Abstract
Acinetobacter baumannii (American Type Culture Collection strain 19606) acquires mutations in the pmrB gene during the in vitro development of resistance to colistin. The colistin-resistant strain has lower affinity for colistin, reduced in vivo fitness (competition index, .016), and decreased virulence, both in terms of mortality (0% lethal dose, 6.9 vs 4.9 log colony-forming units) and survival in a mouse model of peritoneal sepsis. These results may explain the low incidence and dissemination of colistin resistance in A. baumannii in clinical settings.
Acinetobacter baumannii is a gram-negative coccobacillus producing nosocomial infections such as pneumonias, bacteremias, urinary tract infections, surgical site infections, and meningitis, with an attributed mortality of 7.8%–23% [1]. The emergence of multidrug-resistant strains, impervious to most commonly used antibiotics, has thwarted current treatment [1]. Currently, colistin is one of the last treatments available. Despite its extensive use against A. baumannii [2], colistin-resistant outbreaks [3] are quite sporadic [4] when compared with the widespread dissemination of resistance for other antibiotics, such as carbapenems [1].
Recently, variation in the levels of 35 proteins between A. baumannii American Type Culture Collection (ATCC) strain 19606 and its in-vitro-derived colistin-resistant mutant, RC64 (colistin minimal inhibitory concentration, 2 and 64 μg/mL, respectively), was shown by differential proteomics [5]. This included changes in the expression of outer membrane proteins, chaperones, translation factors, and metabolic enzymes in RC64. In addition, this strain showed impaired in vitro fitness and longer duplication times compared with the parental strain [5].
Colistin resistance in gram-negative bacteria is most commonly due to decreased binding to the bacterial outer membrane because of lipopolysaccharide remodeling that is caused by changes in PhoPQ and PmrAB, both two-component regulatory systems, which results in a less anionic lipid A. A recent report addressing colistin resistance in A. baumannii strains demonstrated that mutations in the pmrA and pmrB genes were associated with colistin resistance [6]. Similar mutations were reported in Salmonella enterica [7] and Pseudomonas aeruginosa [8].
In this study, our goal was to test whether the impaired in vitro fitness described for RC64 [5] is maintained in animal models of infection, thereby linking lower virulence to colistin resistance, and to characterize PmrAB mutations in this strain.
METHODS AND RESULTS
A. baumannii ATCC 19606 and its derived colistin-resistant strain RC64 are described elsewhere [5].
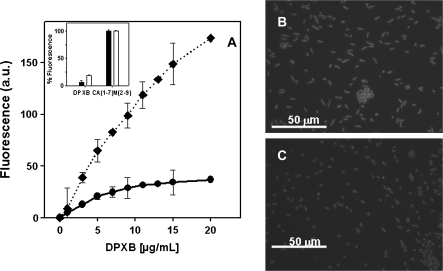
Binding of dansylated polymyxin B (DPXB) to both strains was monitored by increase of its fluorescence (λexc, 380 nm; λem, 485 nm; temperature, 37°C) in bacterial suspensions in phosphate-buffered saline (2 × 106 colony-forming units [CFUs]/mL) [9]. Lack of membrane permeabilization was tested by entrance of the vital dye SYTOX green (λexc, 485 nm; λem, 520 nm; temperature, 37°C), using 5 μmol/L cecropin A(1–7)-melittin(2–9) as a full permeabilization control. Fluorescence microscopy was performed with a Leica microscope at a concentration of 20 μg/mL, with bacteria incubated at the highest DPXB concentration tested after cell concentration by centrifugation (Figure 1). These results evidenced that the lower binding of DPXB to the RC64 strain compared with its parental ATCC 19606 strain was exclusively due to remodeling of the outer membrane, as the existence of additional intracellular binding sites was excluded by the absence of uptake of the vital dye SYTOX green for both strains by colistin, opposite the massive influx caused by the membrane-active CA(1–7)M(2–9), its positive control (Figure 1, inset).
Figure 1.
Differential binding of dansyl polymyxin B (DPXB) to bacterial cells of American Type Culture Collection (ATCC) strain 19606 and colistin-resistant mutant strain RC64 of Acinetobacter baumannii. A, Variation of DPXB binding with concentration bacterial density of 2 × 106 colony-forming units (CFUs)/mL (diamonds and dotted line, ATCC 19606; filled circles and solid line, RC64). The inset shows the inner membrane permeabilization as assessed by entrance of the vital dye SYTOX green, using 5 μmol/L of CA(1–7)M(2–9) as a positive control (100% permeabilization; black bars, ATCC 19606; white bars, RC64). B, Fluorescence microscopy of ATCC 19606 cells incubated with DPXB (20 μg/mL). C, Fluorescence microscopy of RC64 cells incubated with DPXB (20 μg/mL).
