Abstract
Introduction
Identification of yeast isolated from clinical specimens to the species level has become increasingly important. Ever-increasing numbers of immuno-suppressed patients, a widening range of recognized pathogens, and the discovery of resistance to antifungal drugs are contributing factors to this necessity.
Material and methods
A total of 487 yeast strains were studied for the primary isolation and presumptive identification, directly from clinical specimen. Efficacy of CHROMagar Candida has been evaluated with conventional methods including morphology on Corn meal–tween 80 agar and biochemical methods by using API 20 C AUX.
Results
The result of this study shows that CHROMagar Candida can easily identify three species of Candida on the basis of colonial color and morphology, and accurately differentiate between them i.e. Candida albicans, Candida tropicalis, and Candida krusei. The specificity and sensitivity of CHROMagar Candida for C. albicans calculated as 99%, for C. tropicalis calculated as 98%, and C. krusei it is 100%.
Conclusion
The data presented supports the use of CHROMagar Candida for the rapid identification of Candida species directly from clinical specimens in resource-limited settings, which could be very helpful in developing appropriate therapeutic strategy and management of patients.
Keywords: CHROMagar Candida, resource-limited settings, presumptive identification
Infections due to Candida species and other fungi have increased dramatically in recent years and are of particular importance because of the rising number of immuno-compromised patients (1). Although Candida albicans remains the most common cause of human Candidiasis, the frequency of infection attributed to other members of the genus is also increasing (2, 3). This is primarily due to the increase in the number of at risk individuals, particularly those with impaired immunity, such as transplant recipients, cancer patients receiving chemotherapy, and human immunodeficiency virus-infected patients (4–8). In the 1980s C. albicans accounted for more than 80% of all candidal isolates recovered from nosocomial yeast infection (9). Now, C. albicans accounts for more than 80% of all Candida blood isolates in a recent review (10). The emergence of Candida species other than C. albicans is a matter of concern in several major institutions (3, 11–14). These species are also shown to have reduced susceptibility to antifungal agents. The frequency of isolation of Candida krusei, Candida glabrata, Candida tropicalis, and Candida prapsilosis is steadily increasing globally (15, 16). Fungal infections caused by Candida species are being detected more frequently in clinical laboratories (17). Changing Candida epidemiology and availability of newer antifungal drugs with different antifungal spectra means that physicians can no longer make therapeutic decisions based on broad identification of fungi as yeast and mold (18). The conventional methods of yeast identification, which mainly consist of assimilation and fermentation characteristics, are reported to be cumbersome and beyond the expertise range available in local laboratories. In non-specialized clinical laboratories (19), especially in resource-limited settings, identification of yeast and yeast-like organisms requires evaluation of microscopic morphology and biochemical studies. Some unusual yeasts may require unique morphological and biochemical studies for identification, occasionally requiring up to 21 days of incubation (20). Effective treatment requires both early diagnosis and prompt initiation of therapy against fungal infection (21). As the traditional methods are tedious and time consuming to perform in the routine laboratories, numerous isolation media are available in the market that can identify pathogens within 4–72 hours, depending upon the system (22–26). Several brands of chromogenic media are available for rapid identification of yeast. These special media yield microbial colonies with varying pigmentation secondary substrates that react with enzymes secreted by micro organisms (27). These media are species-specific, allowing the organisms to be identified to the species level by their color and colonial characteristics. The manufacturer of CHROMagar Candida currently advertises its product as able to detect and differentiate three species, C. albicans by growth as light to medium green colonies, C. tropicalis by growth as steel blue colonies accompanied by purple pigmentation diffused into surrounding agar, and C. krusei by growth as large, fuzzy, rose colored colonies with white edges, after incubation for 48 hours at 37°C, as also reported in several studies (28–32). Detection of Candida on CHROMagar Candida from poly fungal specimen also allows direct and more rapid and specific identification of C. albicans and other spp. (29, 32, 33) which could decrease the time required to obtain results. Use of chromogenic media in clinical microbiology laboratories for the isolation and presumptive identification of important Candida species is easy to perform, requires less time and is cost effective too. In this study our goal was to evaluate the usefulness of CHROMagar Candida for detection and identification of major Candida species with accuracy to reduce the time of identification, and its characterization from poly fungal specimens, especially in developing countries in resource-limited settings.
