suppressor functions. In line with this hypothesis, functions of FOXO proteins are often modulated and inactivated by various posttranslational mechanisms, including phosphorylation, acetylation, and ubiquitination.12
The proto-oncogene cyclin D1 is implicated in human cancers because of its frequent amplification, overexpression, mutation, and its well-known function in cell proliferation. It is found amplified or mutated in both hematological malignancies and numerous types of solid tumors, including breast and prostate cancer.13–17 The function of cyclin D1 in cell-cycle progression is to bind and activate CDK4 and CDK6, which ultimately leads to phosphorylation and inactivation of the tumor suppressor retinoblastoma (RB) protein.18 However, emerging evidence suggests that a CDK4/6-independent function of cyclin D1 is also implicated in tumorigenesis.19
Here, we show that FOXO1 and FOXO3a are required for cell detachment-triggered anoikis. We also show that cyclin D1 interacts with both FOXO1 and FOXO3a and inhibits their transcriptional activity and anoikis-promoting function. Moreover, our results indicate that this function of cyclin D1 is mediated through a protein–protein interaction that is independent of CDK4 and CDK6.
Results
FOXO1 and FOXO3a promote anoikis
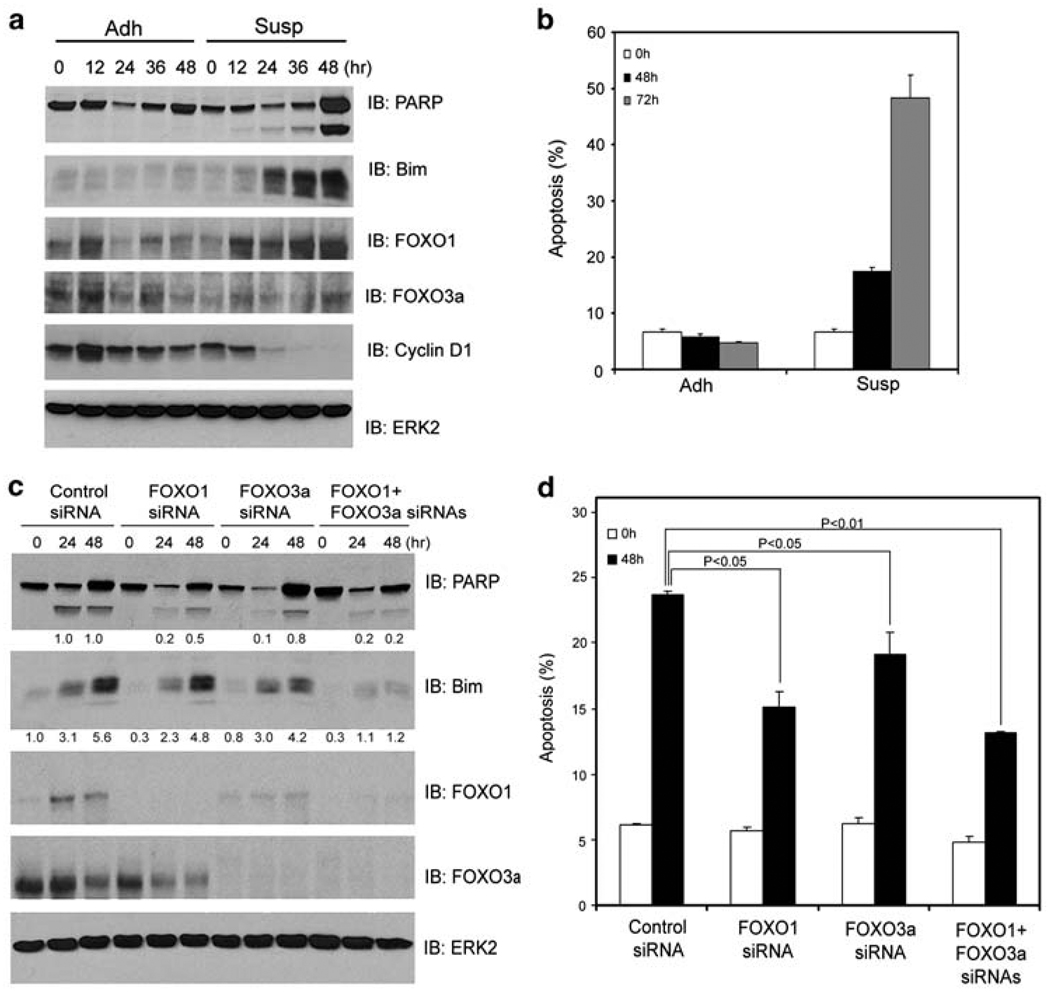
To define the molecular pathways mediating anoikis, we first surveyed anoikis sensitivity among the prostate cancer cell lines DU145, LNCaP, 22Rv1, and PC-3 and the nonmalignant prostatic epithelial cell line BPH-1. DU145 and BPH-1 cells underwent robust anoikis and cleavage of poly(ADP-ribose) polymerase (PARP), an apoptotic marker, when they were detached from ECM (Figure 1a and b; Supplementary Figure 1a and b). Massive cell death was also observed in NIH3T3 fibroblasts when they were cultured in suspension (Figure 2a and b). Under the same conditions, however, little or no anoikis was detected in LNCaP, 22Rv1, and PC-3 cells (Supplementary Figure 1d; data not shown). In agreement with earlier findings that the pro-apoptotic gene, Bim, has an essential function in execution of anoikis,4–7 expression of Bim was substantially upregulated in anoikis-sensitive DU145, NIH3T3, and BPH-1 cells (Figures 1a, 2a; Supplementary Figure 1a) but not in anoikis-resistant LNCaP, 22Rv1 and PC-3 cells (Supplementary Figures 1c, e and f). As Bim is a well-characterized target gene of FOXO transcription factors,9 we investigated whether FOXO proteins are key mediators of anoikis upstream of Bim. Toward this end, FOXO1 and FOXO3a, two members of the FOXO family that are predominantly expressed in prostatic epithelial cells,11,20 were effectively knocked down by FOXO1 and/or FOXO3a-specific small interfering RNAs (siRNAs) in DU145 cells (Figure 1c). Silencing of FOXO1 or FOXO3a alone significantly decreased detachment-induced anoikis, and further reduction in anoikis was detected in cells in which both FOXO1 and FOXO3a were concomitantly silenced (Figure 1d). Silencing of FOXO1 and/or FOXO3a also resulted in reduced cleavage of PARP (Figure 1c). In accordance with these observations, silencing of FOXO1 and/or FOXO3a diminished Bim expression induced by cell detachment (Figure 1c), indicating that FOXO proteins are responsible for upregulation of Bim expression during anoikis. Together, these results suggest that both FOXO1 and FOXO3a are important mediators of anoikis.
Figure 1.
