Abstract
Humans differ from other animals in many aspects of anatomy, physiology, and behavior; however the genotypic basis of most human-specific traits remains unknown1. Recent whole genome comparisons have made it possible to identify genes with elevated rates of amino acid change or divergent expression in humans, and non-coding sequences with accelerated base pair changes2-5. Regulatory alterations may be particularly likely to produce phenotypic effects while preserving viability, and are known to underlie interesting evolutionary differences in other species6-8. Here we identify molecular events particularly likely to produce significant regulatory changes in humans: complete deletion of sequences otherwise highly conserved between chimpanzees and other mammals. We confirm 510 such deletions in humans, which fall almost exclusively in non-coding regions and are enriched near genes involved in steroid hormone signaling and neural function. One deletion removes a sensory vibrissae and penile spine enhancer from the human ANDROGEN RECEPTOR (AR) gene, a molecular change correlated with anatomical loss of androgen-dependent sensory vibrissae and penile spines in the human lineage9,10. Another deletion removes a forebrain subventricular zone enhancer near the tumor suppressor gene GROWTH ARREST AND DNA-DAMAGE-INDUCIBLE, GAMMA (GADD45g)11,12, a loss correlated with expansion of specific brain regions in humans. Deletions of tissue-specific enhancers may thus accompany both loss and gain traits in the human lineage, and provide specific examples of the kinds of regulatory alterations6-8 and inactivation events13 long proposed to play an important role in human evolutionary divergence.
To discover human-specific deletions (hDELs) on a genome-wide scale (Fig. 1a), we identified regions of the chimpanzee genome14 with a clear macaque ortholog, but no closely related sequence in humans (Supplementary Information). This identified 37,251 ancestral primate sequences lost in humans, spanning 34.0 Mb (1.17%) of the chimpanzee genome. To find deletions most likely to produce functional consequences, we also identified highly conserved sequences spanning 70.0 Mb (2.41%) of the chimpanzee genome, producing a conservative estimate of pan-mammalian sequence under purifying selection in chimpanzee14. The intersection of these deletion and conservation surveys identified 583 regions with high sequence conservation that are surprisingly deleted in humans, which we term hCONDELs (Supplementary Table 2).
Fig. 1. Hundreds of sequences highly conserved between chimpanzee and other species are deleted in humans.
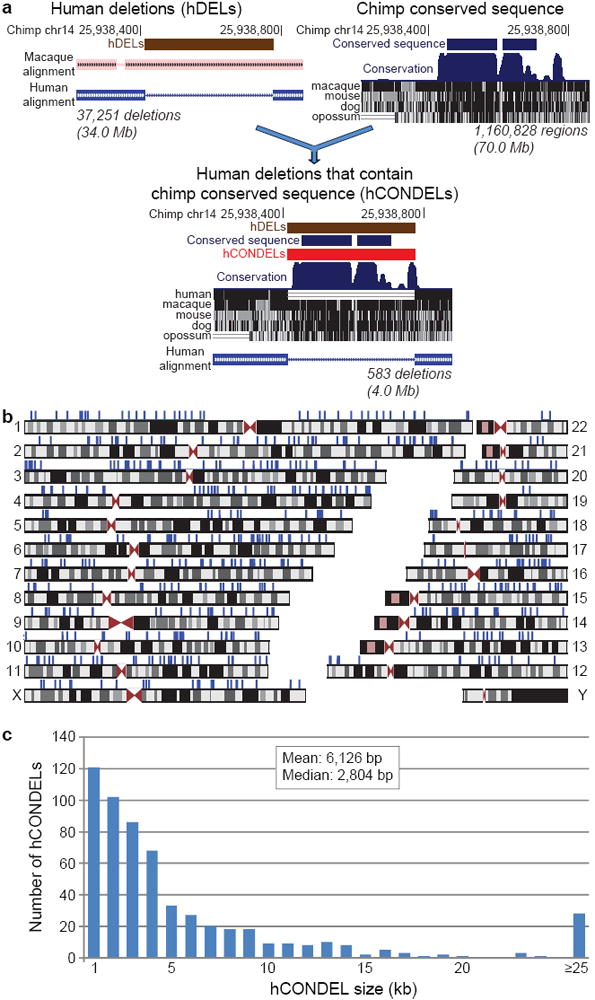
a, Computational approach used to discover human-specific deletions of functional DNA: identification of ancestral chimpanzee genomic sequences deleted in human; discovery of chimpanzee genomic sequences highly conserved in other species; and detection of human-specific deletions that remove one or more chimpanzee conserved sequences. Total chimpanzee sequence identified in each step is displayed beneath each graphic. b, Human genomic locations of the 583 hCONDELs. hCONDELs are displayed as blue ticks above the many locations where they are missing. c, Size distribution of hCONDELs.
