Abstract
Purpose
We review recent advances in the biologic understanding and treatment of childhood acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML), identify therapeutically challenging subgroups, and suggest future directions of research.
Methods
A review of English literature on childhood acute leukemias from the past 5 years was performed.
Results
Contemporary treatments have resulted in 5-year event-free survival rates of approximately 80% for childhood ALL and almost 60% for pediatric AML. The advent of high-resolution genome-wide analyses has provided new insights into leukemogenesis and identified many novel subtypes of leukemia. Virtually all ALL and the vast majority of AML cases can be classified according to specific genetic abnormalities. Cooperative mutations involved in cell differentiation, cell cycle regulation, tumor suppression, drug responsiveness, and apoptosis have also been identified in many cases. The development of new formulations of existing drugs, molecularly targeted therapy, and immunotherapies promises to further advance the cure rates and improve quality of life of patients.
Conclusion
The application of new high-throughput sequencing techniques to define the complete DNA sequence of leukemia and host normal cells and the development of new agents targeted to leukemogenic pathways promise to further improve outcome in the coming decade.
INTRODUCTION
The treatment outcome for children with acute myeloid leukemia (AML) and especially acute lymphoblastic leukemia (ALL) has improved substantially with the use of risk-directed treatment and improved supportive care. The 5-year event-free survival rates for ALL now range between 76% and 86% in children receiving protocol-based therapy in the developed countries (Table 1),1–14and those for AML range between 49% and 63% in some of the more successful clinical trials (Table 2).15–30 The improved treatment has diminished or eliminated the impact of many conventional prognostic factors in ALL, such as male sex and black race.6,10 Thus current ALL trials have focused on improving not only the outcome of a few subtypes that remain refractory to treatment (eg, infant ALL with MLL rearrangement, hypodiploid ALL, and poor early responders), but also the quality of life of the patients. By contrast, with the exception of core-binding factor (CBF) leukemias [t(8;21)(AML1-ETO) and inv(16)(CBFβ-MYH11)] and AML in Down syndrome, most of the AML cases continue to pose a therapeutic challenge.
Table 1.
Characterizations of the Patients and Treatment Results From Selected Clinical Trials for Childhood ALL
| Study | Years of Study | No. of Patients | Age Range (years) | T Cell (%) | WBC Count ≥ 100 × 109/L (%) | DNA Index ≥ 1.16 (%) | ETV6 – RUNX1 (%) | Ph+ (%) | Event-Free Survival at 5 Years |