FOXO1 and FOXO3a promote anoikis. DU145 cells grown in adhesion (Adh) or suspension (Susp) were collected at the indicated time points and subjected to immunoblotting analysis for expression of PARP, Bim, FOXO1, FOXO3a, and cyclin D1 proteins (a) and measurement of apoptosis (b). (c, d) DU145 cells were transfected with control siRNA, or siRNAs for FOXO1, FOXO3a, or both. At 48 h after transfection, cells were plated in adhesion or suspension and at the indicated time points cells were subjected to immunoblotting (c) and apoptosis (d) analysis. ERK2 was used as a loading control. Results from three independent experiments were quantified for apoptosis. Error bars indicate S.D. among three individual experiments
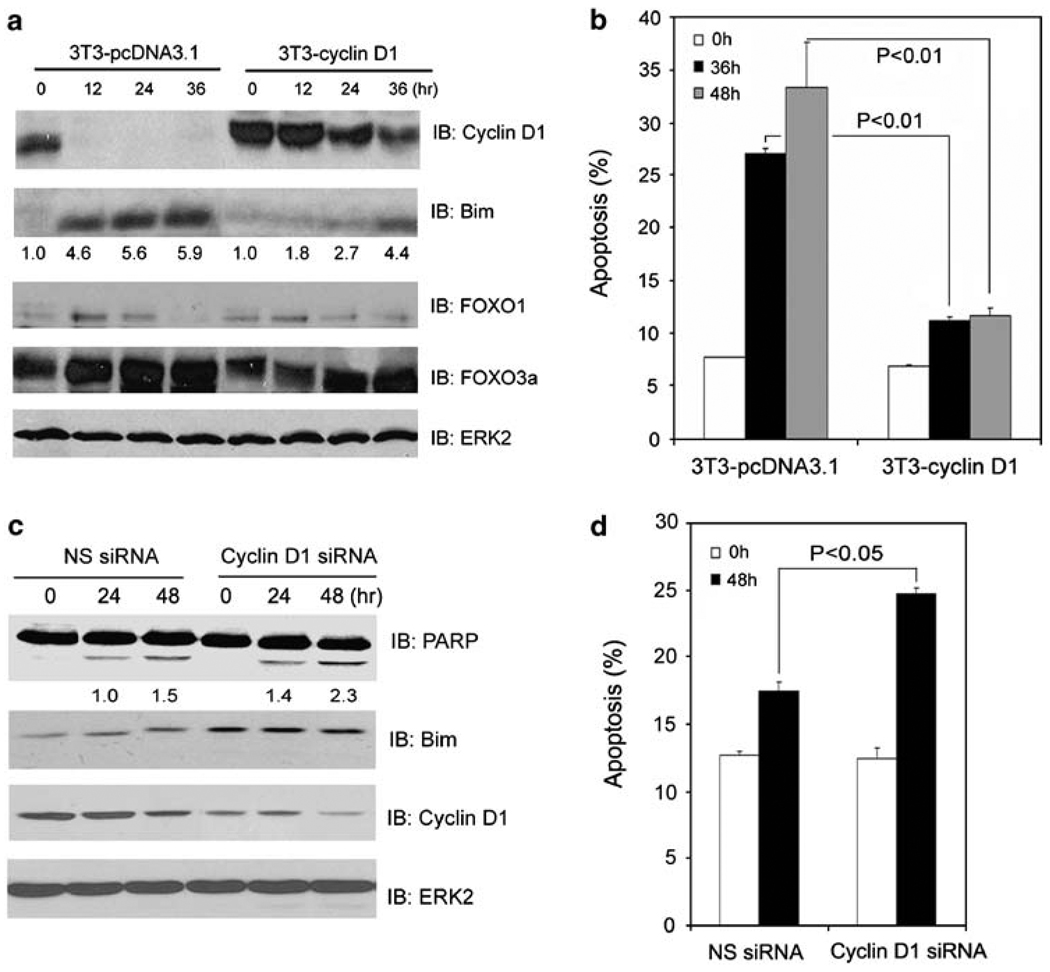
Figure 2.
Cyclin D1 inhibits anoikis. (a, b) NIH3T3 cells stably expressing either an empty vector (3T3-pcDNA3.1) or cyclin D1 (3T3-cyclin D1) were cultured in suspension and at the indicated time points cells were subjected to immunoblotting (a) and apoptosis (b) analysis. (c, d) LNCaP cells were transfected with a pool of cyclin D1-specific siRNAs or nonspecific control siRNAs. At 48 h after transfection, cells were cultured in suspension and at the indicated time points cells were subjected to immunoblotting (c) and apoptosis (d) analysis. The number underneath each band in the immunoblot indicates the relative intensity of the corresponding band. Error bars indicate S.D. among three individual experiments
Given that nuclear translocation is a major indicator of activation of FOXO proteins,12 we examined nuclear/cytoplasmic localization of FOXO1 and FOXO3a proteins in DU145 cells on detachment from ECM. FOXO1 proteins were detected equivalently in both the cytoplasm and the nucleus during the course of cells cultured in suspension (Supplementary Figure 2). FOXO3a proteins were mainly detected in the nucleus of detached DU145 cells. However, no translocation of this protein between the cytoplasm and the nucleus was observed (Supplementary Figure 2). Thus, no nuclear translocation of FOXO1 and FOXO3a proteins was observed after cell detachment, excluding the role of this mechanism in activation of FOXO proteins during anoikis.
Cyclin D1 suppresses anoikis
Resistance to anoikis is a hallmark of cancer cells and yet the underlying molecular mechanism is far from elucidated. Early studies have shown that many factors, including phorbol ester, galectin-3, and β-catenin, cause anoikis resistance in various types of cells. Most importantly, all these factors invariably induce expression of cyclin D1,21–24 suggesting that cyclin D1 may function as an important inhibitor of anoikis. In support of this hypothesis, we showed that no cyclin D1 protein was detected in anoikis-sensitive DU145, NIH 3T3, and BPH-1 cells when they underwent anoikis; however, the level of cyclin D1 protein remained unchanged in anoikis-resistant LNCaP, 22Rv1, and PC-3 cells under the same conditions (Figures 1a, 2a; Supplementary Figures 1a, c, e and f). To further determine the causal role of cyclin D1 in promoting anoikis resistance, we generated a cyclin D1 stable (3T3-cyclin D1) and a control cell line (3T3-pcDNA3.1) from NIH3T3 cells. As shown in Figure 2a, cyclin D1 protein remained at high levels in 3T3-cyclin D1 cells while they were detached from ECM. Importantly, little or no anoikis was detected in these cells relative to control 3T3-pcDNA3.1 cells (Figure 2b). Accordingly, PARP cleavage was readily detected in 3T3-pcDNA3.1 cells during anoikis. However, this event was largely inhibited in 3T3-cyclin D1 cells (Supplementary Figure 3). In contrast, cyclin D1 knocking down in anoikis-resistant LNCaP cells increased PARP cleavage, Bim expression, and anoikis (Figure 2c and d). Therefore, we conclude that elevated cyclin D1 confers cellular resistance to anoikis.
