Recently, there have been studies that indicate that Xenotropic Murine Leukemia Virus (MLV)-related Virus (XMRV), a newly described human gammaretrovirus, and other related viruses, may be associated with both prostate cancer and myalgic encephalomyelitis (ME) / chronic fatigue syndrome (CFS)1–4. It has also been suggested that these viruses have the potential to be transmitted by blood transfusion5. However, a number of studies have failed to support these associations, or indeed detect significant evidence of XMRV in the human population6–9. Currently, there is insufficient information to determine whether or not XMRV and related viruses are a threat to blood safety. Accordingly, the Department of Health and Human Services (HHS) has established a Scientific Research Working Group (SRWG) to explore the following questions: What is the prevalence of XMRV in the donor population? Is XMRV transmissible by blood transfusion? And if XMRV is transmissible by transfusion, are there any pathologic consequences for the infected recipient? As a starting point, the SRWG has focused on standardizing the various tests used to detect XMRV in blood samples and has facilitated the sharing of clinical samples between laboratories. This commentary discusses background information relating to blood safety and XMRV and related viruses and outlines the specific actions that the SRWG has taken and plans to take.
Identification and potential disease correlates of XMRV
XMRV was initially identified in 2006 as a result of a search for an infectious cause or co-factor in prostate cancer. Using a microarray of highly conserved sequences from known viruses, Urisman and colleagues detected the presence of a gammaretrovirus within some prostate cancer patients2. Upon full sequencing, this virus appeared to be closely related to xenotropic murine leukemia viruses (X-MLV) and was thus termed XMRV.
Subsequent studies using PCR-based methods and immunohistochemical staining for XMRV, revealed viral nucleic acid or proteins in 6–23% of prostate cancers, primarily in malignant epithelial cells from higher-grade tumors1,10. Controversially, a number of subsequent studies have either failed to detect XMRV entirely in prostate cancer11,12, or have only detected a few cases and at the same prevalence in cancerous and non-cancerous prostates13–15. Thus, the precise association of XMRV with prostate cancer, as well as any direct role in tumorigenesis, remains to be defined.
ME/CFS are poorly defined diseases, currently with no well-accepted cause. Thus, Mikovits and colleagues at the Whittemore Peterson Institute (WPI) in Reno, Nevada launched a study of the association between XMRV and CFS3. 67% of patients with CFS were found to be XMRV positive by various polymerase chain reaction (PCR) techniques3. Out of 57 patients with CFS, 21% were positive for XMRV proviral DNA in unstimulated peripheral blood mononuclear cells (PBMC), while 54% and 72% were positive for XMRV RNA in unstimulated and stimulated PBMC respectively16. Eighty-two percent of patients also had serological evidence of antibodies cross-reactive to the MLV envelope. Furthermore, nearly 4% of PBMC samples from normal healthy subjects were also found to harbor XMRV DNA3. Critically, the finding that XMRV nucleic acid was present in blood was supported by the fact that in 89% of cases the virus could be directly cultured from stimulated PBMC and/or patient plasma by co-culture with an XMRV-sensitive cell line3.
Controversies in XMRV detection in ME/CFS patients and normal donors
As noted above, a number of laboratories have failed to detect XMRV in prostate cancer patients. Subsequently, using molecular and serological methods a series of studies have also failed to detect XMRV in patients with ME/CFS or other populations from the US, Europe, China and now Africa6–9,17–23. What is particularly interesting is the fact that the majority of the studies do not simply fail to detect an association with disease, but rather find little or no evidence of any XMRV infection whatsoever, including in blood donors9,17 and potentially at-risk populations such as patients with HIV/AIDS6,19,21,23.
However, a recent study from FDA and NIH laboratories reported that over 80% of patients with ME/CFS and nearly 7% of NIH blood donors tested positive using nested PCR of proviral DNA in PBMC or whole blood samples4. The major difference between these findings and those of Lombardi et al were that the majority of viral DNA amplicons detected in samples in the FDA/NIH study, when partially sequenced, appeared to be more closely related to polytropic MLV (P-MLV) rather than XMRV, and thus were simply termed MLV-related viruses, or MRVs. Until full-length virus sequences are obtained and virus isolated, it is not clear whether these represent a group of potential human infectious viruses, whether they be zoonotic infections by polytropic MLVs, or perhaps chimeras between XMRV and P-MRV. Thus far, XMRV is the only ‘human’ gammaretrovirus to be fully sequenced and shown to be infectious in cell lines and animal models2,3,17 and also found to be integrated in human tissue24.
There are numerous possible explanations for why multiple laboratories have published discordant findings on XMRV detection. Patient selection and geographical considerations are likely confounding factors. Likewise, the discovery of other MRVs, in addition to XMRV, may suggest that at least some assays could have lacked sensitivity to detect all potential MRVs in clinical samples. For example, PCR primers or antigens for serological assays may have been designed to be specific for the initial strains of XMRV that were published2,24, and consequently failed to detect divergent MRVs. Contamination with murine DNA or cells, or other nucleic acid sources such as plasmids, at any stage of the process from sample collection to amplification and detection remains a possibility whenever sensitive methods such as nested PCR are employed. Indeed, a number of recent publications have highlighted the potential for mouse DNA contamination in human samples resulting in the detection of XMRV and other MRV sequences25–29. However, many laboratories are utilizing sensitive assays for mouse mitochondrial DNA4 and other tests to rule out gross contamination. Also the contamination argument is less convincing for the Lombardi et al study which documented culture of infectious virus and serological responses that correlated with molecular findings. However, caution must be taken in both the description of MRVs as human viruses and their potential association with disease30. The intricacies of many of the techniques, including sample type, preparation and storage, nucleic acid extraction, use of RNA and/or DNA, amplification method and detection, are all confounding factors and until either multiple laboratories deploy the same techniques, or alternately share large numbers of samples, the exact cause of the discordant results will remain unclear.