Survival at 5 Years |
Cumulative CNS Relapse at 5 Years |
Cumulative Secondary Neoplasm at 10 Years |
Data Source (first author) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | SE | % | SE | % | SE | % | SE | ||||||||||
| AIEOP-95 | 1995–2000 | 1,743 | 0–17 | 11 | 10.2 | 19.2 | 22.4 | 2.3 | 75.9 | 1.0 | 85.5 | 0.8 | 1.2 | 0.3 | 0.4 | 0.2 | Conter1 |
| BFM-95 | 1995–2000 | 2,169 | 0–18 | 13.3 | 11.1 | 20.3 | 21.5 | 2.2 | 79.6 | 0.9 | 86.3 | 0.6 | 1.8 | 0.3 | 1.7 | 0.3 | Möricke2 |
| CCG-1900 | 1996–2002 | 4,464 | 0–21 | 15.9 | 14.1 | 21.5* | NA | 2.9 | 76.0 | 0.7 | 86.3 | 0.6 | 4.6 | 0.3 | 1.0 | 0.2 | Gaynon3 |
| COALL-97 | 1997–2003 | 667 | 0–18 | 14.1 | 12.0 | NA | 23.9 | 1.9 | 76.7 | 1.7 | 85.4 | 1.4 | 2.1 | 0.6 | 1.1 | 0.4 | Escherich4 |
| CPH-95 | 1996–2002 | 380 | 0–18 | 14.8 | 12.6 | NA | 22.3 | 2.3 | 72.1 | 2.3 | 83 | 1.9 | 1.2 | 0.6 | 0.6 | 0.4 | Stary5 |
| DCOG-9 | 1997–2004 | 859 | 1–18 | 11.4 | 12.4 | 21.5 | 15.0 | 1.6 | 80.6 | 1.4 | 86.4 | 1.2 | 2.6 | 0.6 | 0.1 | 0.1 | Kamps14 |
| DFCI 95-01 | 1996–2000 | 491 | 0–18 | 10.6 | 11.0 | 18.0 | 25.8 | NA | 81.6 | 1.8 | 89.6 | 1.4 | 0.7 | 0.4 | 0.6 | Silverman6 | |
| INS 98 | 1998–2003 | 315 | 0–18 | 19.4 | 12.1 | 19.9* | 13.7 | 3.3 | 78.7 | 2.3 | 83.8 | 2.1 | 1.9 | 0.8 | 1.0 | 0.5 | Stark7 |
| NOPHO-2000 | 2002–2007 | 1,023 | 1–15 | 11.3 | 11.5 | NA | 23.2 | 1.1 | 79.4 | 1.5 | 89.1 | 1.1 | 2.7 | 0.6 | NA | Schmiegelow8 | |
| SJCRH-13B | 1994–1998 | 247 | 0–18 | 17.4 | 15.4 | 18.6 | 15.8 | 4.0 | 80.1 | 2.6 | 85.7 | 2.2 | 1.7 | 0.8 | 3.3 | 1.2 | Pui9 |
| SJCRH-15 | 2000–2007 | 498 | 1–18 | 15.3 | 12.7 | 24.3 | 19.3 | 2.0 | 85.6 | 2.9 | 93.5 | 1.9 | 2.7 | 0.8 | 0.3 | 0.3 | Pui10 |
| TCCSG-95-14 | 1995–1999 | 597 | 1–15 | 9.7 | 11.8 | 22.3 | NA | 4.0 | 76.8 | 1.8 | 84.9 | 1.5 | 1.7 | 0.6 | 0.7 | Tscuhida11 | |
| TPOG-2002 | 2002–2007 | 788 | 0–18 | 9.7 | 14.0 | NA | 13.1 | 2.1 | 77.4 | 1.7 | 83.5 | 1.6 | 3.8 | 0.8 | NA | Liang12 | |
| UKALL-97/99 | 1999–2002 | 938 | 1–18 | 10.0 | 13.1 | NA | NA | 2.3 | 80 | 1.2 | 88 | 1.1 | 3.0 | 0.5 | NA | Mitchell13 | |
Abbreviations: ALL, acute lymphoblastic leukemia; Ph+, Philadelphia chromosome positive; AIEOP, Associazione Italiana di Ematologia ed Oncologia Pediatrica; BFM, Berlin-Frankfurt-Münster ALL Study Group; CCG, Children's Cancer Group; COALL, Cooperative ALL Study Group; CPH, Pediatric Hematology in the Czech Republic; DCOG, Dutch Childhood Oncology Group; DFCI, Dana-Farber Cancer Institute ALL Consortium; INS, Israeli National Studies of Childhood ALL; NOPHO, Nordic Society of Pediatric Hematology and Oncology; SJCRH, St Jude Children's Research Hospital; TCCSG, Tokyo Children's Cancer Study Group; TPOG, Taiwan Pediatric Oncology Group; UKALL, UK Medical Research Council Working Party on Childhood Leukaemia; NA, not available.
Ploidy > 50.
Table 2.
