Abstract
Accurate and rapid cell counts of mycobacterial species in culture are difficult to obtain. Here, a method using modified Kinyoun acid-fast staining was adapted for use with a Petroff-Hausser sperm and bacteria cell counting chamber by using a liquid suspension staining technique. Cell counts obtained by this method were compared to viable cell counts by agar plate counting, revealing accurate correlation.
Keywords: acid-fast staining, mycobacteria, microscope, hemacytometer
Unit Introduction
The slow growth rate of mycobacterial species and the absence of expensive fluorescent microscopes or flow cytometers in many labs impedes accurate, instantaneous bacterial counts from mycobacterial cell suspensions. Acid-fast staining techniques are the most rapid way to verify the presence of mycobacteria, however, cells are smeared onto a glass slide and thus, are difficult to accurately quantify (Weyer, 1998; Selvaraju et al., 2008). While microscopic enumeration of bacteria must be verified by more dependable colony forming unit (CFU) counts, a reliable, inexpensive, and rapid cell counting method is necessary and desirable for studies whereby accurate cell enumeration of actively growing mycobacteria is critical. By combining simple Kinyoun staining with the accuracy of hemacytometer counting, we have developed a reliable, precise method for rapidly (within 1 to 2 hours) staining and counting pure mycobacterial cells in liquid suspension.
Biosafety cautions
Caution
Mycobacterium tuberculosis is a Biosafety Level 3 (BSL-3) pathogen, according to the Biosafety in Microbiological and Biomedical Laboratories manual published by the Centers for Disease Control and Prevention. Follow all appropriate guidelines and regulations for the use and handling of BSL-3 pathogenic microorganisms. See UNIT 1A.1 and other pertinent resources (APPENDIX 1B) for more information and instructions on safe handling of microorganisms.
Basic Protocol 1: Carbolfuchsin Staining and Cell Counting of Acid-Fast Stained Mycobacteria in Liquid Suspension
Materials
Mycobacterial liquid culture
TB Carbolfuchsin KF (BD Diagnostics)
TB Decolorizer TM (BD Diagnostics)
Sterile water
5-μm syringe filters
1.5 ml microcentrifuge tubes
Microcentrifuge
Petroff-Hausser sperm and bacteria counting chamber
Light microscope with 100× oil immersion objective
Generate single mycobacterial cell suspension
-
1
Collect the liquid mycobacterial culture to be counted and filter through a 5-μm syringe filter to generate a single-cell suspension.
Mycobacteria in single cell suspensions will yield more accurate counts.If experiments include the use of M. tuberculosis, incubate the M. tuberculosis cells in a 95°C water bath for at least 60 min to kill the bacteria. This heat-killing step is required for the safe removal of BSL-3 pathogens, such as M. tuberculosis, from the BSL3 facility. This additional step will increase the procedure time by at least 1 h. -
2
Transfer 1 ml of the mycobacterial single cell suspension to a 1.5-ml microcentrifuge tube and centrifuge the cells at maximum speed for 2 min (or until all cells are pelleted) at maximum speed, room temperature.
Stain cells with carbolfuchsin
-
3
Remove the supernatant and resuspend the cell pellet in 100 μL of a 0.3% (v/v) carbolfuchsin solution. Mix gently by inverting the tube 4 to 5 times to ensure full staining of the cells.
-
4
Centrifuge the cells for 2 min (or until all cells are pelleted) at maximum speed, room temperature, and carefully remove the carbolfuchsin supernatant, revealing a dark pink to black cell pellet.
-
5
Resuspend the pellet in 1 ml of sterile water to wash off the excess carbolfuchsin.
-
6
Centrifuge the cells for 2 min at maximum speed, room temperature, and discard the supernatant.
Decolorize the cell suspension
-
7
Resuspend the pellet in 100 μl of acid decolorizer solution.
-
8
Centrifuge the cells for 2 min at maximum speed, room temperature, and discard the supernatant.
-
9
Repeat steps 7 and 8 two additional times for a total of three acid decolorizer washes.
The supernatant after the third wash should be pale pink. -
10
Resuspend the cell pellet in 100 μl of sterile deionized water.
Note: The final product should still appear pale pink.