A careful study of the literature and published protocols does not reveal any obvious consistent differences between protocols from laboratories that do or do not detect MRVs. For example, an early suggestion was that use of heparin collection tubes3 for blood may introduce mouse material into assays; however, at least one study has detected MRVs in blood collected into EDTA tubes4. Lombardi et al, utilized both genomic and cDNA for detection, often from activated leukocytes3, while many of the negative studies used just genomic DNA isolated directly from purified leukocytes9, followed by PCR without an RT step. Thus, it could be supposed that virus was present at low copy number and predominantly as non-integrated RNA and hence was not detectable with PCR of PBMC. However, Lo et al used only genomic DNA from freshly purified leukocytes, or whole blood and did not employ an RT step, yet detected MRV at high rates. Thus, in order to attempt to standardize samples and assays in an effort to obtain ‘gold standard assays’ for future studies, the Blood XMRV SRWG has launched a series of collaborative studies to address these questions.
Blood safety concerns leading to formation of the Blood XMRV Scientific Research Working Group
Early in the description of XMRV and ME/CFS, the potential for these gammaretroviruses to be transmitted by blood transfusions or by other blood products was raised5, leading to the important question - could XMRV pose a threat to the safety of the US blood supply? Faced with the possibility of a potential risk of infectious virus in the blood supply, HHS facilitated the establishment and coordination of collaborative groups, both internal and external to HHS, with clearly delineated responsibilities and the necessary expertise in risk assessment, risk management, and risk communication. Thus, in November 2009, the HHS Blood XMRV SRWG was formed and convened for their first meeting in December. At the same time, the AABB formed an XMRV Interorganizational Task Force (see accompanying commentary in this issue of Transfusion).
The Blood XMRV SRWG is charged with the design and coordination of research studies to evaluate whether XMRV poses a threat to blood safety. The Blood XMRV SRWG group is led by the National Heart, Lung, and Blood Institute (NHLBI) and is comprised of experts in transfusion medicine, blood banking, retrovirology, and ME/CFS research, as well as representatives from DHHS, the FDA, CDC, and NIH (see roster provided in Appendix A). The group reports to the Public Health Service Blood Organ and Tissue Safety (PHS BOTS) Committee and to the PHS Blood Organ and Tissue Senior Executive Council (BOTSEC), and liaises with the AABB XMRV Interorganizational Task Force, thus ensuring that the latter is aware of the results of its deliberations and studies.
Lack of evidence for an association between transfusion and ME/CFS or prostate cancer
The SWRG determined that an evaluation of the evidence that transfusion may or may not be associated with the development of ME/CFS and prostate cancer should be undertaken early on because it could potentially impact risk assessment (e.g., research study prioritization) and risk management decisions. The lack of an association between transfusion history and ME/CFS or prostate cancer would be indirect evidence that there does not appear to be transfusion-transmission of a causative disease agent. Conversely, the presence of an association with CFS and/or prostate cancer could be indirect evidence for transmission of a causative disease agent by transfusion and would potentially reinforce the possibility that XMRV or MRVs could be transfusion-transmitted. Only four relevant reports for prostate cancer and one report for CFS were identified; the findings in these papers are summarized below.
Inoue et al. followed a cohort of 10,451 Japanese men and women for a mean of 12.8 years, from 1990 to 2003; the cohort included 4,401 men who were 40–79 years old31. Blood transfusion before 1990 was associated with an increase in overall cancer mortality (liver and non-liver cancer, hazard ratio 1.75, 95% CI 1.32–2.18) and non-liver cancer mortality (HR 1.68, 95% CI 1.25–2.26). This was secondary to an increase in stomach, liver, and pancreatic cancer mortality but not because of an increase in “Other” cancer mortality for men (1.32, 95% CI 0.67–2.57); the “Other” category would include prostate cancer mortality. Transfusion history was assessed solely by a questionnaire and medical record reviews to corroborate transfusion histories were not performed. These results differed from those obtained in an earlier analysis of the same cohort which revealed an association between prostate cancer mortality and a blood transfusion history (hazard ratio of 1.71, 95% CI 1.1–2.66, adjusted for age and area of study)32. The authors did not provide an explanation for these discrepant results. Another study conducted by Blomberg and colleagues in Sweden included 28,338 non-transfused patients and 1,572 transfused patients followed from 1981–1982 until 1991 and used a population based regional tumor registry to identify incident cancers33. They found that there was no excess risk of prostate cancer in either group (there was a trend towards less cancer in the transfused patients than expected but this finding was not significant). Conversely, these investigators found an increase in the incidence of malignant lymphoma and non-melanoma skin cancer in patients receiving a transfusion as compared to patients who did not. Finally, Hjalgrim et al. used the Scandinavian Donations and Transfusions (SCANDAT) database to evaluate cancer incidence in transfusion recipients followed from 1968 in Sweden and 1982 in Denmark, to 200234. About 888,800 transfusion recipients were followed. The standardized incidence ratio for prostate cancer was increased at 1–5 months after transfusion (2.64, 95% CI 2.51–2.77) but not afterwards with a standardized incidence ratio of 0.98 at ≥ 20 years after transfusion. Based on the natural history of prostate cancer, the authors attributed the increased incidence at 1–5 months to the clinical recognition at 1–5 months of prostate cancers that were subclinical at the time of transfusion. Our interpretation of the data from this large study is that there was no evidence of an association between transfusions and the development of prostate cancer.