Characteristics and Treatment Results From Selected Clinical Trials for Childhood AML
| Study* | Years of Study | No. of Patients†‡ | Early Deaths (%) | CR Rate (%) | Time of CR Evaluation | Anthracyclines (mg/m2)§ | Cytarabine (g/m2) | Etoposide (g/m2) | 5-Year EFS |
5-Year OS |
Data Source (first author) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | SE | % | SE | ||||||||||
| POG 8821 | 1988–1993 | 511 | 3.9 | 77 | 2 courses | 360 | 55.7 | 2.25 | 31 | 2 | 40.4 | 2 | Ravindranath25 |
| CCG 2891 | 1989–1995 | 750 | 4 | 77 | 2 courses | 350 | 28.3 | 1.9 | 34 | 3 | 45 | 3 | Smith21 |
| MRC-AML 10 | 1988–1995 | 303 | 4 | 93 | 4 courses | 550 | 10.6 | 0.5–1.5 | 49 | 58 | Gibson27 | ||
| PINDA-92 | 1992–1998 | 151 | 21 | 74 | Not specified | 350 | 7.64 | 0.45 | 36 | 37 | Quintana29 | ||
| LAME- 91∥ | 1991–1998 | 247 | 6.4 | 91 | 2 courses | 460 | 9.8–13.4 | 0.4 | 48 | 4 | 62.3 | 4 | Perel24 |
| TCCSG M91-13/M96-14 | 1991–1998 | 192 | 3.6 | 88 | Not specified | 495 | 69.4–99.4 | 3.75–5.75 | 56 | 62 | Tomizawa17 | ||
| BFM-93 | 1993–1998 | 427 | 7.4 | 82 | 4 courses | 300–400 | 23–41 | 0.95 | 50 | 2 | 57 | 2 | Creutzig20 |
| CCG 2961 | 1996–1999 | 901 | 6 | 83 | 2 courses | 360 | 15.2–19.6 | 1.6 | 42 | 3 | 52 | 4 | Lange15 |
| EORTC-CLG 58,921 | 1993–2000 | 166 | 1 | 84 | 2 courses | 380 | 23–29 | 1.35 | 49 | 4 | 62 | 4 | Entz-Werle23 |
| GATLA-AML90 | 1993–2000 | 179 | 20 | 70 | Not specified | 300 | 41.1 | 1.45 | 31 | 4 | 41 | 4 | Armendariz18 |
| AIEOP LAM-92 | 1992–2001 | 160 | 6 | 89 | 2 courses | Not provided | Not provided | Not provided | 54 | 4 | 60 | 4 | Pession22 |
| NOPHO-AML 93 | 1993–2001 | 223 | 2 | 92 | 3 courses | 300–375 | 49.6–61.3 | 1.6 | 50 | 3 | 66 | 3 | Lie et al19 |
| MRC-AML 12 | 1994–2002 | 455 | 4 | 92 | 4 courses | 300–610 | 4.6–34.6 | 1.5 | 56 | 66 | Burnett16/Gibson27 | ||
| AML99 | 2000–2002 | 260 | 1.7 | 94 | 2 courses | 300–375 | 59.4–78.4 | 3.15–3.2 | 61 | 3 | 75 | 3 | Tsukimoto28 |
| BFM-98 | 1998–2003 | 473 | 3 | 88 | 4 courses | 420 | 41–47 | 0.95 | 49 | 3 | 62 | 3 | Creutzig20 |
| SJCRH AML02¶ | 2002–2008 | 230 | 1 | 94 | 2 courses | 300–550 | 34–48 | 1–1.5 | 63 | 4 | 71 | 4 | Rubnitz26 |
| COG AAML03P1¶# | 2003–2005 | 350 | 2.6 | 83 | 2 courses | 300–480 | 21.6–45.6 | 1–1.75 | 53 | 6 | 66 | 5 | Cooper30 |
Abbreviations: AML, acute myeloid leukemia; CR, complete response; EFS, event-free survival; OS, overall survival; POG, Pediatric Oncology Group; CCG, Children's Cancer Group; MRC-AML, United Kingdom's Medical Research Council Acute Myelogenous Leukemia study; PINDA, National Program for Antineoplastic Drugs for Children; LAME, Leucemie Aigue Myeloblastique Enfant; TCCSG, Tokyo Children's Cancer Study Group; BFM, Berlin-Frankfurt-Münster; EORTC-CLG, European Organisation for Research and Treatment of Cancer Children's Leukemia Group; GATLA-AML, Argentine Group for the Treatment of Acute Leukemia; AIEOP LAM, Associazione Italiana di Ematologia ed Oncologia Pediatrica Leucemia Acuta Meiloide; NOPHO-AML, Nordic Society of Pediatric Hematology and Oncology—Acute Myeloid Leukemia; SJCRH, St Jude Children's Research Hospital; COG, Children's Oncology Group.