Material and methods
Period of study
From April 2006 to September 2009.
Clinical isolates
A total of 487 yeast strains, including C. albicans (n = 201), C. tropicalis (n = 140), C. prapsilosis (n = 32), C. krusei (n = 30), C. glabrata (n = 21), Candida lusitaniae (n = 21), Candida guilliermondii (n = 21), and Candida famata (n = 21), were isolated from various clinical specimens (n = 435) after direct plating on CHROMagar Candida (109 urine, 11 blood, 81 sputum, 22 wound, 91 vaginal secretion, and 21 peritoneal fluid).
Strains and media
Quality control strains of C. albicans (ATCC90029), C. prapsilosis (ATCC 2201), and C. krusei (ATCC6258) were taken from ATCC (American type culture collection) and IIDRL Laboratory, Department of Microbiology, University of Karachi, Pakistan. These control strains were first confirmed by using conventional methods, including germ tube formation test, and morphological characterization, which includes macroscopic features (morphology, color, size, and texture) on Sabouraud dextrose agar (SDA), Corn meal agar with tween 80 and CHROMagar Candida (CHROMagar Candida, France).
CHROMagar Candida medium comprised per liter peptone (10 g), glucose (20 g), agar (15 g), chloramphenicol (0.5 g), and Chromogenic ix. (2 g), pH 6.1. This medium was prepared according to the manufacturer's instructions, does not require autoclaving and is dispensed into Petri plates after cooling. Culture was inoculated and incubation was done at 37°C. The appearance of colonies, including color, size, and textures on CHROMagar Candida was analyzed.
Identification methods
All samples were first plated on SDA and CHROMagar for 48 hours at 37°C. The production of color and morphology as described by the manufacturer were recorded and the photographs were recorded. For C. albicans the germ tube test was also performed to differentiate between C. albicans and non albicans Candida (NAC). The colonies from CHROMagar and SDA were plated on corn meal agar with Tween 80 for morphological examination of the production of chlamydospores, blastospores, true hyphae, and branched pseudohyphae. The results were again compared with the colonies that were grown on CHROMagar. These colonies from SDA were subjected to biochemical analysis by API 20C Aux (Biomuriex). Finally the results were again compared with the culture results and speciation was done. The data suggested that species which were identified by CHROMagar and Corn meal–Tween 80 agar were the same as were confirmed by API 20C Aux. It means that in the presence of CHROMagar it is not necessary to perform germ tube test for C. albicans to confirm, as was also reported by Odds and Bernaerts (31).
Results
We evaluated 487 yeast isolates from 435 clinical specimens during April 2006 to September 2009 (Table 1). These samples were directly plated on CHROMagar Candida (CHROMagar Candida, France), SDA (Oxoid), Corn meal agar (Oxoid) with Tween 80, and on CHROMagar Candida and SDA. Colony growth was observed after 24 and 48 hours of incubation. The development of the colony color was noted on CHROMagar Candida, and we observed that the medium better differentiated the color of colonies after 48 hours of incubation, while after 72 hours of incubation a distinct deep color of the colonies was observed. It was also noted that colony colors of isolated strains were deepened with the passage of time and persistence of pigment was also observed on CHROMagar plates after more than 15 days, as also reported by Pfaller, Houston and Coffmann (32).
Table 1.
