Abstract
Background. Otitis media, for which antibiotic treatment failure is increasingly common, is a leading pediatric public health problem.
Methods. In vitro and in vivo studies using the chinchilla model of otitis media were performed using a β-lactamase-producing strain of nontypeable Haemophilus influenzae (NTHi 86-028NP) and an isogenic mutant deficient in β-lactamase production (NTHi 86-028NP bla) to define the roles of biofilm formation and β-lactamase production in antibiotic resistance. Coinfection studies were done with Streptococcus pneumoniae to determine if NTHi provides passive protection by means of β-lactamase production, biofilm formation, or both.
Results. NTHi 86-028NP bla was resistant to amoxicillin killing in biofilm studies in vitro; however, it was cleared by amoxicillin treatment in vivo, whereas NTHi 86-028NP was unaffected in either system. NTHi 86-028NP protected pneumococcus in vivo in both the effusion fluid and bullar homogenate. NTHi 86-028NP bla and pneumococcus were both recovered from the surface-associated bacteria of amoxicillin-treated animals; only NTHi 86-028NP bla was recovered from effusion.
Conclusions. Based on these studies, we conclude that NTHi provides passive protection for S. pneumoniae in vivo through 2 distinct mechanisms: production of β-lactamase and formation of biofilm communities.
Otitis media (OM) is one of the leading public health problems in pediatrics, with most children experiencing at least 1 episode by 3 years of age and 40% of older children having 6 or more total episodes [1]. Common causes of OM include nontypeable Haemophilus influenzae (NTHi), Streptococcus pneumoniae (pneumococcus), and Moraxella catarrhalis; NTHi and pneumococcus account for ∼75% of infections [2]. In recent years, epidemiologic studies have shown that co-infections with multiple bacterial species are an important part of both acute and chronic OM, particularly co-infections with NTHi and pneumococcus [3–5].
As with many upper airway infections, treatment failure and antibiotic-resistant organisms are common problems in OM, with failure rates as high as 50% in some populations [6–9]. The majority of strains of both NTHi and M. catarrhalis produce a β-lactamase [10–12]. β-Lactam resistance among pneumococcal strains is much less common, although some strains (∼20%) have developed resistance through mutation of the penicillin-binding proteins [10].
Common theories proposed to explain OM treatment failure involving antibiotic-susceptible bacterial strains are the formation of biofilm communities and passive protection by other bacterial pathogens that produce a β-lactamase [13–16]. Bacteria in biofilms are known to be more resistant to antibiotic killing than are planktonic bacteria [17–20]. Previous studies have also shown that M. catarrhalis can passively protect pneumococcus from antibiotic killing in a mouse pneumonia model and in biofilms in vitro, which is likely mediated by β-lactamase as protection was abolished by β-lactamase inhibitor [21, 22].
In this study, we used a β-lactamase-producing strain of NTHi and an isogenic mutant deficient in β-lactamase to distinguish between the roles of biofilm formation and β-lactamase production in NTHi antibiotic resistance. In addition, these strains were utilized to determine whether NTHi provides passive protection for pneumococcus and, if so, the mechanism behind this protection.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
Nontypeable H. influenzae 86-028NP (NTHi 86-028NP) is an OM isolate that has been fully sequenced and is known to cause OM featuring biofilms in the chinchilla infection model [23–28]. Bacteria were grown on brain heart infusion (BHI) agar supplemented with hemin (ICN Biochemicals) and nicotinamide adenine dinucleotide (Sigma) and 3 μg/mL of vancomycin (Sigma). S. pneumoniae TIGR4 is a well-studied clinical isolate for which a complete genomic sequence is available, and which we have recently shown to form biofilms during experimental OM infection [29, 30]. Pneumococci were grown on trypticase soy agar (BD) with 5% sheep blood (Hemostat Laboratories) or in supplemented BHI (sBHI) medium with 10% horse serum.
Generation of β-Lactamase-Deficient Mutant (NTHi 86-028NP bla)
A ∼2-kb DNA fragment containing the β-lactamase triethylene melamine open reading frame (allele designated NTHi 2055) was amplified from H. influenzae 86-028NP genomic DNA using βlac forward (TGG TTA CGC TCG GGT CTC AA) and βlac reverse (ATG GCA CAA GTT ACA CGA TTC AA) primers with an annealing temperature of 55.9°C and extension time of 2.5 min. The fragment was ligated into pCR2.1 (Invitrogen) and transformed into Escherichia coli according to the manufacturer’s instructions to generate plasmid pCR-βlac. The pCR-βlac was digested with HincII, a unique site within the β-lactamase gene, then dephosphorylated with calf intestinal phosphatase (New England Biolabs). This fragment was then ligated with a chloramphenicol resistance cassette [31] cut with SmaI, and transformed into E. coli, generating pCR-βlacCm. The pCR-βlacCm was linearized with Not1 and introduced into NTHi 86-028NP via natural transformation, as described previously [23, 24], to generate NTHi 86-028NP bla::Cm (NTHi 86-028NP bla). Colonies that grew on sBHI containing chloramphenicol (1.5 μg/mL) were screened using primers flanking the site of insertion, forward (GAT GCT GAA GAT CAG TTG GG) and reverse (GTA TGG CTT CAT TCA GCT CC) with an annealing temperature of 52°C and extension time of 2.5 min.