Results are reported for only those trials that had ≥ 150 patients and information provided for each of the column headings. The AIEOP LAM-92 did not allow for definitive dose calculations of drugs because consolidation therapy was given based on the treating physician's judgement, but usually included anthracyclines, etoposide, and high-dose cytarabine.
No. of patients excludes patients with Down syndrome.
Ages include patients from 0 up to and including age 15 years: BFM-98 included patients from 0 to less than 17 years of age; CCG-2961 included patients from 0 to < 21 years of age.
Anthracycline conversions were according to daunorubicin equivalents, including idarubicin 5×, mitoxantrone 5×, doxorubicin 1×. Another conversion factor for idarubicin and mitoxantrone that has been used is 3×.
LAME-91 also used 450 mg/m2 of amsacrine; MRC-10 and MRC-12 both also included 500 mg/m2 of amsacrine.
SJCRH AML02 and the COG AAML03P1 have EFS and OS at 3-year follow-up.
COG AAML03P1 enrolled patients ≥ 1 month and ≤ 21 years of age.
Childhood acute leukemias have long been the best characterized malignancies from a genetic viewpoint. Despite remarkable progress in cataloging the molecular lesions, our understanding of how such changes cooperate to produce overt leukemia or to induce drug resistance is still rudimentary. Recent genome-wide studies have begun to enlighten our understanding of leukemogenesis and prognosis and, in some instances, to stimulate the development of target therapy. Because of the space constraints, we review here only some of the most recent advances in the biology and treatment of childhood leukemias.
MOLECULAR GENETICS OF ALL
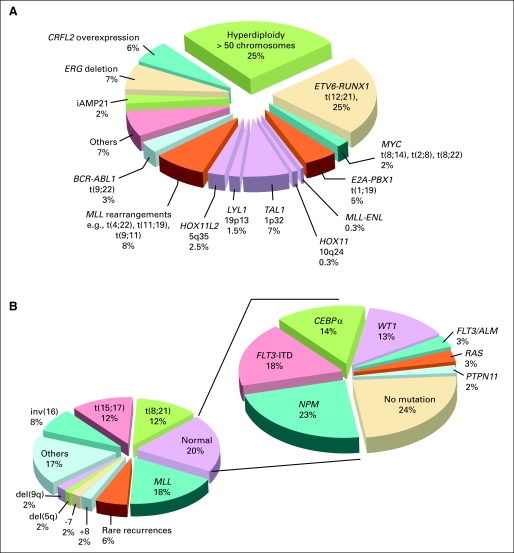
Standard genetic analyses can detect primary genetic abnormalities in more than 75% of the ALL cases, but they cannot identify the full repertoire of the genetic alterations.31 The advent of high-resolution genome-wide analyses of gene expression, DNA copy number alterations (CNA) and loss of heterozygosity, epigenetic changes, and whole-genome sequencing have led to the detection of many novel genetic abnormalities; to date, virtually all patients with ALL can be classified according to specific genetic abnormality (Fig 1A). These studies also provided new insights into the complex interactions of multiple genetic alterations in leukemogenesis and response to therapy.34
Fig 1.
Estimated frequency of specific genotypes in childhood leukemias. (A) Genetic abnormalities in acute lymphoblastic leukemia (ALL). Data were modified from Pui et al32 by including recently identified genotypes. The genetic lesions that are exclusively seen in cases of T-cell ALL are indicated in purple. (B) Genetic abnormalities in acute myeloid leukemia (AML). Panel to the left demonstrates the most common karyotypic alterations. Eighty percent of all children have disease-associated genomic structural alterations. Mutation profile in those without cytogenetic abnormalities (normal karyotype) is shown in the right panel. Seventy-six percent of those in the normal karyotype population have one of the known mutations; thus, more than 95% of all children with AML have at least one known genomic abnormality. (With permission from Reaman and Smith33).