Cyclin D1 inhibits both the transcriptional and anoikis-promoting functions of FOXO1 and FOXO3a
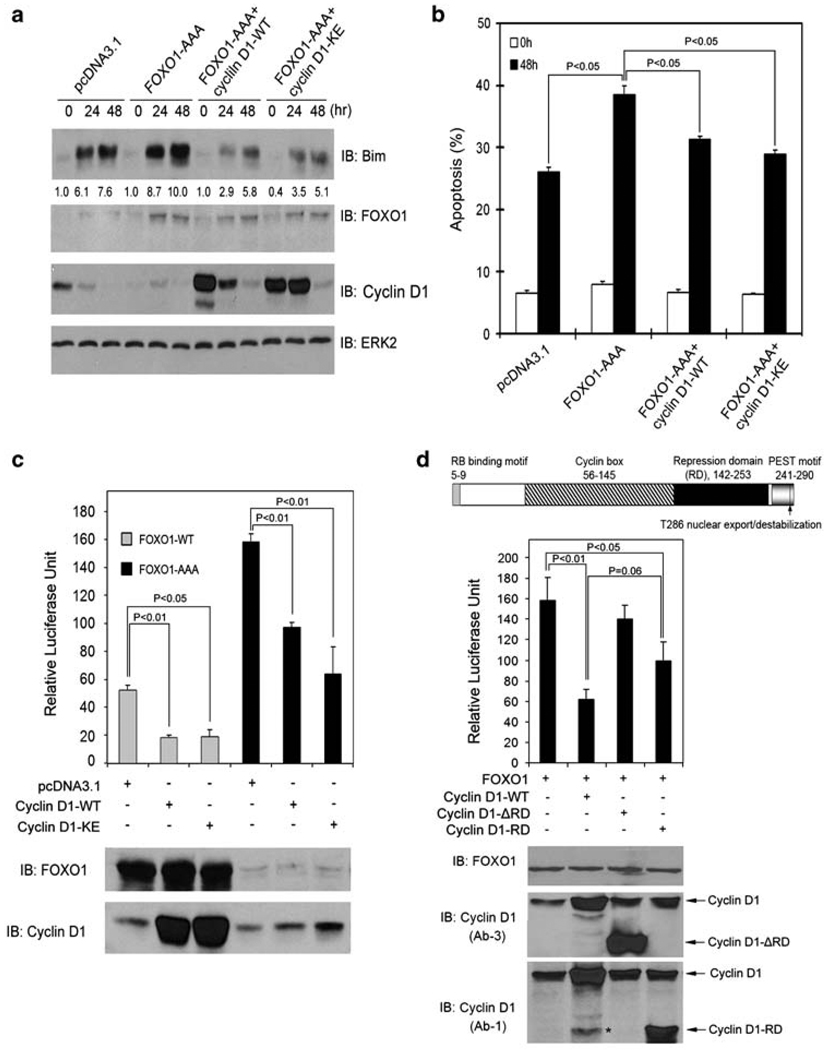
Given that FOXO1 and FOXO3a are responsible for the induction of Bim expression during anoikis (Figure 1), the finding that stable expression of cyclin D1 blocked Bim induction in cells grown in suspension (Figure 2a) suggests that cyclin D1 may protect cells from anoikis by inhibiting the function of FOXO proteins. To test this notion, we attempted to determine whether forced expression of FOXO1 enhances anoikis and this effect of FOXO1 can be blocked by cyclin D1. In line with our earlier observation that FOXO1 silencing inhibits anoikis (Figure 1c and d), ectopic expression of a constitutively active mutant of FOXO1 (FOXO1-AAA), in which three Akt phosphorylation residues were converted into alanines, sensitized DU145 cells to anoikis (Figure 3a and b). However, this effect of FOXO1 was largely abrogated by cotransfection of cells with wild-type cyclin D1 (Figure 3a and b). In agreement with these results, we further showed that cotransfection of cyclin D1 also diminished FOXO1-enhanced expression of endogenous Bim (Figure 3a). This effect of cyclin D1 on the level of Bim protein does not seem to be direct because overexpression of cyclin D1 had no effect on the levels of exogenously expressed Bim (Supplementary Figure 4). These results indicate that cyclin D1 may be able to inhibit FOXO’s transcriptional activity. Next, we sought to test this hypothesis by using a generic FOXO luciferase reporter gene that contains three copies of forkhead response elements. As shown in Figure 3c, forced expression of cyclin D1 inhibited the transcriptional activity of both wild-type (FOXO1-WT) and the active FOXO1 (FOXO1-AAA). Moreover, silencing of endogenous cyclin D1 increased FOXO’s transcriptional activity (Supplementary Figure 5). Thus, our results show that overexpression of cyclin D1 inhibits both the transcriptional and anoikis-promoting functions of FOXO1. Similarly, elevated cyclin D1 also inhibits FOXO3a’s transcriptional activity and FOXO3a-induced Bim expression and anoikis (Supplementary Figure 6a, b; data not shown).
Figure 3.
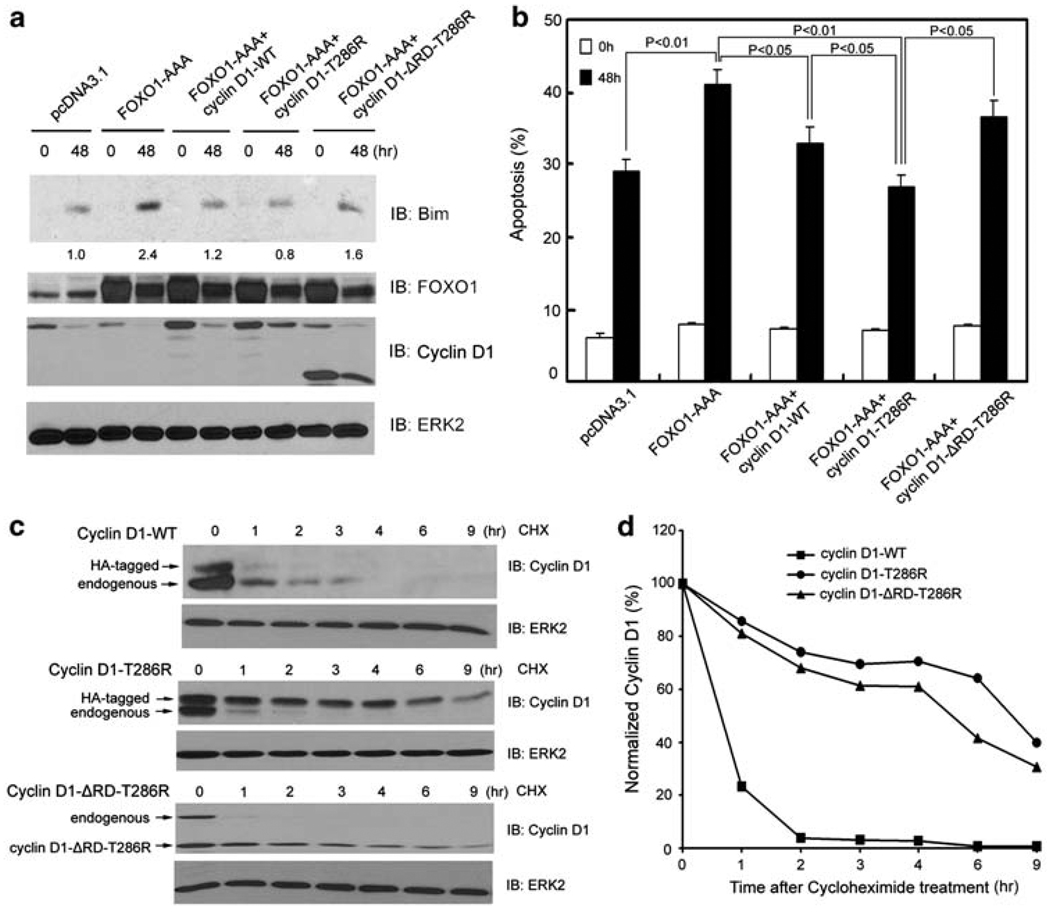
Cyclin D1 inhibits FOXO1-augmented anoikis and FOXO1’s transcriptional activity. (a, b) DU145 cells were transfected with plasmids as indicated. At 36 h after transfection, cells were cultured in suspension and at the indicated time points cells were subjected to immunoblotting (a) and apoptosis (b) analysis. The number underneath each band in the immunoblot indicates the relative intensity of the corresponding band. (c) LNCaP cells were transfected with firefly and Renilla luciferase reporter constructs and plasmids as indicated. At 36 h after transfection, cells were subjected to luciferase activity measurement as described in ‘Materials and Methods’ (upper panel) or western blot analysis (lower panel). Error bars indicate S.D. among three individual experiments. (d) Top, a schematic diagram of the cyclin D1 protein shows its different functional domains. Middle and bottom, LNCaP cells were transfected with the indicated plasmids and luciferase activities and western blots were analyzed as described in (c). The asterisk indicates a nonspecific immunoreactive band
Cyclin D1 inhibits FOXO1 and FOXO3a independent of CDK4 and CDK6 but dependent on its transcription repression domain
A well-known function of cyclin D1 is to bind to and activate CDK4 and CDK6, leading to phosphorylation and inhibition of cyclin D1/CDK substrates such as the RB protein.18 CDK1 and CDK2 are known to inhibit FOXO1 through phosphorylation of FOXO1 at serine residue 249.20,25 However, little or no phosphorylation of FOXO1 by CDK4 or CDK6 was detected with in vitro kinase assays (Supplementary Figure 7a, b and c). Accordingly, cotransfection of CDK4 or CDK6 with cyclin D1 failed to augment cyclin D1-mediated inhibition of FOXO1’s transcriptional activity (Supplementary Figure 7d). In addition, expression of the K112E mutant of cyclin D1 (cyclin D1-KE), which is unable to activate CDK4 and CDK6,26 also inhibited the transcriptional activity of