Amoxicillin Susceptibility
Bacteria were harvested from overnight sBHI plates and resuspended in phosphate-buffered saline (PBS) to a density of ∼108 colony-forming units (CFUs)/mL as determined by optical density. To determine planktonic minimum inhibitory concentrations (MICs), ∼106 CFUs of NTHi 86-028NP or NTHi 86-028NP bla were suspended in 5 mL of sBHI with varying concentrations of amoxicillin (Sigma). Cultures were grown for 16–20 h at 37°C at 150 rpm in a shaking incubator (New Brunswick C24 Incubator Shaker). The turbidity of the medium was used to determine bacterial growth and survival. To determine biofilm susceptibility to amoxicillin, ∼107 CFUs of NTHi 86-028NP or NTHi 86-028NP bla were plated in 1.5 mL of sBHI in 24-well plates (Falcon) and incubated at 37°C and 5% CO2. After 24 h, the supernatant was removed from each well and replaced with 1.5 mL of sBHI containing various concentrations of amoxicillin (Sigma). After 48 h, the supernatant was removed and the surface-attached bacteria (biofilm) were collected by scraping, resuspended in .2 mL of sterile PBS, diluted, and plated for bacterial counts.
Biofilm Protection Assay
NTHi 86-028NP or NTHi 86-028NP bla (∼107 CFUs) was plated in each well of a 24-well plate (Falcon) in a total volume of 1.5 mL of sBHI and incubated at 37°C and 5% CO2. After 24 h, supernatants were removed and replaced with 1.5 mL of sBHI plus 10% horse serum. S. pneumoniae TIGR4 (∼107 CFUs) was then added to co-infection and S. pneumoniae–alone wells. After 48 h, the supernatant was removed and replaced with sBHI with or without amoxicillin. After 72 h, the supernatant was removed and the surface-attached bacteria (biofilm) were collected by scraping, resuspended in .2 mL of PBS, diluted, and plated for bacterial counts.
Chinchilla Infections
Healthy adult chinchillas (Chinchilla lanigera) were purchased from Rauscher’s chinchilla ranch and allowed to acclimate to the vivarium for 1 week prior to infection. All animals were examined by otoscopy prior to infection, and none had any clinical signs of middle ear infection or other overt disease. The chinchilla infection protocols were performed essentially as described elsewhere [23, 24, 28, 29]. On day 0, NTHi 86-028NP and NTHi 86-028NP bla were harvested from a plate, and a freezer stock of known CFUs/mL of S. pneumoniae TIGR4 was thawed; both were diluted using sterile PBS, and bacterial density was confirmed by plate count. Chinchillas (3–5 animals per group per time point) were anesthetized with isofluorane and inoculated via transbullar injection with .1 mL of bacterial suspension containing NTHi 86-028NP, NTHi 86-028NP bla, S. pneumoniae TIGR4, NTHi 86-028NP and S. pneumoniae TIGR4 or NTHi 86-028NP bla and S. pneumoniae TIGR4, as indicated for each experimental group. Infectious doses ranged from 102 to 104 CFUs, as indicated for each experiment. On days 4, 5, and 6, groups of animals were injected with either 20 μg of amoxicillin (.1 mL of 200 μg/mL; Sigma) or .1 mL of sterile PBS directly in the middle ear. Oral administration of the antibiotic was contraindicated due to the risk of developing a Clostridium difficile infection [32]. All animals were euthanized 7 d after infection. Animals exhibiting overt symptoms of systemic disease were euthanized prior to day 7. After euthanasia, the superior bullae were opened to expose the middle ear cavity as described elsewhere [24], and the presence of visible biofilm formation was assessed. If present, middle ear effusion fluids were collected. The middle ear cavity was lavaged with 1 mL of sterile PBS. Effusion and lavage fluids were combined, serially diluted, and assessed by plate count. For animals that received both bacteria, fluid was plated on 2 separate plates, 1 selective for NTHi (sBHI plus vancomycin) and 1 selective for S. pneumoniae (TSA plus sheep blood plus gentamicin). Middle ear bullae were aseptically removed and homogenized using a PowerGen 1000 homogenizer (Fisher Scientific); the bullar homogenates were plated to assess tissue-associated bacterial load. All of the chinchilla infection protocols were approved by the Wake Forest University Health Sciences Institutional Animal Care and Use Committee.