Of the 583 predicted hCONDELs, 510 (87.5%) were independently validated by single human sequence reads spanning both sides of the deletions (Supplementary Information). 85% of sequence-validated hCONDELs appeared fixed in human trace archives. Experimental amplifications across human diversity panels confirmed human deletion of 39/39 randomly chosen hCONDELs, and showed fixation of 31/32 hCONDELs also fixed in the trace archives (Supplementary Tables 3-5). 88% of sequence-validated hCONDELs are missing from the draft Neandertal genome15 (Supplementary Information), in agreement with the length of the common lineage between humans and Neandertals following the divergence with chimpanzee.
The 583 hCONDELs cover 3.96 Mb (0.14%) of the chimpanzee genome, remove an average of 95 base pairs (bp) of conserved sequence, and are found on every nuclear chromosome except Y, where macaque sequence is unavailable (Fig. 1b). hCONDELs have a median size of 2,804 bp, and show a skew towards G/C poor regions compared to chimpanzee genome-wide averages (Fig. 1c, Supplementary Fig. 2). We find no evidence for enrichment in areas with high recombination (P>0.9) or pericentromeric and subtelomeric regions (P>0.5) (Supplementary Information). Thus, loss of sequences is unlikely to be a secondary consequence of higher mutation rates in such regions, a possibility that has been debated for the accelerated sequence changes seen in other human non-coding elements4,5,16.
Only 1 of 510 sequence-validated hCONDELs removes a protein-coding region, corresponding to a previously known 92 bp deletion in CMAH (ref. 17). All other sequence-validated hCONDELs map to non-protein coding regions of the genome (355 intergenic and 154 intronic). Similar highly conserved non-coding sequences frequently correspond to regulatory elements controlling expression of nearby genes18. While hCONDELs cannot be directly tested for evidence of positive human selection, we can examine whether hCONDELs are preferentially located near genes with particular functions, using the Genomic Regions Enrichment of Annotations Tool (GREAT)19. We performed enrichment analysis using simulations to explicitly account for the overrepresentation of highly conserved elements near particular classes of genes (Supplementary Information). This analysis suggests that hCONDELs are significantly enriched near genes involved in steroid hormone receptor signaling and neural function (Table 1), and near genes encoding Fibronectin-type-III or CD80-like immunoglobulin C2-set domains (Supplementary Table 10).
Table 1.
Summary of annotation enrichments of hCONDELs.
| Ontology: Term | Expected # of hCONDELs | Observed # of hCONDELs | Fold enrichment | Binomial p-value19 |
|---|---|---|---|---|
| GO Molecular Function: Steroid hormone receptor activity | 4.7 | 14 | 2.96 | 3.7×10-4 |
| Entrez Gene: Neural genes | 141.3 | 180 | 1.27 | 1.1×10-4 |
| MGI Expression in Theiler Stage 21: Hindbrain | 49.9 | 79 | 1.58 | 3.4×10-5 |
| Cerebral cortex | 42.1 | 68 | 1.62 | 7.0×10-5 |
| Brain; ventricular layer | 29.9 | 52 | 1.74 | 9.4×10-5 |
| Midbrain | 30.5 | 52 | 1.70 | 1.6×10-4 |
| InterPro Protein Domains: Fibronectin, type III | 16.7 | 34 | 2.03 | 1.0×10-4 |
| CD80-like, immunoglobulin C2-set | 2.1 | 8 | 3.84 | 1.4×10-3 |
Showing only non-redundant terms that are enriched after accounting for both multiple testing and for the tendency of conserved elements to be found near particular classes of genes (Supplementary Information).