Add cells to hemacytometer and enumerate using a light microscope
-
11
Load the stained cell sample directly onto the hemacytometer or cell counting chamber, as per manufacturer instructions.
To facilitate efficient counting, dilutions of the stained cell suspension may be generated. Based on this protocol, we recommend 1:10 or 1:100 dilutions. -
12
Visualize the acid-fast stained bacteria using the 100× oil immersion objective on a light microscope.
The acid-fast bacteria should appear pink, while other bacteria (if present) will remain unstained and likely invisible to the naked eye. -
13
Count 10 to 20 fields and calculate the total bacteria/ml according to hemacytometer or cell counter chamber manufacturer instructions.
This step determines the total number of acid-fast stained mycobacteria in a given volume. Step 14 must be performed to verify cell viability and determine the actual number of CFU.
Inoculate appropriate solid medium and determine CFU of original mycobacterial culture
-
14
In parallel, make dilutions and plate the original unstained culture for accurate CFU/ml counts to be obtained after several days/weeks, depending on the replication rate of the mycobacterial strain.
Commentary
Background Information
Mycobacteria are among the few bacteria capable of resisting decolorization by mineral acids after being stained with arylmethane dye (Allen, 1992). Mycobacterial staining is attributed to mycolic acids in the cell envelope that are able to bind to the dye, basic fuchsin. The mycolic acid-fuchsin complex is acid-alcohol resistant, and therefore remains intact after decolorization, whereas eukaryotic cells and other bacteria will not retain the dye (Allen, 1992). Both the classical Ziehl-Neelsen staining method using hot carbolfuchsin (Neelsen, 1883), as well as the modified Kinyoun method using cold carbolfuchsin (Kinyoun, 1915), have been additionally modified numerous times (Allen, 1992; Selvakumar et al, 2002), but have always been described as a dried, glass slide staining procedure.
Mycobacterial enumeration by cell counting has commonly been described using one of the following techniques: i) fluorescent acid-fast auramine-O-rhodamine staining of heat-fixed and dried bacterial smears on glass slides (Selvaraju, 2008; Gilkerson and Kanner, 1963), ii) live/dead fluorescent staining using commercially-available kits (Zahrt and Deretic, 2000), or iii) use of a hemacytometer (also spelled hemocytometer) and a simple light microscope to visualize unstained mycobacterial cells in liquid samples (Gao and Manoranjan, 2005). Some experimental protocols require precise calculations of pure mycobacterial cells in suspension, so accurate determination of the number of viable bacterial cells prior to initiating these experiments is critical. Since actively-growing mycobacterial cells taken from a 37°C culture environment appear more virulent than cells thawed and collected from frozen, −80°C stocks (Lukey and Hooker, 2001), the use of actively-growing cells in macrophage cell infection and virulence studies is pertinent. Therefore, a rapid counting procedure of stained mycobacterial cells in liquid suspension is greatly relevant in the field of mycobacteriology, especially since classical agar plate count methods to determine CFU counts for certain mycobacterial species (e.g., Mycobacterium tuberculosis and Mycobacterium ulcerans) can take four to eight weeks. This counting procedure is not intended to replace viable CFU counts, but rather, to establish the number of cells in suspension within 1 to 2 hours versus days, weeks, or months waiting for the formation of colonies on a solid medium.
Counting acid-fast-stained bacilli also reduces the ambiguity associated with counting unstained cells, especially for those who lack experience using hemacytometers or other counting chambers. Additionally, the described procedure can be performed quickly using a standard light microscope without the need for expensive computers, cameras, or fluorescence microscopes. Compared to other methods of mycobacterial enumeration, this method is simple, rapid, accurate, and easily accessible to all laboratories with a need to count mycobacterial cells in a liquid suspension. Enumeration of mycobacterial cells in clinical or environmental samples has not been attempted using this protocol. Appropriate sample processing would need to be incorporated and optimized to remove and minimize debris contamination.
Critical Parameters and Troubleshooting
See Table 1 for a list of commonly encountered problems, their possible causes, and solutions.