RESULTS
Generation and Confirmation of NTHi β-Lactamase Mutant
To define the role of β-lactamase in NTHi antibiotic resistance, an isogenic β-lactamase-deficient mutant of NTHi 86-028NP was generated (NTHi 86-028NP bla) by insertional disruption of the NTHi 2055 gene (National Center for Biotechnology Information Reference Sequence NC_007146) with a chloramphenicol resistance cassette. No difference in growth was observed between NTHi 86-028NP and NTHi 86-028NP bla in the absence of antibiotic (data not shown). In addition, no difference in in vitro biofilm formation was observed between the 2 strains in terms of biomass, mean thickness, maximum thickness, or surface-to-biovolume ratio, as determined by COMSTAT (MATLAB, Version 5.1) analysis of confocal laser scanning microscopy Z-series of 24-h biofilms (Supplementary Figure 1). To confirm the β-lactamase-negative phenotype of the mutant, the MIC of amoxicillin (a β-lactam antibiotic) for both NTHi 86-028NP and NTHi 86-028NP bla was determined. The MIC for NTHi 86-028NP was <32.0 μg/mL of amoxicillin (data not shown). The MIC of NTHi 86-028NP bla was <1.0 μg/mL, <32-fold that of the parental strain (data not shown). The inactivation of the β-lactamase gene rendered an amoxicillin-resistant strain of NTHi amoxicillin susceptible, confirming the phenotype of NTHi 86-208NP bla.
Role of β-Lactamase in NTHi Biofilm Antibiotic Resistance In Vitro
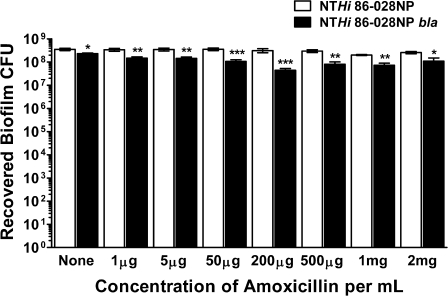
To define the role of β-lactamase in NTHi antibiotic resistance in biofilm, the susceptibility of NTHi 86-028NP and NTHi 86-028NP bla biofilms to amoxicillin was assessed. Various concentrations of amoxicillin were added for 24 h to preformed NTHi biofilms of each strain (0–2 mg/mL). NTHi 86-028NP was resistant to killing at all concentrations of amoxicillin tested (Figure 1). Interestingly, although NTHi 86-028NP bla was much more susceptible than NTHi 86-028NP to amoxicillin killing in planktonic culture (MIC 32-fold less), biofilms formed by this strain were also resistant to all concentrations of amoxicillin tested (Figure 1). There was a significant decrease in the recovered CFUs of NTHi 86-028NP bla from biofilms compared with the parental strain at all antibiotic concentrations tested; however, this difference was <1 log and ∼108 CFUs were still recovered at all concentrations tested. Based on these data, the production of a β-lactamase is necessary for resistance of NTHi to amoxicillin in planktonic culture, but it does not appear to be required for biofilm resistance in vitro.
Figure 1.
Antibiotic susceptibility of nontypeable Haemophilus influenzae (NTHi) in vitro biofilms. Varying concentrations of amoxicillin (0–2 mg/mL) were added to preformed NTHi 86-028NP (white bars) and NTHi 86-028NP bla (black bars) biofilms. The amounts of amoxicillin noted on the x-axis indicate the concentration per milliliter added to each well. After 24 h, biofilms were scraped, serially diluted, and plated for bacterial counts. Bars represent the mean (± standard error of the mean). The graph represents data combined from 3 experiments (8–9 replicates total). Statistical significance was assessed by Mann-Whitney nonparametric analysis. *P < .05; **P < .005; ***P < .001. CFU, colony-forming unit.
Role of β-Lactamase in NTHi Amoxicillin Resistance in Chinchilla Model of Otitis Media
Whereas the previous experiment showed that β-lactamase production was not necessary for biofilm resistance to amoxicillin in vitro, we wished to confirm this result in vivo using the chinchilla model of experimental OM. NTHi 86-028NP establishes a chronic infection in the middle ears of chinchillas, with biofilm formation occuring by day 3 in most animals [23, 28]. On day 0, ∼103 CFUs of NTHi 86-028NP or NTHi 86-028NP bla were inoculated directly into the middle ears of chinchillas via transbullar injection. On days 4, 5, and 6 after infection, 20 μg of amoxicillin or sterile PBS was injected directly into the middle ear. The concentration of amoxicillin in the middle ear following oral antibiotic treatment in children is 1–8 μg/mL [6]. The dose in our experiment is a higher concentration than these bacteria would normally experience, allowing for a stringent test of NTHi resistance in vivo. On day 7, all of the animals were euthanized, and the effusion and bullae were harvested and plated for bacterial counts.