The one small study of transfusions and ME/CFS was a case-control study conducted by Bell and colleagues involving a cluster of 21 students, 10–16 years old, with symptoms of CFS in a farming community in upstate New York (Lyndonville) in 1985; these students were designated as cases35. Two controls were then identified and matched for age and sex to each case. None of the cases received a blood transfusion while 2 of the 42 controls did. Additional to this published study, Vernon and McCleary recently conducted a survey to evaluate the proportion of patients with CFS reporting a history of transfusion: 8.1% (124/1529) of patients reported receiving a blood transfusion prior to their diagnosis, whereas 3.3% (50/1529) reported receiving a transfusion after being diagnosed with CFS36. By comparison, Wang et al. found that 5.3% of 92,581 blood donors surveyed in 1998 and 4.2% of 2.9 million donors giving blood at 5 geographically-disperse U.S. blood centers between 1991–2000 reported a history of transfusion37.
After a careful review of these data, the Blood XMRV SRWG concluded that there was currently no convincing peer-reviewed evidence that transfusion of blood products was associated with the development of prostate cancer or ME/CFS. However, the group recognized that data were sparse, especially for ME/CFS. Thus, the consensus of the group was that additional epidemiological studies designed to evaluate the association between transfusion history and ME/CFS or prostate cancer should be conducted if feasible, particularly if XMRV is found to be prevalent in blood donors and transmissible by transfusion. However, it was recognized that conducting rigorous epidemiologic studies with adequate power will be extremely difficult, and the conduct of such protocols was likely outside the mandate and beyond the capacity of the working group.
Co-ordination of laboratory assays
The Blood XMRV SRWG was faced with three basic questions when it first convened in December of 2009: Is XMRV in the blood supply, and if so, what is the prevalence of viremia and seroreactivity in donors? Is it transfusion-transmitted? If transfusion transmitted, what is the clinical impact on blood recipients. These questions, starting with the simplest one: “what is the prevalence of XMRV in blood donors?” remain unanswered. Lombardi et al found evidence of XMRV DNA in 8/218 (3.7%) PBMC samples from healthy controls3. Similarly, Lo et al detected MRV proviral DNA sequences in PBMC or whole blood samples from 3/44 (6.8%) of healthy NIH blood donors4. In contrast, Switzer et al found no evidence of proviral DNA in PBMCs from the 56 healthy controls matched to their CFS cases and 41 blood donors, as well as no serological evidence by western blot or enzyme-linked immunoassay (EIA) of XMRV infection in 53 of the same healthy controls (3 had no plasma/serum) or 121 separate blood donors9. Although at the time other MRVs had not been described in human samples, it is likely that the assays employed in this study would detect these viruses. Multiple serological assays employed by Qiu et al also detected only inconclusive evidence of XMRV antibody responses in 3 individuals out of over 1500 blood donations screened17. The discordance over prevalence rates in blood donors and similar healthy populations is troublesome, and reaching a consensus on rates of XMRV/MRV infection in donors is a major focus of the SRWG.
The challenge has resided with the identification of sensitive and specific assays that can reliably detect XMRV and MRV nucleic acids and/or immunological responses to these viruses in blood samples. The discordance described above strongly suggested the need for the development of XMRV analytical reference panels, as well as panels of samples from clinical cases and controls, to be used to validate the performance characteristics of assays for subsequent use to investigate the prevalence of XMRV/MRVs in blood donors and recipients. Thus, the development of XMRV/MRV analytical and clinical panels for characterization of nucleic acid tests (NAT) and serological assays was identified as a first priority by the Blood XMRV SRWG. Accordingly, a four phase approach was adopted to achieve these goals, culminating in achieving a consensus for an initial estimate of XMRV/MRV prevalence in US blood donors.
Phase I - Analytical sensitivity panels
As described above, because of the large array of different assays employed by researchers, the interpretation of discordant XMRV results has been difficult. Thus, in phase I of the working group study, which is being funded through the NHLBI Retrovirus Epidemiology Donor Study-II (REDS-II) program, Blood Systems Research Institute (BSRI), the REDS-II Central Laboratory, developed coded analytical performance panels comprised of XMRV virions or XMRV-infected cells serially diluted into plasma or whole blood (WB), respectively. These panels were designed to allow the participating laboratories to compare XMRV assay sensitivity. Although the majority of studies that have investigated XMRV detection in blood have focused on purified and generally cryopreserved PBMC preparations, it was decided that for the purposes of blood screening, plasma and WB were much more practical alternative sample types. Panels were prepared using the human prostate cell line, 22Rv1, which harbors at least 10 proviral DNA copies/cell of XMRV and produces high titers of infectious XMRV virions in the culture supernatant38. The approximate XMRV DNA levels in infected cells and RNA viral load in culture supernatant were established by consensus results from three laboratories with quantitative XMRV detection techniques. The diluents for the panels were obtained after several of the participating laboratories pre-screened a small number of laboratory volunteers to ensure they were XMRV negative by PCR, serology and virus culture.
Phase I panels were distributed to five laboratories (CDC, WPI, NCI, and the independent laboratories of Drs. Lo and Hewlett at FDA) involved in the Working Group. In addition, panels were supplied to Gen-Probe Inc, which although not a part of the SRWG, had recently developed a high-throughput target-capture transcription mediated amplification (TC-TMA) assay for XMRV on the TIGRIS® platform. Thus, their participation was deemed valuable for later stages of the XMRV SRWG research agenda when large numbers of donor and recipient samples would require testing.