Growth and colonial characteristics of 487 yeast isolates incubated for 2 days on CHROMagar Candida and Corn meal–tween 80 agar at 37°C and identification of clinical yeast isolates by API 20C Aux Candida kits (Biomerieux)
| Species | Total number. of isolates | Colony characteristics on CHROMagar Candida | Morphologic features on Corn meal–tween 80 agar | Identification by API 20C AUX |
|---|---|---|---|---|
| Candida albicans | 201 | Apple green colonies; consistent | Chlamydospores present; abundant pseudohyphae, and true hyphae, clusters of blastospores are present | Identified as C. albicans |
| Candida tropicalis | 140 | Dull blue, to purple color that diffused into surrounding agar with pale pink edges | Abundant pseudohyphae with blastoconidia | Identified as C. tropicalis |
| Candida parapsilosis | 32 | White to pale pink colonies | Clusters of blastospores were seen occasionally giant cells | Identified as C. parapsilosis |
| Candida krusei | 30 | Large, flat, spreading, pale pink colonies with matt surfaces | Branched pseudo mycelium with clusters and chains of blastospores | Identified as C. krusei |
| Candida glabrata | 21 | White large glossy pale pink to violet colonies | Pseudohyphae not present | Three isolates were not C. glabrata |
| Candida guilliermondii | 21 | Small pink to purple colonies | Pseudohyphae with clusters of blastospores | Identified as C. guilliermondii |
| Candida lusitaniae | 21 | Pink gray purple | Branched pseudohyphae present | Identified as C. lusitaniae |
| Candida famata | 21 | White to light pink colonies | Pseudohyphae not present | Identified as C. famata |
The yeast cells were identified according to morphology and color of colonies on CHROMagar Candida. Germ tube formation for C. albicans was done with microscopic morphology on Corn meal–Tween 80 agar and confirmed by API 20C Aux yeast identification panel (Biomuriex, France), based on assimilation of carbohydrates.
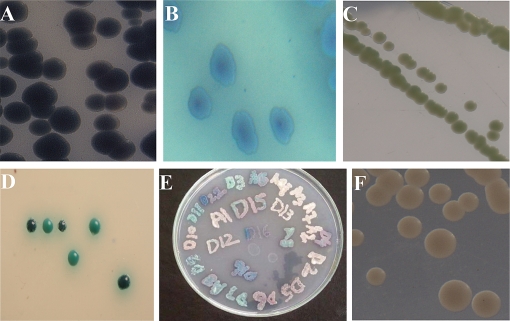
The color of colonies on CHROMagar Candida was similar as given by the manufacturer, i.e. green colonies of C. albicans as indicated in Fig. 1C and D, steel blue colonies of C. tropicalis (Fig. 1A and B) accompanied by purple pigmentation which diffuses into surrounding agar by growth, and large, fuzzy, rose colored colonies with white edges of C. krusei, slightly white pale pink edges and small pink to purple colonies of C. guilliermondii, can easily be differentiated from Candida norvegensis which also produces purple color colonies. The edges of all colored colonies were paler and dark centered and the colony size was between 1 and 5 µm in diameter, also described by Odds and Bernaerts (31). The smooth white to light pink colonies of C. glabrata which later became pink (Fig. 1) and others spp. of Candida appear off-white including C. lusitaniae, C. famata, C. prapsilosis, and Candida rugosa.
Fig. 1.
(A) Candida tropicalis bluish purple colonies. (B) The appearance of C. tropicalis after 72 hours of incubation on CHROMagar at 37°C. (C) Apple green color colonies of Candida albicans grown for 48 hours on CHROMagar Candida at 37°C. (D) Isolated green color colonies of C. albicans grown for 48 hours on CHROMagar Candida at 37°C. (E) Direct plating of sample on CHROMagar Candida for 48 hours at 37°C. Mix colonies of different species can also seen on this plate. (F) Smooth pink colonies of Candida glabrata grown for 48 hours on CHROMagar Candida at 37°C.
Comparative specificity and sensitivity of CHROMagar Candida (CHROMagar Candida, France) in terms of colony growth showed maximum sensitivity and specificity with 95% confidence level in the range of 95.3–100% (Table 2).
Table 2.
Comparative specificity and sensitivity of three different cultural media for Candida spp.
| Name of media | Specificity (N = 487) | Sensitivity (N = 487) | 95% Confidence level | P-value |
|---|---|---|---|---|
| SDA | 464 (95.3%) | 467 (95.9%) | 94.5–100% | <0.05 |
| CHROMagar Candida | 479 (98.4%) | 478 (98.2%) | 95.3–100% | <0.05 |
| Corn meal agar with tween 80 | 470 (96.5%) | 463 (95.0%) | 92–99.4% | <0.05 |
CHROMagar Candida correctly identified 99% of C. albicans, 98% C. tropicalis, 100% C. krusei, 94% of C. glabrata but for other Candida species other tools of identification, for example, API 20C Aux along with CHROMagar Candida (CHROMagar Candida, France) should be used for the correct identification. For example, conventional methods, which includes macroscopic/microscopic characteristic, and biochemical profile by API system of identification (gold standard) to facilitate the diagnosis with efficiency and accuracy, should be possible simultaneously, so that the identification should be accomplished.