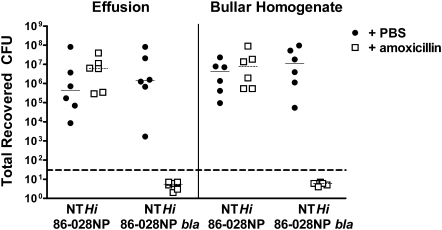
In animals infected with NTHi 86-028NP, no difference was shown in recovered CFUs from animals treated with amoxicillin or PBS in either the effusion (planktonic bacteria) or bullar homogenate (tissue-associated/biofilm bacteria) (Figure 2). This confirms the in vitro results, where a β-lactamase-producing strain of NTHi was resistant to amoxicillin killing. However, in animals infected with the β-lactamase-deficient NTHi 86-028NP bla, no bacteria were detectable in either the effusion or the bullar homogenate of the group treated with amoxicillin, whereas the group treated with PBS had bacterial counts equivalent to those of animals infected with the parental strain (Figure 2). These results demonstrate that β-lactamase production is required for NTHi resistance to amoxicillin in the chinchilla model of OM. Biofilm formation in the PBS-treated animals was within the normal range seen at this time postinfection for animals infected with either the parental strain (5 of 6) or the β-lactamase-deficient mutant (3 of 6) of NTHi. Based on these results, we conclude that either biofilm formation by NTHi is not protective in this model, or the biofilms formed by NTHi 86-028NP bla were not sufficient to provide protection against this concentration of amoxicillin.
Figure 2.
Nontypeable Haemophilus influenzae (NTHi) amoxicillin resistance in chinchilla model of otitis media. Chinchillas were infected with either ∼103 colony-forming units (CFUs) of NTHi 86-028NP or NTHi 86-028NP bla on day 0. On days 4, 5, and 6, animals were treated with either 20 μg of amoxicillin (white squares) or sterile phosphate-buffered saline (PBS) (black circles) directly into the middle ear. All animals were harvested on day 7 after infection. The left half of the graph represents counts from the effusion (planktonic bacteria); the right half is from bullar homogenates (biofilm/tissue-associated bacteria). The long dashed line represents the limit of detection. The short lines represent the mean CFUs for each group.
NTHi Passive Protection of S. pneumoniae In Vitro
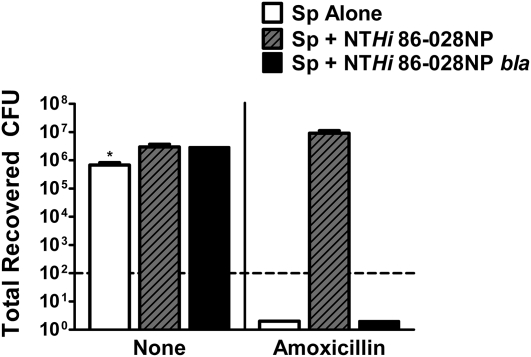
Because NTHi and S. pneumoniae are frequently found together in the middle ear, we wanted to determine whether NTHi could provide passive protection for pneumococcus against amoxicillin. S. pneumoniae is extremely susceptible to killing by β-lactam antibiotics [33]. Both S. pneumoniae planktonic and biofilm bacteria were completely killed by as little as .064 μg/mL of amoxicillin in vitro (data not shown and Figure 3). To determine whether NTHi provides passive protection for S. pneumoniae within a biofilm in vitro, biofilms containing NTHi 86-028NP and S. pneumoniae were treated with amoxicillin. NTHi 86-028NP completely protected pneumococcus from amoxicillin killing, with no difference in recovered pneumococcal CFUs between wells treated with amoxicillin and those treated with medium alone (Figure 3). This passive protection extended to at least 500 μg/mL of amoxicillin (data not shown). Interestingly, although NTHi 86-028NP bla viability was not affected by treatment with amoxicillin and its biofilms have a similar structure to that of the parental strain (Supplementary Figure 1), NTHi 86-028NP bla provided no protection for pneumococcus with as little as .064 μg/mL of amoxicillin (Figure 3). These data clearly demonstrate that NTHi provides passive protection for pneumococcus in vitro within a biofilm through the production of a β-lactamase.
Figure 3.
Nontypeable Haemophilus influenzae (NTHi) passive protection of Streptococcus pneumoniae in vitro. Medium alone or .064 μg/mL of amoxicillin was added to preformed biofilms containing S. pneumoniae (Sp) alone (white bars), S. pneumoniae and NTHi 86-028NP (gray bars with black lines), or S. pneumoniae and NTHi 86-028NP bla (black bars). Twenty-four hours after the addition of antibiotics, the biofilms were scraped, serially diluted, and plated for S. pneumoniae bacterial counts. Bars are the mean (±standard error of the mean) of quadruplicate wells. The graph is representative of 3 independent experiments.