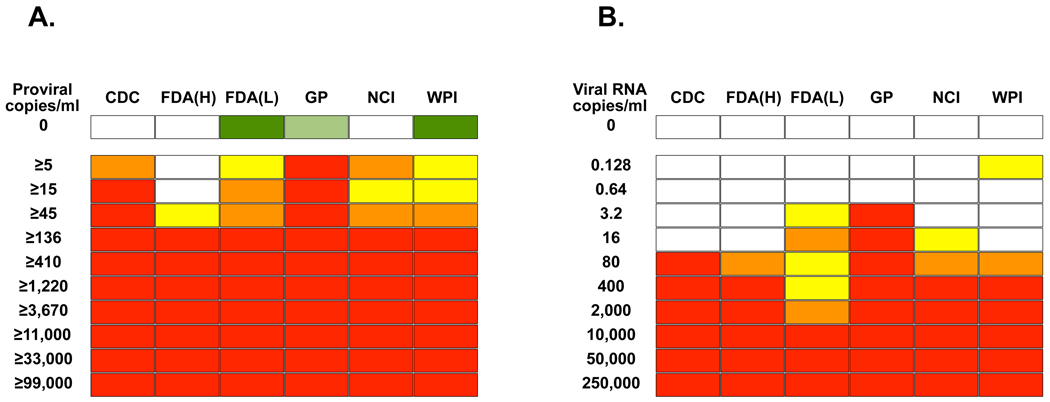
Tables 1 and 2 summarize the different sample input levels and methods for nucleic acid extraction,target amplification and detection employed by the laboratories for WB and plasma, respectively. Three distinct assay types were employed – nested PCR, quantitative real-time PCR and TMA. Furthermore, a number of the laboratories performed secondary confirmation – either sequencing of product or performance of southern blots in order to confirm specificity of positive results. Two laboratories employed more than one assay (CDC and FDA [Lo]). For the sake of simplicity in Figure 1 both laboratories employed the criterion that a positive result in only one assay was sufficient to call a sample positive. Results were reported back to the central laboratory at BSRI, where the blinded code was broken, and data analyzed. As seen in Figure 1, all laboratories detected at least 13 XMRV-infected cells (136 proviral copies/ml), and four out of six assays demonstrated even more sensitive limits of detection of 0.5 – 4.5 cells/ml, unfortunately reaching close to the end of the titration series. Four out of six plasma RNA assays had limits of detection of 80 or fewer RNA copies/mL; the Gen-Probe assay detected 3.2 or fewer RNA copies/mL.
Table 1.
Methods used by participating laboratories for detection of XMRV nucleic acids in whole blood samples
| Lab | Extraction method |
Blood Volume (µl) |
Input DNA (ng) |
Assay # |
Target | Assay type |
Quant/ nested |
Primer Reference |
|---|---|---|---|---|---|---|---|---|
| CDC | QIAamp mini blood |
400 | 1000 | 1 | gag | PCR southern |
Nested | 9 |
| 2 | pol | PCR southern |
Nested | 9 | ||||
| FDA (H)a | QIAamp mini blood |
200 | 500–1000 | gag | PCR | Nested | 2 | |
| FDA (Lo) | Qiagen DNeasy blood |
200 | 30–50 | 1 | gag | PCR | Nested | 4 |
| 30–50 | 2 | gag | PCR | Nested | 4 | |||
| GPb | Magnetic-based target capture |
50 | N/A | Duplex | Proprietaryc | TMAc | N/A | Unpublished |
| NCI | Promega Wizard Genomic DNA |
500 | 4000 | 5’ UTR of gag |
PCR SCAd |
Quant | Unpublishedd | |
| WPI | QIAamp DNA mini blood |
250 | 100 | 5’ UTR of gag |
qPCR | Quant | 24 | |
H=Hewlett
GP=Gen-Probe Inc
Gen-Probe employs a duplex transcription-mediated amplification (TMA) assay based on multiple proprietary probes
SCA=single-copy assay.
Forward primer: 5-TGTATCAGTTAACCTACCCGAGT-3’; Reverse primer: 5-AGACGGGGGCGGGAAG-3’; XMRV probe 5’fam-TGGAGTGGCTTTGTTGGGGGACGA- tamra3’
Table 2.
Methods used by participating laboratories for detection of XMRV nucleic acids in plasma specimens
| Lab | Extraction method |
Volume plasma (µl) |
Assay # |
Target | Assay type |
Quant/ Nested |
Primer Reference |
|---|---|---|---|---|---|---|---|
| CDC | Ultracentrifuge/ QIAamp viral RNA mini |
500 | 1 | pro | qRT-PCR | Quant | 15 |
| 2 | gag | RT-PCR southern |
Nested | 9 | |||
| FDA (H) | Qiagen viral RNA mini |
140 | 1 | gag | RT-PCR | Nested | 2 |
| FDA (Lo) | Ultracentrifuge/ TRIzol |
250 | 1 | gag | RT-PCR | Nested | 4 |
| 2 | gag | RT-PCR | Nested | 4 | |||
| GP | Magnetic- based target capture |
500 | Duplex | Proprietary | TMA | N/A | Unpublished |
| NCI | Ultracentrifuge including internal standard Guanidinium Isothicyanate |
~200– 500ul plasma |
1 | 5’ UTR of gag |
RT-PCR SCA |
Quant | Unpublished |
| WPI | QIAamp viral RNA mini |
140 | 1 | 5’ UTR of gag |
qRT-PCR | Quant | 24 |
Figure 1. Sensitivity of detection of XMRV by multiple laboratories.