Genomic Analysis of ALL
Primary somatic genetic abnormalities have important prognostic and therapeutic implications and play a critical role in leukemogenesis (Table 3).10,31,34–50 However, experimental models have established that cooperative mutations are necessary to induce leukemia and contribute to the development of drug resistance. Although high-throughput analyses of global gene expression have identified distinct subgroups and may in the future be used to stratify patients, these profiles do not distinguish pathways that are “drivers” of leukemogenesis from “passengers.” Using single-nucleotide polymorphism (SNP) array, Mullighan et al34 identified an average of six CNAs per case of childhood ALL. In general, deletions outnumbered amplifications by a ratio of 2:1. The lesions target genes regulating lymphoid differentiation, tumor suppression, cell cycle, apoptosis, signaling, microRNAs, and drug responsiveness. There were substantial differences in the frequency of CNAs among various subtypes. MLL-rearranged cases had less than one CNA per case, suggesting that MLL is a potent oncogene that requires very few cooperating lesions to induce leukemia transformation, whereas ETV6-RUNX1 (formerly known as TEL-AML1) and BCR-ABL1 leukemias demonstrated more than six lesions per case.34 These findings are compatible with the known behavior of these leukemias. MLL-rearranged leukemias often present during infancy and have a concordance rate close to 100% in identical twins, indicating in utero development and transplacental metastasis from one fetus to the other.31 By contrast, ETV6-RUNX1 leukemias present after infancy and have a concordance rate of only 10% in identical twins, and this gene fusion can be found in as many as 1% of normal newborn babies, a frequency 100 times higher than the prevalence of this subtype of leukemia, suggesting that additional postnatal mutations are necessary for malignant transformation.31
Table 3.
Characteristics and Clinical Outcomes of Selected Subtypes of Childhood ALL
| Subtype | Frequency (%) | Clinical Implication | Estimated 5-Year Event-Free Survival (%) | Data Source(first author) |
|---|---|---|---|---|
| B-cell precursor | ||||
| Hyperdiploidy > 50 | 20–30 | Excellent prognosis with antimetabolite-based therapy | 85–95 | Pui10,31 |
| t(12;21)(p13;q22) ETV6-RUNX1 | 15–25 | Expression of myeloid-associated antigens CD13 and CD33; excellent prognosis with intensive asparaginase therapy | 80–95 | Pui10,31 |
| Trisomies 4 and 10 | 20–25 | Excellent prognosis with antimetabolite therapy | 85–90 | Salzer35 |
| t(1;19)(q23;p13) TCF3-PBX1 | 2–6 | Increased incidence in blacks; excellent prognosis with high-dose methotrexate treatment; increased risk of CNS relapse in some studies | 80–85 | Pui10,31 |
| Intrachromosomal amplification of chromosome 21 | 2–3 | More common in older children and adolescents; poor prognosis; benefit from intensive induction and early re-intensification therapy | 30–40 | Attarbaschi36 |
| t(4;11)(q21;q23) MLL-AF4 | 1–2 | Poor prognosis and predominance in infancy, especially those < 6 months of age; overexpression of FLT3 | 30–40 | Pui10,31 |
| t(9;22)(q34;q11.2) BCR-ABL1 | 2–4 | Imatinib plus intensive chemotherapy improve early treatment outcome | 80-90 at 3 years | Schultz37 |
| t(8;14)(q23;q32.3) | 2 | Favorable prognosis with short-term intensive therapy with high-dose methotrexate, cytarabine, and cyclophosphamide | 75–85 | Pui38 |
| Hypodiploidy < 44 chromosomes | 1–2 | Poor prognosis | 35–40 | Nachman39 |
| CRLF2 overexpression | 6–7 | Poor prognosis; common in patients with Down syndrome (55%) | ? | Mullighan,40 Cario,41 Harvey42 |
| T-cell | ||||