FOXO1 and FOXO3a, FOXO1/FOXO3a-induced expression of endogenous Bim, and FOXO1-enhanced anoikis (Figure 3a, b, and c; Supplementary Figure 6a and b). These findings suggest that cyclin D1 inhibits FOXO1 and FOXO3a in a manner independent of CDK4 or CDK6. To define the molecular mechanism underlying cyclin D1 inhibition of FOXO1 and FOXO3a, we focused on the transcription repression domain (RD) of cyclin D1, which has been shown to be important in cyclin D1-mediated repression of certain transcription factors.27 Deletion of RD (cyclin D1-RD) almost completely abolished cyclin D1-mediated inhibition of FOXO1’s transcriptional activity (Figure 3d). Expression of RD alone also considerably inhibited the transcriptional activity of FOXO1 (Figure 3d). No significant difference in inhibition of FOXO1 activity was detected between the full-length cyclin D1 and the RD fragment (Figure 3d). Thus, cyclin D1’s transcription RD is required for its inhibition of FOXO1. Similar to the inhibition of FOXO1, cyclin D1-mediated inhibition of FOXO3a is also RD-dependent (Supplementary Figure 6c).
Cyclin D1 interacts with FOXO1 and FOXO3a
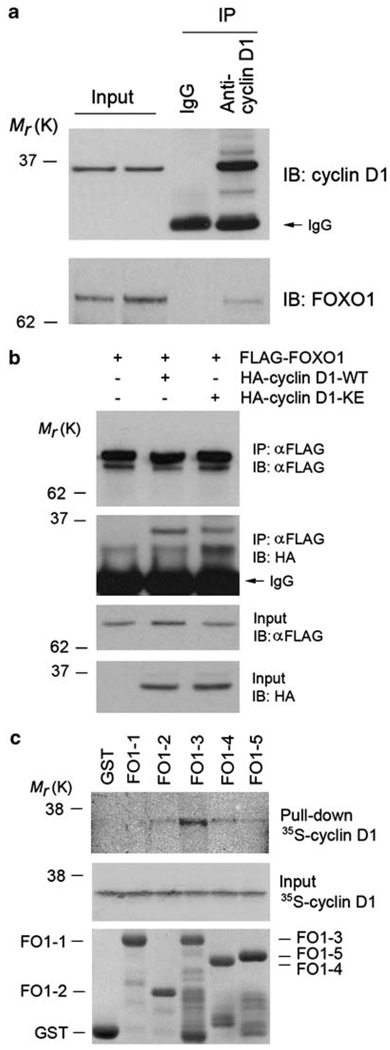
To further examine the molecular mechanism responsible for cyclin D1 inhibition of FOXO1, we sought to determine whether cyclin D1 forms a protein complex with FOXO1. Co-immunoprecipitation assays showed that endogenous cyclin D1 forms a protein complex with endogenous FOXO1 in anoikis-resistant LNCaP cells (Figure 4a). Ectopically expressed hemagglutinin (HA)-tagged cyclin D1 also formed a complex with FLAG-tagged FOXO1 in LNCaP cells (Figure 4b). To further examine the cyclin D1–FOXO1 interaction, both genes were transfected into LNCaP cells and cellular localization of the ectopically expressed FOXO1 and cyclin D1 proteins was assessed by immunofluorescent chemistry. In agreement with earlier reports,28,29 cyclin D1 and FOXO1 proteins were found in both the cytoplasm and nucleus of LNCaP cells. Importantly, these two ectopically expressed proteins were co-localized (Supplementary Figure 8). Consistent with our findings that cyclin D1-KE inhibits FOXO1’s transcriptional and pro-anoikis functions, the HA-cyclin D1-KE protein also interacted with FOXO1 in vivo (Figure 4b). In accordance with the finding that ectopic expression of the cyclin D1 RD diminished the transcriptional activity of FOXO1 (Figure 3d), co-immunoprecipitation assays showed that the RD of cyclin D1 interacts with FOXO1 (data not shown). To determine whether cyclin D1 interacts directly with FOXO1, we performed in vitro protein-binding assays using bacterially purified GST–FOXO1 recombinant proteins and 35S-labeled cyclin D1 produced from in vitro transcription and translation. The strongest interaction was detected between cyclin D1 and FO1-3 (Figure 4c), a region of FOXO1 encompassing amino acids 211–419 (Supplementary Figure 7a). As a negative control, no interaction between cyclin D1 and GST proteins was observed (Figure 4c). These data indicate that cyclin D1 interacts directly with FOXO1. To further explore the mechanism underlying the inhibition of FOXO transcriptional activity mediated by cyclin D1, we examined whether forced expression of cyclin D1 affects the binding of FOXO1 to the Bim promoter using chromatin immunoprecipitation (ChIP) assays. As shown by both semi-quantitative and real-time PCR, the binding of ectopically expressed FOXO1 to the promoter of Bim gene was largely reduced in cells transfected with cyclin D1 (Supplementary Figure 9). These data suggest that the cyclin D1–FOXO1 interaction may impair the binding of FOXO1 to the promoter of its target genes. In line with the finding that cyclin D1 inhibits the transcriptional activity of FOXO3a, co-immunoprecipitation assays showed that cyclin D1 forms a protein complex with FOXO3a in vivo (Supplementary Figure 6d), although their interaction does not seem to be as strong as the cyclin D1–FOXO1 interaction. Nonetheless, our data suggest that cyclin D1 interacts with both FOXO1 and FOXO3a and impairs their role in anoikis. We chose to examine the functional interaction between cyclin D1 and FOXO1 in further studies.
Figure 4.