To compare sequences lost in other lineages, we used a similar computational approach to identify conserved sequences lost specifically in chimpanzee or mouse (termed cCONDELs and mCONDELs, respectively). We identified 344 cCONDELs and 350 mCONDELs validated by sequence reads spanning predicted deletions (Supplementary Information). cCONDELs and mCONDELs show enrichment for synapse and glutamate receptors (Supplementary Table 11) and metal ion binding (Supplementary Table 12), respectively, but not for the categories found near hCONDELs.
To further explore the possible functions of hCONDELs near steroid hormone signaling genes, we examined a 60.7 kb human deletion flanking the ANDROGEN RECEPTOR (AR) locus (Fig. 2a) in detail. AR is required for response of tissues to circulating androgens, and thus represents a strong candidate locus for characteristic changes of secondary sexual traits known in the human lineage20,21. Within this human-specific deletion lies a ~5 kb region containing highly conserved non-coding sequences. We cloned the corresponding 4,839 bp chimpanzee and 7,654 bp mouse regions and tested their capacity to drive expression of an hsp68 basal promoter-lacZ reporter gene during normal mouse development. Chimpanzee and mouse constructs both drove consistent lacZ expression in the facial vibrissae and genital tubercle of five or more independent transgenic embryos (Fig. 2b-g), and the mouse sequence also drove expression in hair follicles.
Fig. 2. Transgenic analysis of a chimpanzee and mouse AR enhancer region missing in humans.
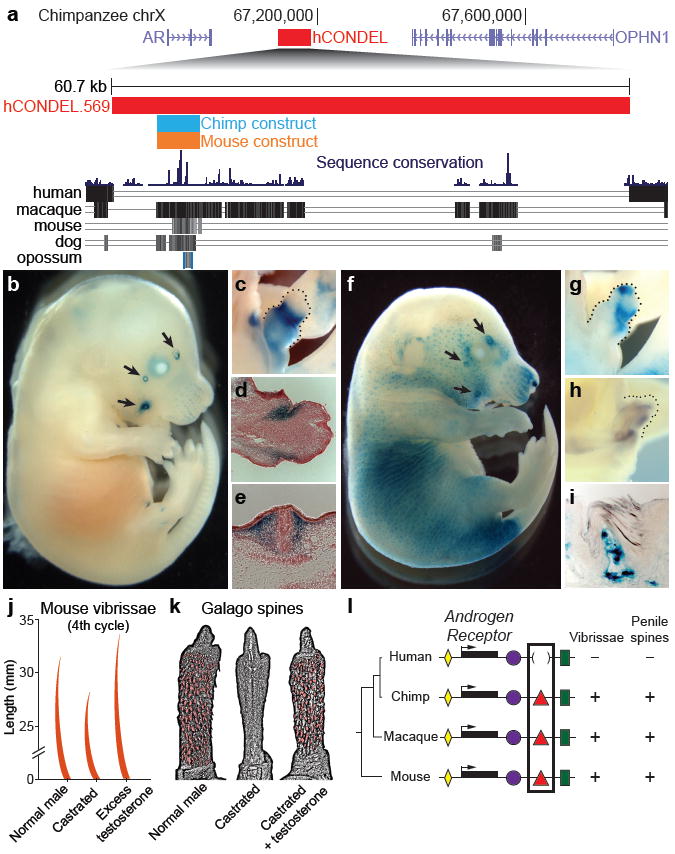
a, Upper panel: 1.1 Mb region of the chimpanzee X chromosome. The red bar shows the position of a 60.7 kb human deletion removing a well-conserved chimpanzee enhancer between the AR and OPHN1 genes. Lower panel: Multiple species comparison of the deleted region, showing sequences alignable between chimpanzee and other mammals. Blue and orange bars represent chimpanzee and mouse sequences tested for enhancer activity in transgenic mice. The chimpanzee sequence drives lacZ expression in, b, facial vibrissae (arrows); and c, genital tubercle (dotted line) of E16.5 mouse embryos. Histological sections reveal strongest staining in superficial mesenchyme of d, the prospective glans of the genital tubercle; and e, dermis surrounding the base of sensory vibrissae. The mouse enhancer also drives consistent expression in f, facial vibrissae; g, genital tubercle; and hair follicles of E16.5 embryos. h, Endogenous AR is expressed in the genital tubercle (dotted line) as demonstrated by in situ hybridization. i, Histological section of a 60-day-old transgenic mouse penis showing postnatal lacZ expression in dermis of penile spines. Vibrissae and penile spines are androgen-dependent, as shown by, j, changes in vibrissae length in castrated and testosterone-treated mice and, k, loss and recovery of penile spines of a castrated and testosterone-treated primate (Galago crassicaudatus) (modified from refs. 23 and 24). l, Model depicting multiple conserved tissue-specific enhancers (colored shapes) surrounding AR coding sequences (black bars) of different species. Loss of an ancestral vibrissae/penile spine enhancer in humans is correlated with corresponding loss of sensory vibrissae and penile spines.