Table 1. Troubleshooting Guide for Staining and Counting Mycobacterial Cells.
| Problem | Possible Cause | Solution |
|---|---|---|
| No detectable pink bacteria. |
|
|
| Large clumps of pink bacteria that cannot be accurately counted. |
|
|
| Stained cell count is not within reasonable error of the determined CFU/ml count. |
|
|
Anticipated Results
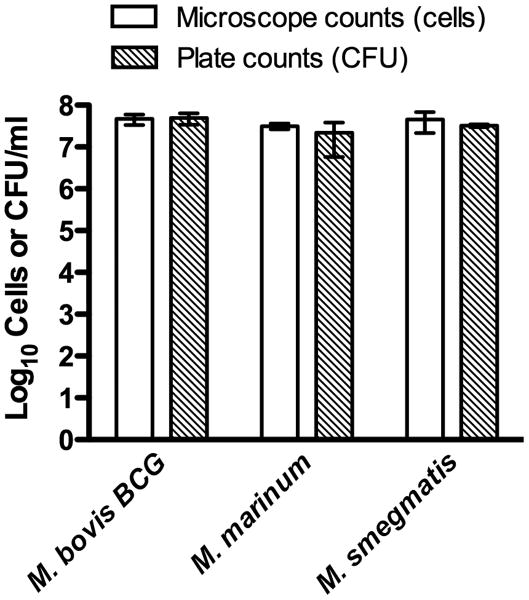
For proof of concept testing, several different mycobacterial strains were subjected to testing: Mycobacterium marinum ATCC 927, Mycobacterium smegmatis ATCC 19420, and Mycobacterium bovis BCG Pasteur. Additionally, to ensure that contaminant non-mycobacterial cells would not be acid-fast stained, we tested non-mycobacterial species including Gram-negative Escherichia coli ATCC 25922 and Gram-positive Staphylococcus aureus ATCC 29213. For the purposes of this study, we counted cells on 10 chamber grids for each bacterial sample and followed the calculations provided by the hemacytometer manufacturer (Hausser Scientific) to determine the total cell count. All samples were performed in triplicate and compared to the CFU counts for the original filtered culture. Results of the stained mycobacterial cell direct microscope counting compared to viable CFU plate counting are shown in Table 1 and Figure 1. The log10 difference for each replicate was calculated by determining the log10 difference between accepted value (CFU count) and the measured value (direct microscope count), followed by calculating the average of the three replicates for each species (Table 1).
Figure 1.
Comparison of microscope-based counting of stained cells and viability CFU counts of slow-growing (M. bovis BCG and M. marinum) and fast-growing (M. smegmatis) mycobacterial species. Values represent the mean cell or CFU count and SD of three independent experiments.
As shown in Table 1, the average log10 difference between direct microscopic counts and accepted CFU/ml counts of each tested mycobacterial species was less than 0.2, with a cross species average of 0.126 log10 difference. Therefore, this modified acid-fast staining method in liquid suspension represents a quick, accurate, and dependable technique to quantify mycobacteria. Moreover, the microscope-counted cells correlate with actual CFU plate counts with analyses revealing that the differences are not statistically significant (Table 1 and Figure 1). Therefore, this approach can be employed for accurately calculating the number of mycobacterial cells from a liquid suspension or an actively growing culture and could be useful for generating the multiplicity of infection (MOI) for cell infection studies and for preparing cellular suspensions for animal infection studies. As a proof of concept, this counting procedure was used to count individual M. tuberculosis cells in suspension prior to performing intracellular infection studies. After heat-inactivating and enumerating the mycobacterial cells in suspension, the microscope cell counts were used to calculate an MOI of 1:1 for cell infectivity. Upon obtaining the CFU counts after a three-week incubation at 37°C, the actual MOIs were calculated as 3.7:1, 3.2:1, and 3.1:1 (bacterial cell:mammalian cell) for triplicate experiments, which was considered within range of acceptability for the experiment (data not shown).
Time Considerations
Once proficiency of the technique is established, an investigator can generally stain and count the mycobacteria within 30 to 60 min. When working with M. tuberculosis cells, an additional heat-inactivation step of 60 min is required, and thus, the protocol will take approximately 2 hours to complete.