Cyclin D1 interacts with FOXO1 in vitro and in vivo. (a) Interaction of endogenous cyclin D1 and FOXO1 proteins in LNCaP cells as shown by co-immunoprecipitation using an antibody against cyclin D1. (b) LNCaP cells were transfected with plasmids as indicated. At 48 h after transfection, cells were subjected to immunoprecipitation and western blot analysis. (c) Binding of 35S-labeled cyclin D1 proteins, produced by in vitro transcription and translation, to GST–FOXO1 recombinant proteins. Top, autography of in vitro-produced cyclin D1 proteins bound by GST–FOXO1 recombinant proteins. Middle, input of 35S-labeled cyclin D1 proteins. Bottom, GST and GST–FOXO1 recombinant proteins used for in vitro protein-binding assays indicated by Coomassie blue staining
Cancer-derived mutants of cyclin D1 possess a greater inhibitory effect on FOXO1-mediated anoikis than does wild-type cyclin D1
Mutation, amplification, or cancer risk-associated polymorphisms of cyclin D1 have been documented in numerous human cancers.27,30 Consistent with the earlier reports that cancer-derived cyclin D1 mutants act as potent oncogenes in vitro and in vivo,31,32 ectopic expression of a cancer-derived mutant of cyclin D1, cyclin D1-T286R, in which the threonine 286 (T286) residue is mutated to arginine, had much greater activity in inhibiting anoikis, Bim expression, and PARP cleavage than that of wild-type cyclin D1 (Supplementary Figure 10). Next, we set out to determine whether the mutated cyclin D1 possesses a higher inhibitory effect on FOXO1-induced anoikis relative to wild-type cyclin D1. To this end, anoikis-sensitive DU145 cells were co-transfected with an active form of FOXO1 (FOXO1-AAA) and wild-type (cyclin D1-WT) or mutated cyclin D1 (cyclin D1-T286R), and the inhibitory effects of cyclin D1-WT and cyclin D1-T286R on FOXO1-AAA-enhanced anoikis were compared. As shown in Figure 5a and b, cyclin D1-T286R had a greater inhibitory effect on both FOXO1-induced Bim expression and anoikis in comparison to wild-type cyclin D1. A similar inhibitory effect on FOXO1-promoted anoikis was observed in cells transfected with cyclin D1-T286A, a T286R-mimicking mutant of cyclin D1 (data not shown). Consistent with the finding that cyclin D1 inhibition of FOXO’s transcriptional activity requires RD, inhibition of FOXO1-enhanced anoikis by cyclin D1-T286R and cyclin D1-T286A is also RD-dependent (Figure 5b; data not shown). Moreover, we found that the cyclin D1-T286R and cyclin D1-T286A proteins remained at higher levels than wild-type cyclin D1 protein under anoikic conditions (Figure 5a, lane 6 versus lane 8; data not shown). By measuring the half-life of cyclin D1 proteins, we showed that under anoikic conditions, the half-life of the cyclin D1-T286R mutant (>4 h) was much longer than that of the wild-type cyclin D1 protein (<1 h) (Figure 5c and d). The half-life of the cyclin D1-T286R mutant was not affected when RD was deleted (Figure 5c and d). Importantly, increased levels of cyclin D1 proteins had no effect on the stability of the FOXO1 protein (Supplementary Figure 11), ruling out the possibility that the inhibition of FOXO1 by cyclin D1 is mediated by FOXO1 protein stability. Taken together, our findings show that under anchorage-independent conditions the cancer-derived mutants of cyclin D1 are not only much more stable than the wild-type counterpart, but they also confer a greater inhibitory effect on FOXO1-mediated anoikis.
Figure 5.
Inhibitory effect on FOXO1-mediated anoikis and protein stability of wild type and a cancer-derived mutant of cyclin D1. (a, b) DU145 cells were transfected with plasmids as indicated. At 24 h after transfection, cells were cultured in suspension and at the indicated time points cells were subjected to immunoblotting (a) and apoptosis (b) analysis. The number underneath each band in the immunoblot indicates the relative intensity of the corresponding band. (c) DU145 cells were transfected with HA-tagged wild-type cyclin D1, HA-tagged cyclin D1-T286R, or cyclin D1-ΔRD-T286R mutants. At 48 h after transfection, DU145 cells were cultured in suspension and treated with 20 µg/ ml cycloheximide. Cells were collected at the indicated time points and cell lysates were analyzed by western blots. (d) Quantification of the cyclin D1 protein signal intensity was obtained from exposures in which the signal was not saturated during the entire time course. Signal intensities were normalized to the signal intensity obtained at time zero
Cyclin D1 blocks FOXO1-mediated inhibition of anchorage-independent growth of cancer cells
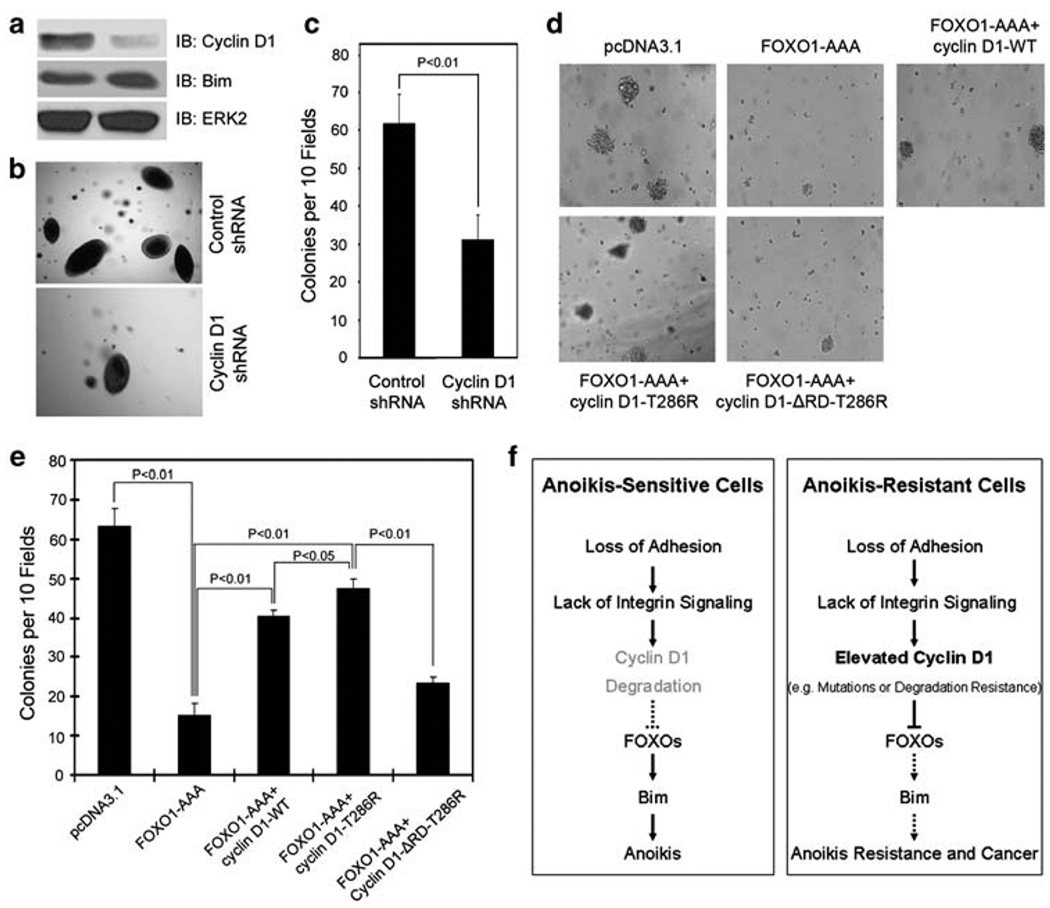
To assess the biological significance of cyclin D1 inhibition of FOXO-mediated anoikis, we specifically sought to determine whether downregulation of cyclin D1 decreases anchorage-independent growth capability of anoikis-resistant cells. Knocking down of cyclin D1 in 22Rv1 cells, which is an anoikis-resistant cell line (Supplementary Figure 1e), markedly decreased anchorage-independent growth (Figure 6a–c). Moreover, cyclin D1 knocking down resulted in an increase in expression of the FOXO target gene Bim (Figure 6a), which is consistent with the finding that knocking down of cyclin D1 augmented the transcriptional activity of FOXO proteins (Supplementary Figure 5). Next, we sought to determine whether forced expression of FOXO1 decreases anchorage-independent growth capability of anoikis-resistant cells and whether this function of FOXO1 can be blocked by cyclin D1. We showed that forced expression of the active FOXO1 (FOXO1-AAA) in anoikis-resistant LNCaP cells diminished their anchorage-independent growth (Figure 6d and e). However, FOXO1-AAA-mediated decrease in anchorage-independent growth of LNCaP cells was markedly diminished by co-transfection of wild-type cyclin D1 (Figure 6d and e). In line with the increased half-life of cyclin D1-T286R protein under anchorage-independent conditions, ectopic expression of this mutant resulted in a greater antagonistic effect on FOXO1-mediated inhibition of anchorage-independent growth of LNCaP cells (Figure 6d and e). This inhibition was almost completely abrogated by deletion of RD (Figure 6d and e). Together, our data indicate that cyclin D1-mediated inhibition of FOXO1 promotes anchorage-independent growth of prostate cancer cells.