NTHi Passive Protection of S. pneumoniae in the Chinchilla Model of Experimental Otitis Media
To determine whether NTHi provides protection against amoxicillin killing for pneumococcus in vivo, and whether this protection was dependent on the production of β-lactamase, a co-infection was performed using the chinchilla model. It has been previously established by our laboratory that NTHi and pneumococcus establish a chronic co-infection in this model with the formation of very large biofilm communities (larger than with either bacterial species alone) that contain both bacterial species [34]. Chinchillas were infected with either NTHi 86-028NP and pneumococcus, NTHi 86-028NP bla and pneumococcus, or pneumococcus alone directly into the middle ear. The same infection/treatment timeline and doses were used as in the previous chinchilla experiment (Figure 2). In brief, animals were infected on day 0, treated with 20 μg of amoxicillin or PBS on days 4, 5, and 6, and euthanized and processed on day 7.
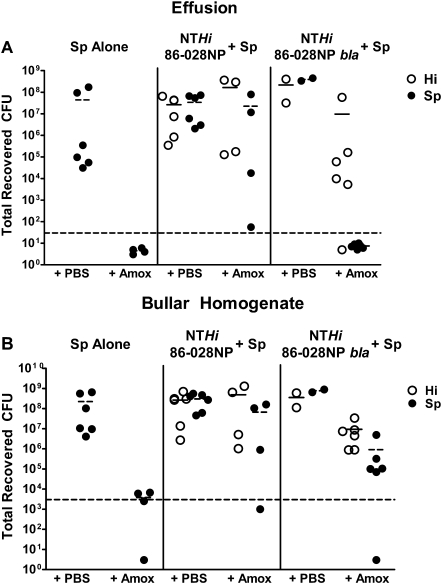
In the animals infected with pneumococcus alone, ∼108 CFUs were recovered from both the effusion (planktonic bacteria) and bullar homogenates (biofilm/tissue-associated bacteria) of PBS-treated animals (Figures 4A and 4B). In contrast, no bacteria were detectable in the effusion, and recovered CFUs were right at the limit of detection in bullar homogenates of animals infected with pneumococcus alone and treated with amoxicillin (Figures 4A and 4B). This confirms that the dose of amoxicillin given essentially clears the infection in animals given pneumococcus alone.
Figure 4.
Nontypeable Haemophilus influenzae (NTHi) passive protection of Streptococcus pneumoniae in chinchilla model of otitis media. Chinchillas were infected with either ∼103 colony-forming units (CFUs) of NTHi 86-028NP and ∼103 CFUs of S. pneumoniae (NTHi 86-028NP + Sp), ∼103 CFUs of NTHi 86-028NP bla and ∼103 CFUs of S. pneumoniae (NTHi 86-028NP bla + Sp), or 102 CFUs of S. pneumoniae alone (Sp alone) on day 0. On days 4, 5, and 6, animals were treated with either 20 μg of amoxicillin (amox) or sterile phosphate-buffered saline (PBS) directly into the middle ear. All animals were harvested on day 7 after infection. The effusion (A) and bullar homogenate (B) were serially diluted and plated for bacterial counts. The white circles represent the recovered CFUs of NTHi, and the black circles represent the recovered CFUs of S. pneumoniae. The long dashed line represents the limit of detection. The short solid lines represent the mean CFUs of NTHi in each group, and the short dashed lines represent the mean CFUs of pneumococcus in each group.
In animals that were co-infected with NTHi 86-028NP and pneumococcus and treated with PBS, equivalent numbers of both bacterial species were recovered from both the effusion and bullar homogenate, as expected (Figures 4A and 4B). Interestingly, in animals in this group that were treated with amoxicillin, pneumococcus was recovered from all 4 effusions and 3 of 4 bullar homogenates, with a mean of ∼107 CFUs recovered (Figures 4A and 4B). The recovered CFUs of NTHi were equivalent to those of PBS-treated animals (Figures 4A and 4B). These results are similar to what was seen in vitro (Figure 3) and clearly demonstrate that NTHi passively protects pneumococcus in the chinchilla OM model.