lacZ expression was specifically located in the mesoderm surrounding vibrissae follicles, and in the superficial mesoderm within the presumptive glans of the developing genital tubercle (Fig. 2). Four stable mouse lines all showed expression in the superficial tissue underlying epidermal spines of the penis (Fig. 2i). Previous studies have shown that AR is expressed in mesenchyme surrounding developing epithelial structures22 and is required for normal development of vibrissae and penile spines (see below). These results suggest that the human deletion removes a conserved enhancer sequence that directs expression in a spatially-restricted subset of the AR expression pattern. The chimpanzee sequence also drives significant reporter gene expression when tested in human foreskin fibroblasts, suggesting that upstream pathways regulating the enhancer are still intact in humans (chimpanzee activity 2.2-5.9-fold greater than human deletion/basal control vector, P<10-7).
Interestingly, humans show obvious morphological differences at the anatomical locations controlled by the enhancer. Sensory vibrissae develop in many mammals including chimpanzees, macaques, and mice9. In contrast, humans lack sensory vibrissae9. Vibrissae development is clearly androgen-responsive, as castration shortens vibrissae in mice, and excess testosterone increases growth23 (Fig. 2j).
Profound changes have also evolved in the genitalia of humans compared to other animals. Many mammals have keratinized epidermal spines overlying tactile receptors in the glans dermis10,20. Penile spine growth is androgen-dependent, as primates lose spines upon castration, and treatment with exogenous testosterone restores spine formation24. Mice with AR protein-coding mutations fail to form penile spines25, confirming an essential role for AR in penile spine development. Our results show that humans have lost an ancestral penile spine enhancer from the AR locus. Humans also fail to form the penile spines commonly found in other animals, including chimpanzees, macaques, and mice10,20,25. Simplified penile morphology tends to be associated with monogamous reproductive strategies in primates20. Ablation of spines decreases tactile sensitivity and increases the duration of intromission20, indicating their loss in the human lineage may be associated with the longer duration of copulation in our species relative to chimpanzees20. This fits with an adaptive suite, including feminization of the male canine dentition, moderate-sized testes with low sperm motility, and concealed ovulation with permanently enlarged mammary glands20, that suggests our ancestors evolved numerous morphological characteristics associated with pair-bonding and increased paternal care21.
Sensory vibrissae and penile spines are examples of morphological structures lost in humans. However, deletion of enhancers could also be associated with tissue expansion, by removing regulatory sequences from genes that control developmental patterns of cell proliferation, death, or migration. One of the most dramatic tissue expansions in human evolution is increased size of the cerebral cortex1. Using a curated resource of gene expression domains during mouse development, we find that hCONDELs are significantly enriched near genes expressed during cortical neurogenesis (P=7×10-5, Table 1). Within this set, hCONDELs are preferentially near genes acting as suppressors of cell proliferation or migration (P=0.003) (Supplementary Information).