Figure 6.
Effect of the functional interaction between cyclin D1 and FOXO1 on anchorage-independent growth of cancer cells. (a–c) 22Rv1 cells were transduced with lentivirus containing expression vectors for cyclin D1 shRNA or nonspecific control shRNA. At 3 days after infection, the cells were plated for growth in medium containing 0.3% agar. The remaining cells were used for western blot analysis (a). After 3 weeks, microscopic colonies were visualized and counted in a blinded fashion. Results from one representative field in each group are shown in (b). Quantification of results of colony formation data is presented in (c). Error bars indicate S.D. among three individual experiments. (d, e) LNCaP cells were transfected with plasmids as indicated. Transfected cells were plated for growth in medium containing 400 µg/ml of G418 and 0.3% agar. After 3 weeks, microscopic colonies were visualized and counted. Results from one representative field in each group are shown in (d). Quantification of results of colony formation data is presented in (e). Error bars indicate S.D. among three individual experiments. (f) A model of the role of cyclin D1 deregulation in tumorigenesis through sustained inhibition of FOXO1-mediated anoikis
Discussion
Acquisition of resistance to anoikis is necessary for cancer cells to survive, proliferate, and metastasize in the absence of appropriate ECM support.33 Thus, identification of molecular events that mediate anoikis resistance in cancer cells not only enhances our understanding of the processes of tumor formation and progression, but also has significant implications in cancer therapy. In this study we provide evidence that FOXO1 and FOXO3a are two key transcription factors mediating detachment-induced anoikis. Therefore, in addition to their known role in apoptosis and cell-cycle arrest in cells attached to ECM, our study shows a previously unrecognized tumor suppressor function of FOXO proteins in promoting anoikis of cells in the absence of ECM support.
To define the molecular mechanism(s) that regulate the function of FOXO proteins during anoikis, we showed that FOXO1 and FOXO3a proteins inevitably decreased in the entire cell lines examined when they were cultured in adhesion. Intriguingly, these proteins remain at high levels in both anoikis-sensitive and -resistant cells detached from the ECM (except the late time points in BPH-1 cells), suggesting that other regulatory mechanism(s), but not the level of these proteins per se, are important for determining the sensitivity of prostate cancer cells to anoikis. In this study, we identified the oncoprotein cyclin D1 as a critical upstream inhibitor of FOXO-mediated anoikis.
Cyclin D1 has long been implicated in human cancers because of its well-known function in regulating cell-cycle progression. Cyclin D1 promotes transit through the cell cycle by binding to and activating CDK4 and CDK6, resulting in phosphorylation and inactivation of the tumor suppressor protein RB.18 However, increasing evidence suggests that a CDK4/6-independent function of cyclin D1 may also be important for tumorigenesis.19 In support of this hypothesis, we showed here that overexpression of cyclin D1 not only inhibits the transcriptional activity of FOXO1 and FOXO3a in prostate cancer cells, but also abolishes FOXO-mediated anoikis. Most importantly, this function of cyclin D1 is independent of CDK4 and CDK6, because unlike for RB protein, we observed no phosphorylation of FOXO1 by cyclin D1–CDK4/6 complexes. Moreover, co-transfection of cyclin D1 with CDK4 or CDK6 failed to enhance cyclin D1-mediated inhibition of the transcriptional activity of FOXO1. Furthermore, cyclin D1-K112E, a mutated form of the protein that is unable to bind to and activate CDK4 and CDK6,26 retains a wild-type-like inhibitory effect on the transcriptional and anoikis-promoting functions of FOXO proteins. Given the importance of anoikis resistance in tumorigenesis and cancer metastasis, our findings suggest that, in a manner independent of CDK4 and CDK6, cyclin D1 may have a pivotal function in tumorigenesis and cancer metastasis by binding to and inhibiting the anoikis-promoting function of FOXO proteins.
Chromosomal rearrangement, amplification, and mutation of the cyclin D1 gene have been reported in many types of human cancers, including mantle cell lymphomas, parathyroid adenomas, breast, prostate, colon, lung, bladder and liver adenocarcinomas, and squamous carcinomas of the esophagus, head, and neck.13–17 It has been shown that in comparison to wild-type cyclin D1, exogenous expression of cancer-derived mutants of cyclin D1 accelerates cell transformation in vitro and tumor formation in mice.32,34 However, the mechanism by which such cancer-derived mutants of cyclin D1 function as potent oncogenes is poorly understood. We showed in this study that under anchorage-independent conditions, wild-type cyclin D1 protein undergoes rapid degradation, a process known to be primarily reliant on phosphorylation of the T286 residue of cyclin D1.35 In contrast, the cancer-derived T286R mutant of cyclin D1 is much more stable under the same conditions. Accordingly, we further showed that the T286R mutant possesses a greater ability to inhibit FOXO-mediated anoikis. On the basis of these findings, we envisage a model whereby unaltered cyclin D1 protein undergoes rapid degradation in anoikis-sensitive cells when they are detached from the ECM. Under this scenario, cyclin D1 inhibition of FOXO-mediated anoikis is abrogated, which leads to anoikis (Figure 6f, left panel). However, in cells in which cyclin D1 is mutated, amplified, or translocated at the genomic level or dysregulated at the protein levels (e.g. resistant to degradation), the cyclin D1 protein remains at a high level, thereby resulting in sustained inhibition of FOXO’s tumor suppression function as well as anoikis (Figure 6f, right panel). Cyclin D1 proteins have been shown to be elevated in human prostate cancer tissues.36 However, no mutation at T286 was detected in four commonly used prostate cancer cell lines including DU145, LNCaP, 22Rv1, and PC-3 (Supplementary Figure 12). Thus, whether or not elevated level of cyclin D1 in prostate cancers is caused by increased protein stability because of dysregulation of T286 phosphorylation-dependent proteasomal degradation of cyclin D135 remains to be determined.