Analytical panels for whole blood (A) and plasma (B) were created by serial dilution of 22Rv1 cells or 22Rv1 cell supernatant in whole blood and plasma respectively. The dilutions were tested in triplicate by six laboratories (CDC, FDA lab of Dr Hewlett [FDA(H)], FDA lab of Dr Lo [FDA(Lo)], Gen-Probe Inc [GP], NCI and WPI). Red represents 3/3 replicates being positive, orange is 2/3, yellow is 1/3 and white is 0/3. Replicates of six negatives were performed and white represents 0/6, while green represents 1/6 replicates being positive. In the case of FDA(Lo) and WPI subsequent sequencing demonstrated in each case that the amplification product in the single false positive result for a negative control sample was of human genomic origin. In the case of GP, a repeat by a separate operator yielded 0/6 controls as positive.
Three negative samples were initially falsely labeled positive - one each in three separate laboratories. However, in the case of two of these results (FDA [Lo] and WPI), subsequent sequencing of the positive bands revealed DNA of human genomic origin, rather than MRV sequences, suggesting non-specific priming of PCR assays. The third false positive (Gen-Probe) was correctly called negative by the submitting group, as a repeat analysis by a different operator did not detect the sample as positive. Thus, while the number of replicates of negative samples tested was low, little evidence of false positivity was observed. However, these findings do highlight the need for some labs to sequence every positive PCR result in order to confirm specificity. Additionally, the overall similarity of results on the analytical panels suggests that major differences in the sensitivity of assays, at least for the XMRV sequence found in 22Rv1 cells, cannot explain the dramatic differences in XMRV detection in clinical samples reported by the participating laboratories.
Phase II – Pilot clinical panels
Although the majority of assays described in Tables 1 and 2, at least in theory, will amplify the polytropic MRVs described by Lo et al.4 in addition to XMRV, a major caveat to the phase I panels is the fact that 22Rv1 cells appear to contain a single strain of XMRV, and thus the ability to detect diverse strains of XMRV, as well as other MRVs, was not directly examined. Consequently, the working group decided that whereas the phase I study demonstrated the relatively comparable sensitivity of all the assays involved, clinical samples need to be included in the three additional phases (Phases II–IV).
Phase II was designed as a pilot study to provide the participating laboratories their first opportunity to test the same clinical samples, as well as to examine whether the sample types in blood donor repositories collected both specifically for these studies, and generally for the study of transfusion-transmitted infections39, were suitable for XMRV detection. These repositories consist primarily of plasma and/or serum and, in some instances, frozen WB aliquots. The SRWG also wanted to ensure that the routine delay between blood collection and sample processing would not adversely affect XMRV detection in clinical samples. Blood was collected under current IRB approval from four patients with CFS found to be XMRV-positive in the 2009 WPI study published in Science3, as well as a XMRV/MRV-negative controls identified in phase I. Specimen tubes were then divided into three groups, with one set immediately processed into PBMC, WB and plasma, while the other two sets were refrigerated and then similarly processed after two and four days. These samples were coded, blinded and distributed to three SRWG laboratories – CDC, NCI, and WPI, as well as to Gen-Probe. Phase II is currently in progress. The findings will be used to optimize the development of the clinical sensitivity and specificity panels for phase III (see below), and will inform sample preparation for future studies under development, such as a new NIH study aimed at further evaluating the association between XMRV/MRVs and ME/CFS40.
Phase III - Clinical sensitivity and specificity panels
Phase III will develop larger clinical panels in order to assess the ability of participant assays to effectively detect XMRV/MRVs in clinical specimens that will be processed by methods optimized in phase II, i.e., a choice will be made between using PBMC or WB for cellular samples, and whether samples should be processed immediately after phlebotomy (day 0) or at one to four days. To attempt to overcome the issues of variation in viral sequences as well as possible sources of non-specificity, the phase III panels will include relatively larger numbers of pedigreed XMRV-positive and -negative clinical samples. These will include samples from ~30 patients with ME/CFS reported as positive for XMRV/MRVs by PCR, serology and/or virus culture in the Lombardi et al. and Lo et al. studies3,4, thus both XMRV- and PMRV-positive clinical samples will be represented. For negative specimens, samples prepared from 10 additional pedigreed XMRV/MRV-negative donors will be included in order to introduce human host genetic variability - these donors have been pre-screened as negative by 1) PCR at the WPI, FDA (Lo), CDC and Gen-Probe laboratories, 2) serology at the WPI, CDC and NCI (Ruscetti) laboratories, and 3) culture at the WPI laboratory. These positive and negative samples will be assembled into coded panels and distributed to all participating laboratories for XMRV/MRV nucleic acid. In addition, samples for serological analyses will also be included, allowing an initial indication of sensitivity and specificity of serological assays developed by some of the participating laboratories, and the correlation between detection of XMRV/MRV antibodies and nucleic acids.
Phase IV - Clinical Panel for Donor Prevalence
Phase IV will be comprised of blinded panels of WB and plasma collected from approximately at least 300 blood donors. Additionally, samples that are known to be XMRV/MRV positive and negative will be included. Blinded panels will be distributed to at least four of the participating laboratories for testing. Test results obtained using WB will be compared to those obtained using plasma for each laboratory; results will also be compared between laboratories. Also, the proportion of donors who are positive for XMRV/MRV DNA, RNA and/or antibodies (XMRV/MRV prevalence) will be estimated based on compiled results from each lab. In addition to providing preliminary information as to the prevalence of XMRV/MRVs in blood donors, it is hoped that this series of studies will yield protocols that can be the basis of a set of ‘gold standard’ assays allowing the standardization of tests across laboratories in order to minimalize the type of controversies thus far seen in XMRV/MRV detection.