Discussion
In resource-limited countries, lack of training and of proper reagents, supplies and equipment makes detection and rapid presumptive identification of yeast species quite difficult. In order to reduce the financial burden of the poor patients, these laboratories do not go beyond the germ tube test and limit their diagnosis to C. albicans or NAC. The biochemical assimilation and fermentation tests are not used in these laboratories due to lack of resources, expertise, and the time required for these tests which increases the cost of mycology cultures (32). As a result, direct selection of appropriate agents for antifungal therapy or prophylaxis becomes almost impossible.
In resource-limited countries, clinicians have little confidence in the accuracy and quality of laboratory test results. They continue to prescribe costly antifungals without knowing the exact antifungal profile of the infectious agent; thereby they increase the economic burden of the society, which also contributes to the emergence of resistant Candida spp.
Without diagnostic tools, safe and effective drug treatment, prevention of resistance to antimicrobial therapy, and monitoring of resistance are not possible. In this setting there is always a need of a medium which helps not only in the isolation but also in the identification of the agent at the species level. The CHROMagar Candida medium was selected as a primary culture medium along with SDA, and we found it an appropriate and affordable diagnostics medium in resource-limiting setting. The reason is that an approximate cost per culture for complete identification of Candida using SDA, Corn meal agar, and API 20C Aux in Pakistan is around US$ 15 (Pak Rs. 1,200), while CHROMagar Candida costs around US$ 3 (Pak Rs. 250) per specimen culture, and thus it is more economical.
We also found that CHROMagar Candida, France easily identifies several species of Candida on the basis of colony color and morphology and accurately differentiates between the three most common species of Candida, i.e. C. albicans, C. tropicalis, and C. krusei, which has also been reported by Murray et al. (20). This medium easily facilitates the detection of more than two species in a single specimen by giving different colored colonies on a plate at the same time, and it was also observed that if the specimen was heavily inoculated, it was difficult to differentiate between mixtures of yeast species on a single agar plate because of the different color reactions (Fig. 1).
The specificity and sensitivity of CHROMagar Candida for green color colony of C. albicans was calculated as 99%, for blue colonies with dark center surrounded by gray halo of C. tropicalis calculated as 98%, and for pink rough and spreading colonies with broad white edges of C. krusei as 100%. The CHROMagar Candida (CHROMagar Candida, France) can be used as culture medium for the primary isolation and presumptive identification of organisms in cases where early diagnosis of infections is needed without doing PCR (31). CHROMagar Candida (CHROMagar Candida, France) can also support the growth of fungi, in some cases where the causative agent might be a mold, as reported by Beck-Sague and Jarvis (9). This has also been observed in our study. Sometimes we found a mixture of yeast and mold in a single specimen, so this ability of the medium to support the growth of mold can easily be utilized in the laboratory setting.
Conclusion
On the whole it was observed that as the CHROMagar Candida (CHROMagar Candida, France) gives a presumptive identification within 48 hours, preliminary antifungal treatment can be administered with confidence while the confirmed identification is being obtained. In resource-limited settings, availability of this type of media not only facilitates the provision of rapid patient care, but may also assist to control the rise in antifungal agent resistance by reducing the time taken for presumptive identification of the organism at species level to start the therapeutic regime.
We can conclude that use of fast and accurate diagnostic methods can help in rapid treatment of patients.
Acknowledgements
We are grateful to CHROMagar Candida, France and Biomerieux international for the generous donation of CHROMagar Candida and API 20C AUX used in this study, Dr. Uzair ul Ghani for his guidance, Prof. Dr. Riaz Ahmed Hashmi and administration of Jinnah University for Women, Karachi, Pakistan, for their financial support, and Ms. Fasiha Saeed, Lecturer, Department of Microbiology, Jinnah University for Women, Karachi for her technical support throughout the study.
Conflict of interest and funding
The authors have not received any funding or benefits from industry to conduct this study.