In summary, this study showed a functional interaction between cyclin D1 and FOXO transcription factors that regulates resistance of cancer cells to anoikis. Specifically, our work shows that FOXO1 and FOXO3a proteins are required for cell detachment-induced anoikis. However, such activity of these proteins is inhibited by the cyclin D1 protein. This inhibitory function of cyclin D1 is mediated by its transcription RD but independent of its association with CDK4 and CDK6. Given that the cyclin D1 gene is often deregulated in human cancers, our finding that cyclin D1 inhibits FOXO-mediated anoikis suggests that besides its role in cell-cycle progression, cyclin D1 may contribute to tumorigenesis and cancer metastasis by promoting anchorage-independent cell survival.
Materials and Methods
Plasmids, small interfering RNA (siRNA), and small hairpin RNA (shRNA)
Plasmids for FLAG-FOXO1-WT, FLAG-FOXO1-AAA, FOXO3a-WT, FOXO3a-AAA, and 3xIRS were described earlier.20 Plasmid for human cyclin D1 was kindly provided by M. Pagano and subcloned into pcDNA3.1 (Invitrogen, Carlsbad, CA, USA). HA-tagged cyclin D1 (HA-cyclin D1) expression vector was constructed in pcDNA3.1/D (Invitrogen) by adding an HA tag to the N-terminal end of cyclin D1 using a PCR-based method. The FLAG-tagged cyclin D1 RD construct (FLAG-cyclin D1-RD) and RD deletion mutant of cyclin D1 (cyclin D1-ΔRD) were described earlier.37 Plasmids encoding the amino acid substitution mutants of cyclin D1 (T286A, T286R, cyclin D1-ΔRD-T286R) were generated by PCR-based mutagenesis (Stratagene, La Jolla, CA, USA). All the mutants were verified by sequencing. Plasmid for human Bim was purchased from Addgene (Cambridge, MA, USA) and the V5-tagged expression vector of Bim was generated in the pcDNA3.1D/V5-His using PCR. Constructs for GST–FOXO1 recombinant proteins were described earlier.20 The Renilla luciferase reporter vector was purchased from Promega (Madison, WI, USA). Lentiviral vectors for cyclin D1 shRNA and control shRNA were kindly provided by Quintanilla-Martinez.38 The smart pool of siRNAs for cyclin D1, FOXO1, FOXO3a, and control siRNAs were purchased from Dharmacon (Lafayette, CO, USA).
Cell culture, transfection, lentiviral infection, luciferase assay, and ChIP assay
The prostate cancer cell lines DU145, LNCaP, and PC-3 were purchased from American Type Culture Collection (Manassas, VA, USA). 22Rv1 cells were kindly provided by CY Young. Immortalized but nonmalignant human prostate epithelial cell line BPH-1 was kindly provided by SW Hayward. These cell lines were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS) (Hyclone, Logan, UT, USA), 100 µg/ml streptomycin, and 100 units/ml penicillin. NIH3T3 cells were maintained in DMEM supplemented with 10% FBS, 100 µg/ml streptomycin, and 100 units/ml penicillin. Transfections by electroporation were performed as described.20 Approximately 75–90% transfection efficiencies were routinely achieved. NIH3T3 stable cell lines 3T3-pcDNA3.1 and 3T3-cyclin D1 were generated by transfection of plasmids pcDNA3.1 and wild-type cyclin D1, respectively, followed by selection with G418 (500 µg/ml). Lentiviral infection was performed as described.38 Luciferase assays were performed as described.20 ChIP analysis of the binding of FOXO1 to the Bim promoter was performed using the method and primers as described.25
Anoikis/apoptotic cell death assay
Six-well tissue culture plates were coated with 18 mg/ml poly-HEMA (Sigma-Aldrich, St Louise, MO, USA) in 95% ethanol and allowed to dry overnight under sterile conditions in a laminar flow hood. Cells were then plated at 200000 per well in 2ml growth medium for the indicated time points. Cells were collected and washed with 1 × PBS. Apoptosis was measured using an Annexin V-PE apoptosis detection kit according to the manufacturer’s instructions (BD Pharmingen, San Diego, CA, USA) and analyzed by a FACScan cytometer equipped with Cell Quest software (BD Biosciences). A quadrant analysis was performed and cells that stained positive for Annexin V-PE and/or 7-AAD were designated as dead and unstained cells were designated as alive.
Nuclear and cytoplasmic protein extraction, immunoprecipitation, immunoblotting, immunofluorescent chemistry, and confocal microscopy and antibodies
Nuclear and cytoplasmic protein extraction was performed as described.39 Protein immunoprecipitations were carried out using an immunoprecipitation kit (Roche Applied Sciences, Indianapolis, IN, USA) as described.20 Immunoblotting, immunofluorescent chemistry, and confocal microscopy were performed as described.20 The antibodies used were anti-FOXO1, anti-PARP (Cell Signaling Technology, Danvers, MA, USA); anti-FOXO3a (Millipore, Billerica, MA, USA); anti-FLAG (M2) (Sigma-Aldrich); anti-HA (HA.11, Covance Innovative), anti-Bim (Chemicon, Billerica, MA, USA); anti-cyclin D1 (Ab-1 and Ab-3) (Lab Visions, Thermo Scientific, Fremont, CA, USA); anti-V5 (Invitrogen); anti-14-3-3ζ, anti-CDK2, anti-CDK4, anti-CDK6, and anti-ERK2 monoclonal (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
GST recombinant protein purification, in vitro protein-binding assay and in vitro kinase assay
GST protein and GST–FOXO1 recombinant proteins were purified from the BL21 Star (DE3) Escherichia coli strain (Invitrogen). 35S-labeled cyclin D1 proteins were synthesized by using TnT Quick Coupled Transcription/Translation Systems (Promega) and subjected to in vitro protein-binding assays using GST–FOXO1 recombinant proteins as described.20 Kinase assays were carried out in the presence of [γ-32P] ATP by using an in vitro kinase buffer system obtained from Cell Signaling Technology as described.20
Soft agar colony formation assay
22Rv1 cells were infected with lentivirus containing cyclin D1 shRNA or nonspecific control shRNA. At 3 days after infection, 10000 cells were suspended in 2ml RPMI 1640 medium containing 10% FBS and 0.3% agar. LNCaP cells were transfected with the indicated plasmids and 10000 cells were mixed with 2ml RPMI 1640 medium containing 10% FBS, 400 µg/ml of G418, and 0.3% agar. Cells were then plated over a layer of solidified RPMI 1640 medium containing 10% FBS and 1% agar in six-well plates. Plates were incubated at 37°C and the medium supplied with (for LNCaP cells) or without (for 22Rv1 cells) 400 µg/ml of G418 was changed every 2 to 3 days for 3 weeks before colonies were photographed and counted.
Statistics
Experiments were carried out with three or four replicates. Statistical analyses were performed by Student’s t test. Values with P < 0.05 are considered significant.