Future studies for potential consideration
Depending on the results of phase IV, additional studies may need to be considered. For example, it likely will be necessary to further evaluate donor prevalence in a larger and more geographically-dispersed donor population using both molecular and serological methods. If the samples stored in the NHLBI donor and donor-recipient repositories are deemed adequate for testing (i.e., if sample preparation and crypreservation methods are not an issue) studies to assess the temporal distribution of XMRV/MRV in blood donors may be warranted; such studies could be done by accessing various NHLBI repositories that span the past four decades39. Phase IV results may also help define the feasibility, sample size, and power of studies to assess infectivity by transfusion. A large enough XMRV/MRV prevalence in blood donors would ensure sufficient statistical power to investigate whether XMRV/MRVs are transfusion-transmissible using samples from the linked NHLBI donor-recipient repositories (and estimate rates of transmission by XMRV-MLV-reactive donations). Further, if XMRV/MRVs are found to be transmissible by transfusion, studies can be conducted to achieve a greater understanding of the correlates of transmission and of the viral and immunological parameters of acute infection. This in turn would help define the optimal approach to blood screening if the latter was deemed necessary, and in particular would highlight the relative value of serological versus NAT screening.
Conclusions
Over the last two decades, there has been a focus within the blood banking community not only on emerging infectious diseases but also on the emerging knowledge of known infectious agents and the risk they may pose to transfusion recipients41. Evaluating emerging infectious agents which may affect the safety of the blood supply includes early recognition of potential threats and development of scientific priorities, as well as having a proactive, flexible and funded strategic research agenda that will effectively and rapidly mobilize available expertise and resources to address pressing questions in a scientifically-sound and timely manner. In this commentary we have presented our methodical approach to address the current scientific knowledge gaps in XMRV/MRV, and through the NHLBI-funded REDS-II program have been responsive by developing and coordinating multi-laboratory studies of XMRV/MRV related to the safety of the nation's blood supply. The creation and combined efforts of the Blood XMRV SRWG and the AABB Interorganizational Task Force are critical to addressing the possibility that XMRV/MRV may turn out to be transfusion-transmitted pathogen(s). We hope that this investigation will not only help evaluate whether XMRV/MRV represent a threat to the safety of the Nation’s blood supply, but also contribute new science related to this potentially important class of retroviruses.
Acknowledgments
The authors acknowledge the tremendous effort contributed by all SRWG members, as well as the AABB and many laboratory members from all of the contributing laboratories, including Leslie Tobler, Ingrid Wilson, Brian Custer and Lubov Pitina at BSRI, HaoQiang Zheng, Hongwei Jia, Shaohua Tang, and Anupama Shankar at CDC, Max Pfost, Cassandra Puccinelli and Kathyrn Hagen at WPI, Kui Gao at Gen-Probe Incorporated, Frank Ruscetti, Mary Kearney and Rachel Bagni at NCI, and Shixing Tang and Jiangqin Zhao in OBRR, FDA. The laboratory work was funded by the NHLBI REDS-II Central Laboratory Contract to Blood Systems Research Institute (N01 HB-57181). The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Department of Health and Human Services.
Appendix A – Roster for the Blood XMRV Scientific Research Working Group
Simone Glynn, MD, MSc, MPH
Chair
Branch Chief, Transfusion Medicine and Cellular Therapeutics Branch
Division of Blood Diseases and Resources, National Heart, Lung, and Blood Institute
Jerry A. Holmberg, PhD
Co-Chair
Senior Advisor for Blood Policy
Executive Secretary of the Advisory Committee on Blood Safety and Availability
U.S.Department of Health and Human Services, Office of the Secretary
Office of Public Health and Science
Celso Bianco, MD
Executive Vice President
America’s Blood Centers
Michael P. Busch, MD, PhD
Director, Blood Systems Research Institute
Vice President, Research/Scientific Affairs, Blood Systems Professor of Laboratory
Medicine, UCSF
Roger Y. Dodd, PhD
Vice President, Research and Development
American Red Cross, Holland Laboratory
Louis M. Katz, MD
Executive Vice President
Medical Affairs, Mississippi Valley Regional Blood Center
Steven H. Kleinman, MD
AABB Liaison
Clinical Professor of Pathology, University of British Columbia
Anthony L. Komaroff, MD
The Simcox-Clifford-Higby Professor of Medicine
Harvard Medical School
Senior Physician, Brigham & Women's Hospital
Judy A. Mikovits, PhD
Research Director, Whittemore Peterson Institute
Dept Microbiology and Immunology Applied Research Facility
Graham Simmons, PhD
Associate Investigator
Blood System Research Institute
Department of Laboratory Medicine
University of California, San Francisco
Susan L. Stramer, PhD
Executive Scientific Officer
American Red Cross, Scientific Support Office
Leslie H. Tobler, DrPH
Senior Scientist
Blood System Research Institute
Suzanne D. Vernon, PhD
Scientific Director
The CFIDS Association of America
NIH
Harvey Alter, MD
Clinical Center
National Institutes of Health
John Coffin, PhD
Contractor
NIH/NCI/DBS/HDRP/RRL
National Institutes of Health
Tufts University, Boston
Dennis F. Mangan, PhD
Chair, Trans-NIH ME/CFS Research Working Group
Senior Research Advisor, Office of Research on Women's Health
Office of the Director
National Institutes of Health
Francis Ruscetti, PhD
Senior Investigator
NIH/NCI/ DBS/CIP/LEI
National Institutes of Health
CDC
William Bower, MD
Medical Epidemiologist
Centers for Disease Control and Prevention
Walid Heneine, PhD
Laboratory Branch,
Division of HIV/AIDS Prevention
National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention
Centers for Disease Control and Prevention
R. Michael Hendry, DSc
Chief, Laboratory Branch
Division of HIV/AIDS Prevention
National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention
Centers for Disease Control and Prevention
Stephan S. Monroe, PhD
Director of the CDC Division of Viral and Rickettsial Diseases
Centers for Disease Control and Prevention
William Switzer, MPH
Laboratory Branch,
Division of HIV/AIDS Prevention
National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention
Centers for Disease Control and Prevention
FDA
Jay Epstein, M.D.