In animals that were co-infected with the β-lactamase-deficient mutant of NTHi and pneumococcus and treated with PBS, equivalent numbers of both species were recovered from the effusion and bullar homogenate, as seen with the parental strain (Figures 4A and 4B). In the animals in this group that were treated with amoxicillin, no pneumococcus was detected in the effusion; however, NTHi 86-028NP bla was recovered from the effusion in 5 of 6 ears, with a mean of ∼107 CFUs (Figures 4A and 4B). In addition, NTHi 86-028NP bla was recovered from the bullar homogenate of 6 of 6 ears and pneumococcus was recovered from the bullar homogenate of 5 of 6 ears in the amoxicillin-treated animals (Figures 4A and 4B). This is in stark contrast to the results in Figure 2, where animals infected with NTHi 86-028NP bla alone and treated with amoxicillin had no detectable bacteria in either the effusion or bullar homogenate. This is also contrary to what was seen in the in vitro passive protection assay (Figure 3), where NTHi 86-028NP bla provided no protection for pneumococcus. These data indicate that although β-lactamase production may be necessary for passive protection in vitro, it is not required for NTHi passive protection of pneumococcus against amoxicillin killing in vivo. In addition, these studies indicate that the presence of pneumococcus in the middle ear may afford protection for NTHi, even though pneumococcus is very susceptible to amoxicillin killing.
DISCUSSION
As antibiotic treatment failure becomes increasingly common, it is important to understand bacterial mechanisms of antibiotic resistance so that this knowledge can be applied to improve treatment. It is known that biofilm formation and the production of β-lactamase both contribute to NTHi antibiotic resistance; however, the contribution of each during an infection and the role in passive protection of other bacterial species had not been elucidated.
We have shown that, in vitro, NTHi 86-028NP and NTHi 86-028NP bla biofilms are both resistant to amoxicillin killing at all concentrations tested, 0–2 mg/mL (Figure 1). Despite this, only NTHi 86-028NP was able to passively protect pneumococcus from amoxicillin killing in biofilms in vitro (Figure 3). When the antibiotic resistance of NTHi 86-028NP was tested in the chinchilla model, equal numbers of CFUs were recovered from the effusion (planktonic bacteria) and bullar homogenate (biofilm/tissue-associated bacteria) regardless of whether the animal was treated with PBS or amoxicillin (Figure 2). However, NTHi 86-028NP bla was cleared from both the effusion and bullar homogenate of every animal. It is possible that NTHi 86-028NP bla did not form sufficient biofilms to protect against the continuous, large doses of amoxicillin given. If this were the case, NTHi 86-028NP bla might persist if a lower antibiotic concentration was given, or if biofilm formation was increased. Alternatively, biofilm formation may not be fully protective in vivo. Taken together, these data indicate that although the production of β-lactamase is not required for NTHi antibiotic resistance in vitro, it is necessary for in vivo NTHi resistance and in vitro passive protection of pneumococcus.
In this study, we also demonstrated that NTHi provides passive protection for pneumococcus in the chinchilla model (Figure 4). When co-infected with NTHi 86-028NP, pneumococcus was recovered from both the bullar homogenate and effusion of the majority of animals. Interestingly, when co-infected with NTHi 86-028NP bla, both NTHi and pneumococcus were recovered from the bullar homogenate, but only NTHi was recovered from the effusion (Figure 4). These data point toward 2 separate mechanisms of NTHi protection of pneumococcus from amoxicillin killing in vivo. The first is through the production of β-lactamase, which provides strong protection in both the effusion and bullar homogenate. The second mechanism is through the formation of biofilm communities. As mentioned previously, NTHi and pneumococcus form a much larger biofilm together than either bacterium does on its own [34]. This larger mixed biofilm appears to have provided protection against amoxicillin killing for both pneumococcus and NTHi 86-028NP bla, both of which were cleared when inoculated on their own and treated with amoxicillin. In contrast, prior work has shown a lack of protection by a β-lactamase-producing strain of NTHi in a rat experimental OM model [35]. Potential differences in the 2 studies that could explain the differences seen are the route and dose of amoxicillin and the animal model used. In the previous study, amoxicillin was added to the animals’ water for oral ingestion and reached a mean serum concentration of ∼4 μg/mL. Although that is more clinically relevant than our method of delivery, giving the antibiotic directly in the middle ear allowed for a consistent concentration of drug in the middle ear. Additionally, the concentration of amoxicillin in our study was much higher, 20 μg, and should provide a more stringent test of protection than the lower dose achieved in the above-mentioned study. The most important difference in the studies is that biofilm formation occurs in the majority of chinchillas with OM, whereas it is unclear if biofilms are formed in the rat model of OM. This could explain the differences in protection seen between the 2 studies and would indicate an even larger role for biofilm formation in passive protection.
Supplementary Data
Supplementary data are available at http://jid.oxfordjournals.org/ online.
Funding
This work was supported by the National Institutes of Health, National Institute on Deafness and Other Communication Disorders (grant numbers DC007444 and DC10051 to W.E.S); and the National Institute of Allergy and Infectious Diseases (grant number T32A107401 to K.E.D.W.).
Acknowledgments
We gratefully acknowledge the expert technical assistance of Gayle Foster. We also thank Sean Reid for providing helpful critique on this work and comments on the manuscript.