To test the function of one such hCONDEL, we further analyzed a deletion removing a 3,181bp region located next to the tumor suppressor gene GROWTH ARREST AND DNA-DAMAGE-INDUCIBLE, GAMMA (GADD45g) (Fig. 3a). This hCONDEL removes a forebrain-specific p300 binding site18, which predicts enhancer sequences with strong specificity. The chimpanzee version of this sequence, and a smaller 546 bp mouse sequence overlapping the p300 enhancer-binding region, both drove lacZ expression in the developing ventral telencephalon and diencephalon in at least five independent E14.5 transgenic embryos (Fig. 3b-i), confirming that the ancestral sequence corresponds to a conserved forebrain-specific enhancer. The chimpanzee sequence also drives significant gene expression when tested in immortalized human fetal neural progenitor cells, suggesting the enhancer would also affect transcription if still present in humans (1.7-2-fold greater expression than human deletion/basal control vector, P<10-7).
Fig. 3. Transgenic analysis of a chimpanzee and mouse forebrain enhancer missing from a tumor suppressor gene in humans.
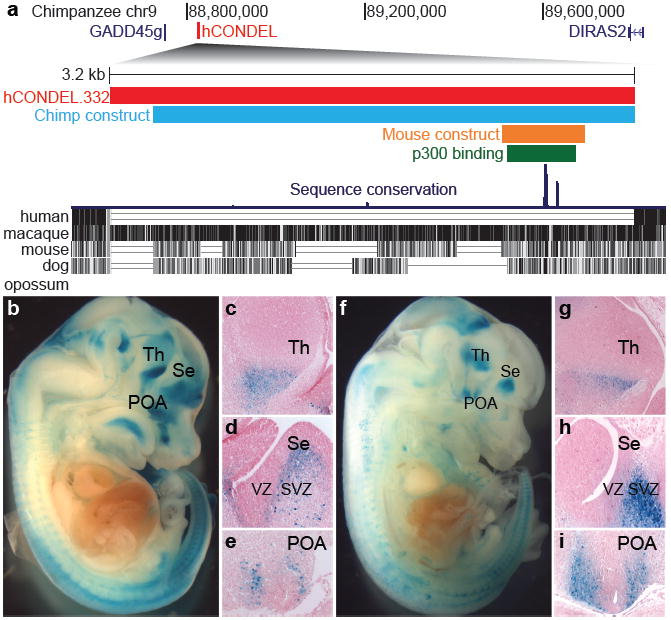
a, Upper panel: 1.3 Mb region of the chimpanzee chromosome 9. The red bar illustrates a 3,181 bp human-specific deletion removing a conserved chimpanzee enhancer located downstream of GADD45g. Lower panel: Multiple species comparison of the deleted region, showing sequences alignable between chimpanzee and other mammals. The green bar represents a mouse forebrain-specific p300 binding site18, and the blue and orange bars represent chimpanzee and mouse sequences tested for enhancer activity in transgenic mice. The chimpanzee (b,c,d,e) and mouse sequence (f,g,h,i) both drive consistent lacZ expression in E14.5 mouse embryos in the ventral thalamus (c, g), the SVZ of the septum (d, h), and the preoptic area (e, i). Increased production of neuronal subtypes from these regions may contribute to thalamic and cortical expansion in humans27-30. All sections are sagittal with anterior to right. Th, thalamus; Se, septum; POA, preoptic area.
Histological sections of transgenic embryos show that the chimpanzee and mouse sequences drive lacZ expression in the subventricular zone (SVZ) of the septum, the preoptic area, and in regions of the ventral thalamus and hypothalamus (Fig. 3c-e,g-i). The lacZ expression matches a sub-domain of the expression pattern of GADD45g, which is normally expressed throughout the telencephalon SVZ and diencephalon26. The preoptic area generates inhibitory interneurons that migrate to the neocortex and ventral telencephalon27. The ventral thalamus domain corresponds to a region that generates inhibitory interneurons, which have notably increased in proportion in human thalamus28,29.