Supplementary Material
Acknowledgements
We thank DJ Tindall, KL Guan, M Pagano, CY Young, and SW Hayward for plasmids and cell lines, Y Shimizu, V Bardwell, KL Schwertfeger, SM Dehm and members of Huang laboratory, especially LR Bohrer for critical reading of the manuscript. This work was supported in part by funds from the Department of Defense (W81XWH-07-1-0137 and PC080591) and the Brainstorm Award from the University of Minnesota Masonic Cancer Center (to HH), the National Institutes of Health (CA82295 to JBM and CA099996 to KEK), and the National Natural Science Foundation of China (30500109 to LG).
Abbreviations
- FOXO
O-class forkhead box transcription factor
- CDK
cyclin-dependent kinase
- ECM
extracellular matrix
- siRNA
small inference RNA
- FasL
Fas ligand
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- PARP
poly(ADP-ribose) polymerase
- RB
retinoblastoma protein
- RD
transcription repression domain
- HA
hemagglutinin
- PCR
polymerase chain reaction
Footnotes
All authors claim no conflict of interest with this study.
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
References
- 1.Schwartz MA. Integrins, oncogenes, and anchorage independence. J Biol Chem. 1997;139:575–578. doi: 10.1083/jcb.139.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frisch SM, Ruoslahti E. Integrins and anoikis. Curr Opin Cell Biol. 1997;9:701–706. doi: 10.1016/s0955-0674(97)80124-x. [DOI] [PubMed] [Google Scholar]
- 3.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 4.Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J, et al. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol. 2003;5:733–740. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- 5.Mailleux AA, Overholtzer M, Schmelzle T, Bouillet P, Strasser A, Brugge JS. BIM regulates apoptosis during mammary ductal morphogenesis, and its absence reveals alternative cell death mechanisms. Dev Cell. 2007;12:221–234. doi: 10.1016/j.devcel.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z, Zhao J, Du Y, Park HR, Sun SY, Bernal-Mizrachi L, et al. Down-regulation of 14-3-3 zeta suppresses anchorage-independent growth of lung cancer cells through anoikis activation. Proc Natl Acad Sci USA. 2008;105:162–167. doi: 10.1073/pnas.0710905105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woods NT, Yamaguchi H, Lee FY, Bhalla KN, Wang HG. Anoikis, initiated by Mcl-1 degradation and Bim induction, is deregulated during oncogenesis. Cancer Res. 2007;67:10744–10752. doi: 10.1158/0008-5472.CAN-07-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 9.Gilley J, Coffer PJ, Ham J. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol. 2003;162:613–622. doi: 10.1083/jcb.200303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 11.Modur V, Nagarajan R, Evers BM, Milbrandt J. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J Biol Chem. 2002;277:47928–47937. doi: 10.1074/jbc.M207509200. [DOI] [PubMed] [Google Scholar]
- 12.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 13.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 14.Weinstein IB. Cancer. Addiction to oncogenes–the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 15.Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 16.Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A, Schmidt EV. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369:669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- 17.Creighton CJ. Multiple oncogenic pathway signatures show coordinate expression patterns in human prostate tumors. PLoS ONE. 2008;3:e1816. doi: 10.1371/journal.pone.0001816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 19.Ewen ME, Lamb J. The activities of cyclin D1 that drive tumorigenesis. Trends Mol Med. 2004;10:158–162. doi: 10.1016/j.molmed.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 20.Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314:294–297. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- 21.Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HR, Lin HM, Biliran H, Raz A. Cell cycle arrest and inhibition of anoikis by galectin-3 in human breast epithelial cells. Cancer Res. 1999;59:4148–4154. [PubMed] [Google Scholar]
- 23.Weng Z, Xin M, Pablo L, Grueneberg D, Hagel M, Bain G, et al. Protection against anoikis and down-regulation of cadherin expression by a regulatable beta-catenin protein. J Biol Chem. 2002;277:18677–18686. doi: 10.1074/jbc.M105331200. [DOI] [PubMed] [Google Scholar]
- 24.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 25.Liu P, Kao TP, Huang H. CDK1 promotes cell proliferation and survival via phosphorylation and inhibition of FOXO1 transcription factor. Oncogene. 2008;27:4733–4744. doi: 10.1038/onc.2008.104. [DOI] [PubMed] [Google Scholar]
- 26.Lamb J, Ramaswamy S, Ford HL, Contreras B, Martinez RV, Kittrell FS, et al. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–334. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 27.Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25:1620–1628. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- 28.Radu A, Neubauer V, Akagi T, Hanafusa H, Georgescu MM. PTEN induces cell cycle arrest by decreasing the level and nuclear localization of cyclin D1. Mol Cell Biol. 2003;23:6139–6149. doi: 10.1128/MCB.23.17.6139-6149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P, Li S, Gan L, Kao TP, Huang H. A transcription-independent function of FOXO1 in inhibition of androgen-independent activation of the androgen receptor in prostate cancer cells. Cancer Res. 2008;68:10290–10299. doi: 10.1158/0008-5472.CAN-08-2038. [DOI] [PubMed] [Google Scholar]
- 30.Benzeno S, Lu F, Guo M, Barbash O, Zhang F, Herman JG, et al. Identification of mutations that disrupt phosphorylation-dependent nuclear export of cyclin D1. Oncogene. 2006;25:6291–6303. doi: 10.1038/sj.onc.1209644. [DOI] [PubMed] [Google Scholar]
- 31.Gladden AB, Woolery R, Aggarwal P, Wasik MA, Diehl JA. Expression of constitutively nuclear cyclin D1 in murine lymphocytes induces B-cell lymphoma. Oncogene. 2006;25:998–1007. doi: 10.1038/sj.onc.1209147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin DI, Lessie MD, Gladden AB, Bassing CH, Wagner KU, Diehl JA. Disruption of cyclin D1 nuclear export and proteolysis accelerates mammary carcinogenesis. Oncogene. 2008;27:1231–1242. doi: 10.1038/sj.onc.1210738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liotta LA, Kohn E. Anoikis: cancer and the homeless cell. Nature. 2004;430:973–974. doi: 10.1038/430973a. [DOI] [PubMed] [Google Scholar]
- 34.Aggarwal P, Lessie MD, Lin DI, Pontano L, Gladden AB, Nuskey B, et al. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev. 2007;21:2908–2922. doi: 10.1101/gad.1586007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 36.Drobnjak M, Osman I, Scher HI, Fazzari M, Cordon-Cardo C. Overexpression of cyclin D1 is associated with metastatic prostate cancer to bone. Clin Cancer Res. 2000;6:1891–1895. [PubMed] [Google Scholar]
- 37.Petre-Draviam CE, Williams EB, Burd CJ, Gladden A, Moghadam H, Meller J, et al. A central domain of cyclin D1 mediates nuclear receptor corepressor activity. Oncogene. 2005;24:431–444. doi: 10.1038/sj.onc.1208200. [DOI] [PubMed] [Google Scholar]
- 38.Klier M, Anastasov N, Hermann A, Meindl T, Angermeier D, Raffeld M, et al. Specific lentiviral shRNA-mediated knockdown of cyclin D1 in mantle cell lymphoma has minimal effects on cell survival and reveals a regulatory circuit with cyclin D2. Leukemia. 2008;22:2097–2105. doi: 10.1038/leu.2008.213. [DOI] [PubMed] [Google Scholar]
- 39.Sauter D, Himmelsbach K, Kriegs M, Carvajal Yepes M, Hildt E. Localization determines function: N-terminally truncated NS5A fragments accumulate in the nucleus and impair HCV replication. J Hepatol. 2009;50:861–871. doi: 10.1016/j.jhep.2008.11.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.