References
- 1.Rovers MM, Schilder AG, Zielhuis GA, Rosenfeld RM. Otitis media. Lancet. 2004;363:465–73. doi: 10.1016/S0140-6736(04)15495-0. [DOI] [PubMed] [Google Scholar]
- 2.Benninger MS. Acute bacterial rhinosinusitis and otitis media: changes in pathogenicity following widespread use of pneumococcal conjugate vaccine. Otolaryngol Head Neck Surg. 2008;138:274–8. doi: 10.1016/j.otohns.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Leibovitz E, Piglansky L, Raiz S, et al. Bacteriologic and clinical efficacy of oral gatifloxacin for the treatment of recurrent/nonresponsive acute otitis media: an open label, noncomparative, double tympanocentesis study. Pediatr Infect Dis J. 2003;22:943–9. doi: 10.1097/01.inf.0000095468.89866.14. [DOI] [PubMed] [Google Scholar]
- 4.Leibovitz E, Piglansky L, Raiz S, Press J, Leiberman A, Dagan R. Bacteriologic and clinical efficacy of one day vs. three day intramuscular ceftriaxone for treatment of nonresponsive acute otitis media in children. Pediatr Infect Dis J. 2000;19:1040–5. doi: 10.1097/00006454-200011000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Hendolin PH, Markkanen A, Ylikoski J, Wahlfors JJ. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J Clin Microbiol. 1997;35:2854–8. doi: 10.1128/jcm.35.11.2854-2858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wald ER, Nash D, Eickhoff J. Effectiveness of amoxicillin/clavulanate potassium in the treatment of acute bacterial sinusitis in children. Pediatrics. 2009;124:9–15. doi: 10.1542/peds.2008-2902. [DOI] [PubMed] [Google Scholar]
- 7.Courter JD, Baker WL, Nowak KS, et al. Increased clinical failures when treating acute otitis media with macrolides: a meta-analysis. Ann Pharmacother. 2010;44:471–8. doi: 10.1345/aph.1M344. [DOI] [PubMed] [Google Scholar]
- 8.Morris PS, Gadil G, McCallum GB, et al. Single-dose azithromycin versus seven days of amoxycillin in the treatment of acute otitis media in Aboriginal children (AATAAC): a double blind, randomised controlled trial. Med J Aust. 2010;192:24–9. doi: 10.5694/j.1326-5377.2010.tb03396.x. [DOI] [PubMed] [Google Scholar]
- 9.Pichichero ME. Recurrent and persistent otitis media. Pediatr Infect Dis J. 2000;19:911–6. doi: 10.1097/00006454-200009000-00034. [DOI] [PubMed] [Google Scholar]
- 10.Harrison CJ, Woods C, Stout G, Martin B, Selvarangan R. Susceptibilities of Haemophilus influenzae, Streptococcus pneumoniae, including serotype 19A, and Moraxella catarrhalis paediatric isolates from 2005 to 2007 to commonly used antibiotics. J Antimicrob Chemother. 2009;63:511–9. doi: 10.1093/jac/dkn538. [DOI] [PubMed] [Google Scholar]
- 11.Hoban D, Felmingham D. The PROTEKT surveillance study: antimicrobial susceptibility of Haemophilus influenzae and Moraxella catarrhalis from community-acquired respiratory tract infections. J Antimicrob Chemother. 2002;50(suppl S1):49–59. doi: 10.1093/jac/dkf810. [DOI] [PubMed] [Google Scholar]
- 12.Tristram S, Jacobs MR, Appelbaum PC. Antimicrobial resistance in Haemophilus influenzae. Clin Microbiol Rev. 2007;20:368–89. doi: 10.1128/CMR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart PS, William Costerton J. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–8. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 14.Wardle JK. Branhamella catarrhalis as an indirect pathogen. Drugs. 1986;31(suppl 3):93–6. doi: 10.2165/00003495-198600313-00020. [DOI] [PubMed] [Google Scholar]
- 15.Brook I. The concept of indirect pathogenicity by beta-lactamase production, especially in ear, nose and throat infection. J Antimicrob Chemother. 1989;24(suppl B):63–72. doi: 10.1093/jac/24.suppl_b.63. [DOI] [PubMed] [Google Scholar]
- 16.Brook I. Direct and indirect pathogenicity of beta-lactamase-producing bacteria in mixed infections in children. Crit Rev Microbiol. 1989;16:161–80. doi: 10.3109/10408418909104470. [DOI] [PubMed] [Google Scholar]
- 17.Anderson GG, O’Toole GA. Innate and induced resistance mechanisms of bacterial biofilms. Curr Top Microbiol Immunol. 2008;322:85–105. doi: 10.1007/978-3-540-75418-3_5. [DOI] [PubMed] [Google Scholar]
- 18.Kaji C, Watanabe K, Apicella MA, Watanabe H. Antimicrobial effect of fluoroquinolones for the eradication of nontypeable Haemophilus influenzae isolates within biofilms. Tohoku J Exp Med. 2008;214:121–8. doi: 10.1620/tjem.214.121. [DOI] [PubMed] [Google Scholar]