References
- 1.Fraser VJ, Jones M, Dunkel J, Storfer S, Medoff G, Dunagan WC. Candidemia in a tertiary care hospital: epidemiology, risk factors, and predictors of mortality. J Clin Infect Dis. 1992;15:415–21. doi: 10.1093/clind/15.3.414. [DOI] [PubMed] [Google Scholar]
- 2.Price MF, La Rocco MT, Gentry LO. Fluconazole susceptibilities of Candida species and distribution of species recovered from blood cultures over a 5-year period. J Antimicrob Agents Chemother. 1994;38:1422–4. doi: 10.1128/aac.38.6.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wingard JR. Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin Infect Dis. 1995;20:115–25. doi: 10.1093/clinids/20.1.115. [DOI] [PubMed] [Google Scholar]
- 4.Beck-Sague CM, Jarvis WR. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. J Infect Dis. 1993;167:1247–51. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 5.McNeil MM, Nash SL, Hajjeh RA, Phelan MA, Conn LA, Plikaytis BD, et al. Trends in mortality due to invasive mycotic diseases in the United States, 1980–1997. J Clin Infect Dis. 2001;33:641–7. doi: 10.1086/322606. [DOI] [PubMed] [Google Scholar]
- 6.Patterson TF. Invasive mycoses: management and unmet medical needs. Curr Opin J Infect Dis. 2001;14:669–71. doi: 10.1097/00001432-200112000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Raad II, Hachem RY, Herbrecht R, Graybill JR, Hare R, Corcoran G, et al. Posaconazole as salvage treatment of invasive fusariosis in patients with underlying hematologic malignancy and other conditions. J Clin Infect Dis. 2006;42:1398–403. doi: 10.1086/503425. [DOI] [PubMed] [Google Scholar]
- 8.Yasuda JM. An update on antifungal therapy: a focus on systemic agents for invasive fungal infections. Calif J Health Syst Pharm. 2001;13:4–12. [Google Scholar]
- 9.Beck-Sague C, Jarvis WR. National nosocomial infections surveillance system: secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. J Infect Dis. 1993;167:1247–51. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 10.Rangel-Frausto MS, Wiblin T, Blumberg HM, Saiman L, Patterson J, Rinaldi M, et al. National Epidemiology of Mycoses Survey (NEMIS): variations in rates of bloodstream infections due to Candida species in seven surgical intensive care units and six neonatal intensive care units. Clin Infect Dis. 1999;29:253–8. doi: 10.1086/520194. [DOI] [PubMed] [Google Scholar]
- 11.Pfaller Michael A. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. J Clin Infect Dis. 1996;22:S89–S94. doi: 10.1093/clinids/22.supplement_2.s89. [DOI] [PubMed] [Google Scholar]
- 12.Pfaller M, Cabezudo I, Koontz F, Bale M, Gingrich R. Predictive value of surveillance cultures for systemic infection due to Candida species. Eur J Clin Microbiol. 1987;6:628–33. doi: 10.1007/BF02013057. 37. [DOI] [PubMed] [Google Scholar]
- 13.Rex JH, Pfaller MA, Barry AL, Nelson PW, Webb CD NIAID Mycoses Study Group and the Candidemia Study Group. Antifungal susceptibility testing of isolates from a randomized, multicenter trial of fluconazole versus amphotericin B as treatment of non neutropenic patients with candidemia. Antimicrob Agents Chemother. 1995;39:40–4. doi: 10.1128/aac.39.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandford GR, Merz WG, Wingard JR, Charache P, Saral R. The value of fungal surveillance cultures as predictors of systemic fungal infections. J Infect Dis. 1980;142:503–9. doi: 10.1093/infdis/142.4.503. [DOI] [PubMed] [Google Scholar]
- 15.Magee BB, Magee PT. Recent advances in the genomic analysis of Candida albicans . Rev Iberoam Micol. 2005;22:187–93. doi: 10.1016/s1130-1406(05)70042-5. [DOI] [PubMed] [Google Scholar]
- 16.Pontion J, Ruckehl R, Clemons KV, Coleman DC, Grillot R, Guarro J, et al. Emerging pathogens. Med Mycol. 2000;38:225–36. doi: 10.1080/mmy.38.s1.225.236. [DOI] [PubMed] [Google Scholar]