The in vitro competition index (CI) was determined from growth cultures that consisted of (1) ATCC 19606, (2) RC64, and (3) a mixed inoculum of equivalent CFUs from both strains. Bacteria at a concentration of 5 × 105 CFUs/mL were grown in 20 mL of Mueller-Hinton broth (Becton Dickinson Microbiology Systems). At 0, 12, 24, 36, 48, 60, and 72 h, 100-μL aliquots were taken and susceptible and resistant CFUs were counted by plating serial log dilutions on blood agar or Mueller-Hinton agar plus colistin (32 μg/mL; Sigma Chemical). The CI was defined as the number of RC64 CFUs recovered/number of ATCC 19606 CFUs recovered, divided by the number of RC64 CFUs inoculated/number of ATCC 19606 CFUs inoculated. Spontaneous reversion of RC64 into the susceptible phenotype was negligible during the time span of experiment.
For bacterial growth and CI experiments in an animal model of infection, 3 groups of 12 C57BL/6 female 18–20-g mice (University of Seville, Seville, Spain) were inoculated intraperitoneally with 7.9 log CFUs (the 100% lethal dose; see below) of each strain separately and with a mixed inoculum (50% of each strain). Subgroups of 3 mice were sacrificed at 2, 4, 8, and 24 h after inoculation. Spleens were aseptically removed, weighed, and homogenized (Stomacher 80 homogenizer; Tekmar), and CFUs were counted as above. To measure the CI, 9 additional mice were inoculated and processed at 24 h after inoculation. For these experiments, approval from the Ethics and Clinical Research Committee of the Virgen del Rocío University Hospitals were obtained.
By genomic mining, A. baumannii lacks PhoPQ; thus, activation of PmrAB is likely of utmost importance for developing colistin resistance [13]. In fact, mutations in the pmrA and pmrB genes leading to a gain-of-function phenotype were recently associated with colistin resistance in A. baumannii isolates [6]. We therefore characterized these genes by locus sequencing in both ATCC 19606 and RC64 strains. To this end, genomic DNA was obtained by resuspending a single colony in 25 μL of water and then lysing by incubation at 100°C for 10 min. After centrifugation, the genomic DNA in the supernatant was used to amplify pmrA and pmrB by polymerase chain reaction using PmrA-B forward and PmrA-B reverse primers (PmrA-B forward, ATGACAAAAATCTTGATGAT; PmrA-B reverse, TCACGCTCTTGTTTCATGTA). Amplified products were cloned into the pGEM-T Easy vector and sequenced using standard methods.
The nucleotide sequence of the A. baumannii RC64 pmrB gene was submitted to the EMBL database (GenBank accession no. GU797240).
In the in vitro experiments (Figure 2A), the growth rates of the 3 cultures were similar after 24 h, including in the experiments in which strains were grown together (CI, 1.05). However, at 72 h, the growth rate of RC64 was lower than that of ATCC 19606 (∼1 log difference), with a CI of .09. In the in vivo experiments, the CFUs in spleens from animals infected with either ATCC 19606 or RC64 (Figure 2B) differed by 1 log at 24 h; the difference increased to 2 log in the mixed infection group (CI, .016). This result suggests that in vivo conditions enhance the difference in fitness between the strains. Whereas the lower in vitro fitness of RC64 required at least 3 d to reach significance, in the in vivo experiments, only 1 d was required. Competition for nutrients and the stress produced by effector systems of the innate immune response in vivo may heighten the impaired metabolic efficiency and capacity to cope with stress for RC64 [5].
Figure 2.
In vitro (A) and in vivo (B) growth of American Type Culture Collection (ATCC) strain 19606 and colistin-resistant mutant strain RC64 of Acinetobacter baumannii, separately and in competition, and time of survival (evaluated every 4 h) with the 100% lethal dose of A. baumannii strains ATCC 19606 and RC64 (C). CFU, colony-forming unit.
The virulence of both strains was further compared in terms of mortality and lethal dose (Reed and Muench method) [10], as well as by measuring the survival time of the infected mice. For each strain, groups of 10 animals were infected intraperitoneally with an inoculum of 250 μL starting at 8.5 log CFUs/mL, and then the inocula were serially 10-fold diluted until 100% survival was attained. Bacteria were mixed with porcine mucin (Sigma) at 5% (weight per volume) final concentration prior to inoculation. Mortality with the ATCC 19606 strain at an inoculum of 8.5, 7.5, 6.5, and 5.5 log CFUs/mL was 100%, 80%, 20%, and 0%, respectively. With the RC64 strain, 100% mortality was observed with an inoculum of 8.5 log CFUs/mL, but no mortality was achieved with the rest of inocula. With these results, lethal doses of 100% (LD100), 50% (LD50), and 0% (LD0) were 7.9, 6.4, and 4.9 log CFUs, respectively, for ATCC 19606 and 7.9, 7.4, and 6.9 log CFUs, respectively, for RC64.
In animals inoculated with LD100 (7.9 log CFUs), the mean time to death for RC64 was twice that for ATCC 19606 (mean time to death [± SD], 38.4 ± 9.3 h vs 20.0 ± 0.0 h; P < .001; Student t test), (Figure 2C). This lower virulence associated with colistin resistance has also been described for S. enterica [11]. In our experiments, the lower virulence of RC64 is in tune with the lower expression of metabolic proteins and of the OmpA porin [5], which is involved in Acinetobacter species virulence [12].