Expression in brain subventricular zones is particularly interesting in light of previous suggestions that increased proliferation of SVZ intermediate progenitors underlies the evolutionary expansion of the neocortex in primates30. GADD45g normally represses cell cycle and can activate apoptosis, and somatic loss of GADD45g expression is clearly linked with excess tissue growth in human pituitary adenomas11,12. Our results show that species-specific loss of an ancestral SVZ enhancer has clearly occurred in the human lineage, providing a plausible molecular basis for increasing production of particular neuronal cell types by regulatory changes in a tumor suppressor gene.
We cannot exclude the possibility that loss of AR and GADD45g enhancers has occurred because of relaxed selection following other genetic changes that have led to anatomical differences in the human lineage. However, based on the previously established role of AR in vibrissae and penile spine development, and of GADD45g in negative regulation of tissue proliferation, we think it likely that deletions of tissue-specific enhancers in these genes have contributed to both loss and expansion of particular tissues during human evolution. The full set of hCONDELs may contain loci associated with other human-specific characteristics, a possibility that can now be tested by further functional studies of these conserved non-coding sequences that are surprisingly missing from the human genome.
Methods Summary
We examined 2,696 Mb (92.67%) of the chimpanzee genome aligning with single regions in humans, removing assembly and alignment artifacts, segmental duplications, and other regions with complex genome histories (Supplementary Information). Chimpanzee sequences aligning to macaque but not human were identified as human-specific deletions. We identified 70.0 Mb (2.41%) of the chimpanzee genome as highly conserved by calculating the fraction of identical base pairs within multiple sequence alignments using a series of sliding window criteria (Supplementary Table 1). hCONDELs were validated by searching for individual human sequence reads in the NCBI Trace Archives spanning predicted deletions; and by testing 39 hCONDELs by experimental amplification from individuals of 23 human populations (Coriell Cell Repositories, Supplementary Information). Gene enrichments were analyzed using GREAT (great.stanford.edu)19. For each ontology we ran 1,000 simulations over 510 size-matched random deletions overlapping one or more chimpanzee conserved elements to derive a multiple test P value threshold for enrichment. Functional enhancer assays were carried out by injecting chimp and mouse expression constructs into FVB embryos (Xenogen Biosciences and Cyagen Biosciences) and staining for lacZ expression activity at different developmental stages; or by transfecting chimp and mouse enhancer constructs into human foreskin fibroblasts (System Biosciences) or ReNcell CX Human Neural Progenitor cells (Chemicon), and comparing expression to human-deletion/control constructs using Galacto-Light Plus (Applied Biosciences) (Supplementary Information).
Supplementary Material
Acknowledgments
We thank D. DeGusta for providing chimpanzee DNA, S. McConnell and P. Buckmaster for useful discussions, and M. Hiller and C. Barr for ontology analysis support. This work was supported in part by a Bio-X graduate fellowship (C.Y.M.), a Ruth L. Kirschstein NRSA post-doctoral fellowship (1 F32 HD062137-01, P.L.R.), a National Defense Science and Engineering Graduate fellowship (A.A.P.), a National Science Scholarship of the Agency of Science, Technology, and Research, Singapore (X.L.), a Stanford Graduate Fellowship (A.M.W.), an Edward Mallinckrodt, Jr. Foundation grant (G.B.), and National Institute of Health grants R01 HD059862 (G.B.), R01 HG005058 (G.B.) and P50 HG002568 (D.M.K.). G.B. is a Packard Fellow, Searle Scholar, Microsoft Faculty Fellow and an Alfred P. Sloan Fellow. D.M.K. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions G.B. and D.M.K. conceived the investigation; C.Y.M. performed the computational analyses; P.L.R, A.A.P., A.I.B., T.D.C, C.G., V.B.I., X.L.,D.B.M., and B.T.S. performed the experiments; C.Y.M., P.L.R., A.A.P, B.T.S, A.M.W., G.B., and D.M.K. analyzed the data; and C.Y.M., P.L.R., A.A.P., G.B., and D.M.K. wrote the paper with contributions from all authors.