- 17.Fridkin SK, Jarvis WR. Epidemiology of nosocomial fungal infections. J Clin Microbiol Rev. 1996;9:499–511. doi: 10.1128/cmr.9.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hospenthal DR, Murray CK, Rinaldi MG. The role of antifungal susceptibility testing in the therapy of candidiasis. J Diagn Microbiol Infect Dis. 2004;48:153–60. doi: 10.1016/j.diagmicrobio.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Freydiere AM, Guinet R, Boiron P. Yeast identification in the clinical microbiology laboratory: phenotypical methods. Med Mycol. 2001;39:9–33. doi: 10.1080/mmy.39.1.9.33. [DOI] [PubMed] [Google Scholar]
- 20.Murray CK, Beckius ML, Green JA, Hospenthal DR. Use of chromogenic medium in the isolation of yeasts from clinical specimens. J Med Microbiol. 2005;54:981–5. doi: 10.1099/jmm.0.45942-0. [DOI] [PubMed] [Google Scholar]
- 21.Maertens JA. History of the development of azole derivatives. J Clin Microbiol Infect. 2004;10:1–10. doi: 10.1111/j.1470-9465.2004.00841.x. [DOI] [PubMed] [Google Scholar]
- 22.Fenn JP, Segal H, Barland B, Denton D, Whisenant J, Chun H, et al. Comparison of updated Vitek yeast biochemical card and API 20C yeast identification systems. J Clin Microbiol. 1994;32:1184–7. doi: 10.1128/jcm.32.5.1184-1187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heelan JS, Sotomayor E, Coon K, D'Arezzo JB. Comparison of the rapid yeast plus panel with the API20C yeast system for identification of clinically significant isolates of Candida species. J Clin Microbiol. 1998;36:1443–5. doi: 10.1128/jcm.36.5.1443-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitch TT, Jacobs MR, McGinnis MR, Applebaum PC. Ability of RapID Yeast Plus System to identify 304 clinically significant yeasts within 5 hours. J Clin Microbiol. 1996;34:1069–71. doi: 10.1128/jcm.34.5.1069-1071.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marler JK, Eriquez LA. Comparison of the IDS RapID Yeast Plus System and the API 20C for the identification of medically important yeast; Proceedings of the 95th General Meeting of the American Society for Microbiology, Washington, DC; 1995. p. 73. abstr. C-418. [Google Scholar]
- 26.Schuffenecker I, Freydiere A, de Montclos H, Gille Y. Evaluation of four commercial systems for identification of medically important yeasts. Eur J Clin Microbiol Infect Dis. 1993;12:255–60. doi: 10.1007/BF01967255. [DOI] [PubMed] [Google Scholar]
- 27. http://www.chromagar.com.
- 28.Baumgartner C, Freydiere A, Gille Y. Direct identification and recognition of yeast species from clinical material by using Albicans ID and CHROMagar Candida plates. J Clin Microbiol. 1996;34:454–6. doi: 10.1128/jcm.34.2.454-456.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernal S, Mazuelos EM, Garcia M, Aller AI, Martinez MA, Gutierrez MJ. Evaluation of CHROMagar Candida medium for isolation and presumptive identification of species of Candida of clinical importance. Diagn Microbiol Infect Dis. 1996;24:201–4. doi: 10.1016/0732-8893(96)00063-6. [DOI] [PubMed] [Google Scholar]
- 30.Huang L, Chen C, Chou C, Lu J, Chi W, Lee W. A comparison of methods for yeast identification including CHROMagar Candida, vitek system YBC and a traditional biochemical method. Chin Med J. 2001;64:568–74. [PubMed] [Google Scholar]
- 31.Odds FC, Bernaerts R. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol. 1994;32:1923–9. doi: 10.1128/jcm.32.8.1923-1929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaller MA, Houston A, Coffmann S. Application of CHROMagar Candida for rapid screening of clinical specimens for Candida albicans, Candida tropicalis, Candida krusei, and Candida (Torulopsis) glabrata . J Clin Microbiol. 1996;34:58–6. doi: 10.1128/jcm.34.1.58-61.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willinger B, Manafi M. Evaluation of CHROMagar Candida for rapid screening of clinical specimens for Candida species. Mycoses. 1999;42:61–5. doi: 10.1046/j.1439-0507.1999.00406.x. [DOI] [PubMed] [Google Scholar]