- 19.Slinger R, Chan F, Ferris W, et al. Multiple combination antibiotic susceptibility testing of nontypeable Haemophilus influenzae biofilms. Diagn Microbiol Infect Dis. 2006;56:247–53. doi: 10.1016/j.diagmicrobio.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Starner TD, Zhang N, Kim G, Apicella MA, McCray PB., Jr Haemophilus influenzae forms biofilms on airway epithelia: implications in cystic fibrosis. Am J Respir Crit Care Med. 2006;174:213–20. doi: 10.1164/rccm.200509-1459OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budhani RK, Struthers JK. Interaction of Streptococcus pneumoniae and Moraxella catarrhalis: investigation of the indirect pathogenic role of beta-lactamase-producing moraxellae by use of a continuous-culture biofilm system. Antimicrob Agents Chemother. 1998;42:2521–6. doi: 10.1128/aac.42.10.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hol C, Van Dijke EE, Verduin CM, Verhoef J, van Dijk H. Experimental evidence for Moraxella-induced penicillin neutralization in pneumococcal pneumonia. J Infect Dis. 1994;170:1613–6. doi: 10.1093/infdis/170.6.1613. [DOI] [PubMed] [Google Scholar]
- 23.Hong W, Mason K, Jurcisek JA, Novotny LA, Bakaletz LO, Swords WE. Phosphorylcholine decreases early inflammation and promotes the establishment of stable biofilm communities of nontypeable Haemophilus influenzae strain 86-028NP in a chinchilla model of otitis media. Infect Immun. 2007;75:958–65. doi: 10.1128/IAI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong W, Pang B, West-Barnette S, Swords WE. Phosphorylcholine expression by nontypeable Haemophilus influenzae correlates with maturation of biofilm communities in vitro and in vivo. J Bacteriol. 2007;189:8300–7. doi: 10.1128/JB.00532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jurcisek JA, Bakaletz LO. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol. 2007;189:3868–75. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jurcisek JA, Bookwalter J, Baker B, et al. The PilA protein of nontypeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol Microbiol. 2007;65:1288–99. doi: 10.1111/j.1365-2958.2007.05864.x. [DOI] [PubMed] [Google Scholar]
- 27.Jurcisek JA, Greiner L, Watanabe H, Zaleski A, Apicella MA, Bakaletz LO. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun. 2005;73:3210–8. doi: 10.1128/IAI.73.6.3210-3218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong W, Juneau RA, Pang B, Swords WE. Survival of bacterial biofilms within neutrophil extracellular traps promotes nontypeable Haemophilus influenzae persistence in the chinchilla model for otitis media. J Innate Immun. 2009;1:215–24. doi: 10.1159/000205937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reid SD, Hong W, Dew KE, et al. Streptococcus pneumoniae forms surface-attached communities in the middle ear of experimentally infected chinchillas. J Infect Dis. 2009;199:786–94. doi: 10.1086/597042. [DOI] [PubMed] [Google Scholar]
- 30.Tettelin H, Nelson KE, Paulsen IT, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 31.Whitby PW, Morton DJ, Stull TL. Construction of antibiotic resistance cassettes with multiple paired restriction sites for insertional mutagenesis of Haemophilus influenzae. FEMS Microbiol Lett. 1998;158:57–60. doi: 10.1111/j.1574-6968.1998.tb12800.x. [DOI] [PubMed] [Google Scholar]
- 32.The Merck Veterinary Manual. 9th ed. Whitehouse Station, NJ: Merck & Co; 2008. [Google Scholar]
- 33.Jacobs MR, Bajaksouzian S, Windau A, et al. Susceptibility of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis to 17 oral antimicrobial agents based on pharmacodynamic parameters: 1998–2001 U S Surveillance Study. Clin Lab Med. 2004;24:503–30. doi: 10.1016/j.cll.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Weimer KD, Armbruster C, Juneau RA, Hong W, Pang B, Swords WE. Coinfection with Haemophilus influenzae promotes pneumococcal biofilm formation during experimental otitis media and impedes the progression of pneumococcal disease. J Infect Dis. 2010;202:1068–75. doi: 10.1086/656046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westman E, Lundin S, Hermansson A, Melhus A. β-lactamase-producing nontypeable Haemophilus influenzae fails to protect Streptococcus pneumoniae from amoxicillin during experimental acute otitis media. Antimicrob Agents Chemother. 2004;48:3536–42. doi: 10.1128/AAC.48.9.3536-3542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]