Table 2. Comparison of Direct Microscope Counts, Using the Modified Liquid Suspension Carbolfuchsin Stain, and CFU/ml Counts for Mycobacterial Cultures.
| Mycobacteria species | Direct microscope countsa (cells/ml) | CFU/ml counts | Log10 differenceb | Average Log10 differencec | p valued |
|---|---|---|---|---|---|
| M. bovis BCG | |||||
| Culture 1 | 6.00E+07 | 6.57E+07 | 0.057 | 0.033 | 0.3498 |
| Culture 2 | 3.40E+07 | 3.70E+07 | 0.030 | ||
| Culture 3 | 4.60E+07 | 4.47E+07 | 0.013 | ||
| M. marinum | |||||
| Culture 1 | 3.60E+07 | 1.16E+07 | 0.223 | 0.184 | 0.5225 |
| Culture 2 | 3.20E+07 | 1.37E+07 | 0.183 | ||
| Culture 3 | 2.60E+07 | 4.07E+07 | 0.147 | ||
| M. smegmatis | |||||
| Culture 1 | 3.40E+07 | 3.36E+07 | 0.004 | 0.161 | 0.4973 |
| Culture 2 | 7.20E+07 | 2.93E+07 | 0.423 | ||
| Culture 3 | 2.80E+07 | 3.36E+07 | 0.056 | ||
All direct microscope counts were computed by counting ten grids on a Petroff-Hausser counting chamber and performing the calculations according to manufacturer instructions.
Log10 differences between direct microscope counted cells and agar plate cultures (CFU/ml).
Average Log10 difference of direct microscope counted cells and viable CFU counts from triplicate experiments for each bacterial strain.
Two-tailed p values (paired Student's t test) revealed that the differences between the microscope and CFU counts were not statistically different.
Acknowledgments
This work was supported by NIH grant AT004690 to S.E.H.
Literature Cited
- Allen JL. A Modified Ziehl-Neelsen Stain for Mycobacteria. Med Lab Sci. 1992;49:99–102. [PubMed] [Google Scholar]
- Gao LY, Manoranjan J. Laboratory maintenance of Mycobacterium marinum. Curr Protoc Microbiol. 2005;10:B.1.1–B.1.8. doi: 10.1002/9780471729259.mc10b01s00. [DOI] [PubMed] [Google Scholar]
- Gilkerson SW, Kanner O. Improved technique for the detection of acid-fast bacilli by fluorescence. J Bacteriol. 1963;86:890–891. doi: 10.1128/jb.86.4.890-891.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinyoun JJ. A note on Uhlenhuths method for sputum examination, for tubercle bacilli. Am J Public Health. 1915;5:867–870. doi: 10.2105/ajph.5.9.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukey PT, Hooker EU. Macrophage virulence assays. In: Parish T, Stoker NG, editors. Methods in Molecular Medicine: Mycobacterium tuberculosis protocols. Humana Press; Totowa, N.J: 2001. pp. 271–280. [DOI] [PubMed] [Google Scholar]
- Neelsen F. Ein casuistischer Beitrag zur Lehre von der Tuberkulose. Centrabl Med Wissenschaften. 1883;28:497–501. [Google Scholar]
- Selvakumar N, Rahman F, Rajasekaran S, Narayanan PR, Frieden TR. Inefficiency of 0.3% carbol fuschin in Ziehl-Neelsen staining for detecting acid-fast bacilli. J Clin Microbiol. 2002;40:3041–3043. doi: 10.1128/JCM.40.8.3041-3043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraju SB, Khan IUH, Yadav JS. Specific detection and quantification of culturable and non-culturable mycobacteria in metalworking fluids by fluorescence-based methods. Lett Appl Microbiol. 2008;47:451–456. doi: 10.1111/j.1472-765X.2008.02419.x. [DOI] [PubMed] [Google Scholar]
- Weyer K. WHO technical bulletin 98.258. World Health Organization; Geneva: 1998. Laboratory Services in Tuberculosis Control. Part II. Microscopy. [Google Scholar]
- Zahrt T, Deretic V. An essential two-component signal transduction system in Mycobacterium tuberculosis. J Bacteriol. 2000;182:3832–3838. doi: 10.1128/jb.182.13.3832-3838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]