In the resistant strain RC64, no mutations were found in pmrA, the response regulator element. In pmrB, the sensor kinase, mutations were found at positions R134C and A227V. The latter was previously reported in a colistin-resistant A. baumannii strain [6] and mapped inside the conserved histidine box motif [14] adjacent to H228, a putative acceptor for the phosphoryl group, and to A226, which is involved in histidine kinase dimerization [14]. A similar mutation in P. aeruginosa is also associated with a polymyxin-resistant phenotype [8]. The other mutation, R134C, was not previously described. This mutation is presumably not innocuous because of the chemical nature of the new side group and also because this arginine was conserved throughout the species of the genus Acinetobacter.
DISCUSSION
The lower in vivo bacterial fitness and decreased virulence of colistin-resistant A. baumannii showed in this study may suggest that the extensive use of colistin for the treatment of A. baumannii infections is a safe and consistently effective strategy from an evolutionary point of view. However, caution must be stressed in this sense. Many resistance mutations are initially maladaptive, but for most of these, compensatory mutations can ameliorate the fitness deficit. Ultimately, this process can lead to fixation of resistance mutations, even those with a high initial fitness cost. Constant exposure to the initial selective agent, such as colistin in A. baumannii, may prevent reversion of resistance, thus providing the time required for the resistant strain to re-adapt and for the resistance to thereby become fixed. Once this process is complete, reversion is very unlikely and resistance can spread without further selection, as occurs with Mycobacterium tuberculosis [15]. Thus, if compensatory mutations are possible, but simply have yet to occur, this could actually be the path by which colistin could be rendered useless for such infections, as extended use could provide the exact environment in which compensatory mutations could arise, become fixed, and then allow the spread and fixation of resistance mutations.
In summary, we have shown for the first time (to our knowledge) that resistance to colistin in A. baumannii is associated with lower in vivo bacterial fitness and decreased virulence, similar to other gram-negative pathogens. This may account for the low incidence of colistin resistance in the clinical setting.
Funding
This work was supported by the European Development Regional Fund “A way to achieve Europe” (ERDF); the Spanish Network for the Research in Infectious Diseases (REIPI RD06/0008), Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación, Spain; and the Autonomous government of Madrid (COMBACT S-BIO-0260/2006, L.R.).
References
- 1.Pachon J, Vila J. Treatment of multiresistant Acinetobacter baumannii infections. Curr Opin Investig Drugs. 2009;10:150–6. [PubMed] [Google Scholar]
- 2.Maviglia R, Nestorini R, Pennisi M. Role of old antibiotics in multidrug resistant bacterial infections. Curr Drug Targets. 2009;10:895–905. doi: 10.2174/138945009789108846. [DOI] [PubMed] [Google Scholar]
- 3.Valencia R, Arroyo LA, Conde M, et al. Nosocomial outbreak of infection with pan-drug-resistant Acinetobacter baumannii in a tertiary care university hospital. Infect Control Hosp Epidemiol. 2009;30:257–63. doi: 10.1086/595977. [DOI] [PubMed] [Google Scholar]
- 4.Livermore DM, Hill RL, Thomson H, et al. Antimicrobial treatment and clinical outcome for infections with carbapenem- and multiply-resistant Acinetobacter baumannii around London. Int J Antimicrob Agents. 2010;35:19–24. doi: 10.1016/j.ijantimicag.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Reyes M, Rodriguez-Falcon M, Chiva C, Pachon J, Andreu D, Rivas L. The cost of resistance to colistin in Acinetobacter baumannii: a proteomic perspective. Proteomics. 2009;9:1632–45. doi: 10.1002/pmic.200800434. [DOI] [PubMed] [Google Scholar]
- 6.Adams MD, Nickel GC, Bajaksouzian S, et al. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother. 2009;53:3628–34. doi: 10.1128/AAC.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun S, Negrea A, Rhen M, Andersson DI. Genetic analysis of colistin resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. 2009;53:2298–305. doi: 10.1128/AAC.01016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moskowitz SM, Ernst RK, Miller SI. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol. 2004;186:575–9. doi: 10.1128/JB.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore RA, Chan L, Hancock RE. Evidence for two distinct mechanisms of resistance to polymyxin B in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1984;26:539–45. doi: 10.1128/aac.26.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O′Reilly T, Cleeland R, Squires EL. Evaluation of antimicrobials in experimental animal infections. In: Lorian V, editor. Antibiotics in laboratory medicine. Baltimore, MD: Williams and Wilkins; 1996. pp. 604–765. [Google Scholar]
- 11.Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar typhimurium. Infect Immun. 2000;68:6139–46. doi: 10.1128/iai.68.11.6139-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi CH, Hyun SH, Lee JY, et al. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell Microbiol. 2008;10:309–19. doi: 10.1111/j.1462-5822.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- 13.Gunn JS. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 2008;16:284–90. doi: 10.1016/j.tim.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–54. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJ. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312:1944–6. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]