Director
Office of Blood Research and Review
Center for Biologics Evaluation and Research
Food and Drug Administration
Diane M. Gubernot, M.P.H.
Senior Regulatory Scientist
Office of Blood Research and Review
Center for Biologics Evaluation and Research
Food and Drug Administration
Indira Hewlett, Ph.D.
Chief, Laboratory of Molecular Virology
Office of Blood Research and Review
Center for Biologics Evaluation and Research
Food and Drug Administration
Shyh-Ching Lo, MD, PhD
Director, Tissue Safety Program Division of Cellular and Gene Therapies & Division of Human Tissues
Center for Biologics Evaluation and Research
Food and Drug Administration
Footnotes
Statement of Disclaimer
The views expressed do not necessarily represent the view of the Department of Health and Human Services, or the U.S. Federal Government
Conflict of Interest: None
References
- 1.Schlaberg R, Choe DJ, Brown KR, et al. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc Natl Acad Sci U S A. 2009;106(38):16351–16356. doi: 10.1073/pnas.0906922106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Urisman A, Molinaro RJ, Fischer N, et al. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2(3):e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Lombardi VC, Ruscetti FW, Das Gupta J, et al. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science. 2009;326(5952):585–589. doi: 10.1126/science.1179052. [DOI] [PubMed] [Google Scholar]
- 4.Lo SC, Pripuzova N, Li B, et al. Detection of MLV-related virus gene sequences in blood of patients with chronic fatigue syndrome and healthy blood donors. Proc Natl Acad Sci U S A. 2010;107(36):15874–15879. doi: 10.1073/pnas.1006901107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Coffin JM, Stoye JP. Virology. A new virus for old diseases? Science. 2009;326(5952):530–531. doi: 10.1126/science.1181349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnes E, Flanagan P, Brown A, et al. Failure to Detect Xenotropic Murine Leukemia Virus-Related Virus in Blood of Individuals at High Risk of Blood-Borne Viral Infections. J Infect Dis. 2010;202(10):1482–1485. doi: 10.1086/657167. [DOI] [PubMed] [Google Scholar]
- 7.Erlwein O, Kaye S, McClure MO, et al. Failure to detect the novel retrovirus XMRV in chronic fatigue syndrome. PLoS One. 2010;5(1):e8519. doi: 10.1371/journal.pone.0008519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groom HC, Boucherit VC, Makinson K, et al. Absence of xenotropic murine leukaemia virus-related virus in UK patients with chronic fatigue syndrome. Retrovirology. 2010;7:10. doi: 10.1186/1742-4690-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Switzer WM, Jia H, Hohn O, et al. Absence of evidence of xenotropic murine leukemia virus-related virus infection in persons with chronic fatigue syndrome and healthy controls in the United States. Retrovirology. 2010;7:57. doi: 10.1186/1742-4690-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danielson BP, Ayala GE, Kimata JT. Detection of xenotropic murine leukemia virus-related virus in normal and tumor tissue of patients from the southern United States with prostate cancer is dependent on specific polymerase chain reaction conditions. J Infect Dis. 2010;202(10):1470–1477. doi: 10.1086/656146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hohn O, Krause H, Barbarotto P, et al. Lack of evidence for xenotropic murine leukemia virus-related virus(XMRV) in German prostate cancer patients. Retrovirology. 2009;6:92. doi: 10.1186/1742-4690-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aloia AL, Sfanos KS, Isaacs WB, et al. XMRV: A New Virus in Prostate Cancer? Cancer Res. 2010;70(24):10028–10033. doi: 10.1158/0008-5472.CAN-10-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer N, Hellwinkel O, Schulz C, et al. Prevalence of human gammaretrovirus XMRV in sporadic prostate cancer. J Clin Virol. 2008;43(3):277–283. doi: 10.1016/j.jcv.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 14.Verhaegh GW, de Jong AS, Smit FP, et al. Prevalence of human xenotropic murine leukemia virus-related gammaretrovirus (XMRV) in dutch prostate cancer patients. Prostate. 2010 doi: 10.1002/pros.21255. epub Sep 28. [DOI] [PubMed] [Google Scholar]
- 15.Switzer WM, Jia H, Zheng H, et al. Identification and low prevelance of distinct Xenotropic Murine Leukemia Virus-Related Virus infection in prostate cancer. 2011 doi: 10.1371/journal.pone.0019065. In preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikovits JA, Lombardi VC, Pfost MA, et al. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Virulence. 2010;1(5):386–390. doi: 10.4161/viru.1.5.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu X, Swanson P, Luk KC, et al. Characterization of antibodies elicited by XMRV infection and development of immunoassays useful for epidemiologic studies. Retrovirology. 