Supplementary Information, including the full list of 583 human conserved deletions, is available online at www.nature.com/nature.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Methods
Detailed methods can be found in the Supplementary Information.
References
- 1.Varki A, Altheide T. Comparing the human and chimpanzee genomes: searching for needles in a haystack. Genome Res. 2005;15:1746–1758. doi: 10.1101/gr.3737405. [DOI] [PubMed] [Google Scholar]
- 2.Bustamante CD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 3.Khaitovich P, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- 4.Pollard KS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 5.Prabhakar S, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 7.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 8.Chan YF, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muchlinski MN. A comparative analysis of vibrissa count and infraorbital foramen area in primates and other mammals. J Hum Evol. 2010;58:447–473. doi: 10.1016/j.jhevol.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Hill WCO. Note on the male external genitalia of the chimpanzee. Proc Zool Soc London. 1946;116:129–133. [Google Scholar]
- 11.Zerbini LF, et al. NF-kappa B-mediated repression of growth arrest- and DNA-damage-inducible proteins 45alpha and gamma is essential for cancer cell survival. Proc Natl Acad Sci U S A. 2004;101:13618–13623. doi: 10.1073/pnas.0402069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, et al. Loss of expression of GADD45 gamma, a growth inhibitory gene, in human pituitary adenomas: implications for tumorigenesis. J Clin Endocrinol Metab. 2002;87:1262–1267. doi: 10.1210/jcem.87.3.8315. [DOI] [PubMed] [Google Scholar]
- 13.Olson M. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 15.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 17.Chou HH, et al. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci U S A. 1998;95:11751–11756. doi: 10.1073/pnas.95.20.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dixson AF. Primate Sexuality. Oxford University Press; Oxford: 1998. [Google Scholar]
- 21.Lovejoy CO. Reexamining human origins in light of Ardipithecus ramidus. Science. 2009;326:74e1–8. [PubMed] [Google Scholar]
- 22.Crocoll A, Zhu CC, Cato AC, Blum M. Expression of androgen receptor mRNA during mouse embryogenesis. Mech Dev. 1998;72:175–178. doi: 10.1016/s0925-4773(98)00007-0. [DOI] [PubMed] [Google Scholar]
- 23.Ibrahim L, Wright EA. Effect of castration and testosterone propionate on mouse vibrissae. Br J Dermatol. 1983;108:321–326. doi: 10.1111/j.1365-2133.1983.tb03971.x. [DOI] [PubMed] [Google Scholar]
- 24.Dixson AF. Effects of testosterone on the sternal cutaneous glands and genitalia of the male greater galago (Galago crassicaudatus crassicaudatus) Folia Primatol. 1976;26:207–213. doi: 10.1159/000155751. [DOI] [PubMed] [Google Scholar]
- 25.Murakami R. A histological study of the development of the penis of wild-type and androgen-insensitive mice. J Anat. 1987;153:223–231. [PMC free article] [PubMed] [Google Scholar]
- 26.Gohlke JM, et al. Characterization of the proneural gene regulatory network during mouse telencephalon development. BMC Biol. 2008;6:15. doi: 10.1186/1741-7007-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gelman DM, et al. The embryonic preoptic area is a novel source of cortical GABAergic interneurons. J Neurosci. 2009;29:9380–9389. doi: 10.1523/JNEUROSCI.0604-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vue TY, et al. Characterization of progenitor domains in the developing mouse thalamus. J Comp Neurol. 2007;505:73–91. doi: 10.1002/cne.21467. [DOI] [PubMed] [Google Scholar]
- 29.Arcelli P, Frassoni C, Regondi MC, De Biasi S, Spreafico R. GABAergic neurons in mammalian thalamus: a marker of thalamic complexity? Brain Res Bull. 1997;42:27–37. doi: 10.1016/s0361-9230(96)00107-4. [DOI] [PubMed] [Google Scholar]
- 30.Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