2010;7(1):68. doi: 10.1186/1742-4690-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henrich TJ, Li JZ, Felsenstein D, et al. Xenotropic murine leukemia virus-related virus prevalence in patients with chronic fatigue syndrome or chronic immunomodulatory conditions. J Infect Dis. 2010;202(10):1478–1481. doi: 10.1086/657168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunstman KJ, Bhattacharya T, Flaherty J, et al. Absence of xenotropic murine leukemia virus-related virus in blood cells of men at risk for and infected with HIV. Aids. 2010;24(11):1784–1785. doi: 10.1097/qad.0b013e32833b76fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Kuppeveld FJ, de Jong AS, Lanke KH, et al. Prevalence of xenotropic murine leukaemia virus-related virus in patients with chronic fatigue syndrome in the Netherlands: retrospective analysis of samples from an established cohort. BMJ. 2010;340:c1018. doi: 10.1136/bmj.c1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cornelissen M, Zorgdrager F, Blom P, et al. Lack of Detection of XMRV in Seminal Plasma from HIV-1 Infected Men in The Netherlands. PLoS One. 2010;5(8):e12040. doi: 10.1371/journal.pone.0012040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong P, Li J, Li Y. Failure to detect Xenotropic murine leukaemia virus-related virus in Chinese patients with chronic fatigue syndrome. Virol J. 2010;7(224) doi: 10.1186/1743-422X-7-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang S, Zhao J, Viswanath R, et al. Absence of Detectable Xenotropic Murine Leukemia Virus-related Virus in Plasma or Peripheral Blood Mononuclear Cells of Human Immunodeficiency Virus Type One Infected Blood donors or Individuals in Africa. Transfusion. 2010 doi: 10.1111/j.1537-2995.2010.02932.x. epub Nov 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong B, Kim S, Hong S, et al. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc Natl Acad Sci U S A. 2007;104(5):1655–1660. doi: 10.1073/pnas.0610291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hue S, Gray ER, Gall A, et al. Disease-associated XMRV sequences are consistent with laboratory contamination. Retrovirology. 2010;7:111. doi: 10.1186/1742-4690-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oakes B, Tai AK, Cingoz O, et al. Contamination of human DNA samples with mouse DNA can lead to false detection of XMRV-like sequences. Retrovirology. 2010;7:109. doi: 10.1186/1742-4690-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MJ, Erlwein OW, Kaye S, et al. Mouse DNA contamination in human tissue tested for XMRV. Retrovirology. 2010;7:108. doi: 10.1186/1742-4690-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato E, Furuta RA, Miyazawa T. An endogenous murine leukemia viral genome contaminant in a commercial RT-PCR Kit is amplified using standard primers for XMRV. Retrovirology. 2010;7:110. doi: 10.1186/1742-4690-7-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith RA. Contamination of clinical specimens with MLV-encoding nucleic acids: implications for XMRV and other candidate human retroviruses. Retrovirology. 2010;7:112. doi: 10.1186/1742-4690-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss RA. A cautionary tale of virus and disease. BMC Biol. 2010;8:124. doi: 10.1186/1741-7007-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue Y, Wada Y, Motohashi Y, Koizumi A. History of blood transfusion before 1990 is associated with increased risk for cancer mortality independently of liver disease: a prospective long-term follow-up study. Environ Health Prev Med. 2009 doi: 10.1007/s12199-009-0125-6. epub Dec 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kikuchi S. Personal past history and mortality in the Japan Collaborative Cohort Study for Evaluation of Cancer (JACC) Asian Pac J Cancer Prev. 2007;(8) Suppl:9–20. [PubMed] [Google Scholar]
- 33.Blomberg J, Moller T, Olsson H, et al. Cancer morbidity in blood recipients--results of a cohort study. Eur J Cancer. 1993;29A(15):2101–2105. doi: 10.1016/0959-8049(93)90042-e. [DOI] [PubMed] [Google Scholar]
- 34.Hjalgrim H, Edgren G, Rostgaard K, et al. Cancer incidence in blood transfusion recipients. J Natl Cancer Inst. 2007;99(24):1864–1874. doi: 10.1093/jnci/djm248. [DOI] [PubMed] [Google Scholar]
- 35.Bell KM, Cookfair D, Bell DS, et al. Risk factors associated with chronic fatigue syndrome in a cluster of pediatric cases. Rev Infect Dis. 1991;(13) Suppl 1:S32–S38. doi: 10.1093/clinids/13.supplement_1.s32. [DOI] [PubMed] [Google Scholar]
- 36.Vernon SD, McKleary KK. Blood donation and transfusion in CFS patients (abstract) Reviews in Antiviral Therapy & Infectious Diseases. 2010;8:30. [Google Scholar]
- 37.Wang B, Higgins MJ, Kleinman S, et al. Comparison of demographic and donation profiles and transfusion-transmissible disease markers and risk rates in previously transfused and nontransfused blood donors. Transfusion. 2004;44(8):1243–1251. doi: 10.1111/j.1537-2995.2004.04034.x. [DOI] [PubMed] [Google Scholar]
- 38.Knouf EC, Metzger MJ, Mitchell PS, et al. Multiple integrated copies and high-level production of the human retrovirus XMRV (xenotropic murine leukemia virus-related virus) from 22Rv1 prostate carcinoma cells. J Virol. 2009;83(14):7353–7356. doi: 10.1128/JVI.00546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Busch MP, Glynn SA. Use of blood-donor and transfusion-recipient biospecimen repositories to address emerging blood-safety concerns and advance infectious disease research: the National Heart, Lung, and Blood Institute Biologic Specimen Repository. J Infect Dis. 2009;199(11):1564–1566. doi: 10.1086/598860. [DOI] [PubMed] [Google Scholar]
- 40.Enserink M. New XMRV Paper Looks Good, Skeptics Admit—Yet Doubts Linger. Science. 2010;329(5995):1000. doi: 10.1126/science.329.5995.1000. [DOI] [PubMed] [Google Scholar]
- 41.Stramer SL, Hollinger FB, Katz LM, et al. Emerging infectious disease agents and their potential threat to transfusion safety. Transfusion. 2009;(49) Suppl 2:1S–29S. doi: 10.1111/j.1537-2995.2009.02279.x. [DOI] [PubMed] [Google Scholar]