| TAL/LMO rearrangement | 15–30 | Good prognosis in some studies; potentially responsive to histone deacetylase inhibitor | ? | Meijerink43 |
| HOX11 rearrangement | 7–8 | Good prognosis | ? | Meijerink43 |
| HOX11L2 (TLX3) rearrangement | 20–24 | Poor prognosis in some studies | ? | Meijerink43 |
| HOXA rearrangement | 4–5 | Poor prognosis; potentially sensitive to histone H3K79 methyltransferases inhibitor | ? | Meijerink43 |
| NUP214-ABL1 | 6 | Sensitive to tyrosine kinase inhibitor | ∼50 (survival) | Graux44 |
| MLL-ENL | 2–3 | Favorable prognosis | 80–90 | Pui31 |
| Early T-cell precursor | 12 | Poor prognosis; expressed myeloid or stem-cell markers | 30–35 | Coustan-Smith45 |
| Cooperation mutations | ||||
| B-cell precursor | Mullighan46 | |||
| 1KZF1 deletions/mutations | 15–30 | Poor prognosis; resistant to asparaginase and daunorubicin | 50–55 | Den Boer47 |
| JAK mutations | ?2–5 | Predominance in high-risk patients; JAK2 mutations in 20% of Down syndrome cases; potentially responsive to JAK2 inhibitors | ∼60 | Mullighan48 |
| T-cell | ||||
| NOTCH/FBXW7 mutations | 50 | Favorable prognosis; potentially responsive to NOTCH inhibitor | 90 | Breit49 |
| PTEN-P13K-AKT pathway | ∼50 | ? Poor prognosis | ? | Gutierrez50 |
| CDKN2A/2B deletions | ∼70 | ? Potentially responsive to DNA methyltransferases inhibitor | ? | Meijerink43 |
Abbreviation: ALL, acute lymphoblastic leukemia.
Genetic Determinants of High-Risk B-Cell Precursor ALL
In Philadelphia chromosome–positive ALL with constitutively active BCR-ABL1 tyrosine kinase, IKZF1 (encoding the lymphoid transcription factor IKAROS) is deleted in approximately 80% of the cases.51 Interestingly, there is a high-risk subgroup of BCR-ABL1–negative ALL that is characterized by IKZF1 deletion and has a genetic profile similar to that of cases with BCR-ABL1 fusion.46,47 In search of activated tyrosine kinase signaling in this leukemic subtype, Mullighan et al48 identified activating mutations in the Janus kinases (JAK1, JAK2, and JAK3) in approximately 10% of high-risk BCR-ABL1–negative cases. The presence of JAK mutations was associated with alteration of IKZF1 and deletion of CDKN2A/B. Recent studies showed CRLF2 over-expression (predominantly resulting from P2RY8-CRLF2 fusion or IGH@-CRLF2 rearrangement) in 6% to 7% of B-cell precursor ALL cases and strikingly in 50% to 60% of patients with Down syndrome–associated ALL, all of whom lacked recurring translocations commonly associated with non–Down syndrome ALL.40–42,52 CRLF2 alteration was associated with activating mutations of JAK1 or JAK2; these two genetic lesions together resulted in constitutive Jak-Stat activation and the growth of cytokine-dependent mouse B-progenitor cell lines in the absence of exogenous cytokine, indicating that they are cooperative mutations in leukemogenesis.40 Importantly, CRLF2 alteration was associated with Hispanic/Latino ethnicity and a poor treatment outcome.40,41
Genomic Analysis of T-Cell ALL
On the basis of gene expression profiling, T-cell ALL cases can be classified into several distinct genetic subgroups that correspond to specific T-cell development stages: HOX11L2, LYL1 plus LMO2, TAL1 plus LMO1 or LMO2, HOX11, and MLL-ENL (Table 3).43 Although HOX11L2 generally confers a poor outcome, HOX11 and MLL-ENL are associated with a favorable outcome.43,53 Using SNP and other genome-wide platforms, many novel genomic alterations have recently been identified, including focal deletions leading to dysregulated expression of TAL1 and LMO2, deletion and mutation of PTEN, mutations of NOTCH1 and FBXW7, deletions of RB1, duplications of MYB, deletions of RB1, and fusion of SET or ABL1 to NUP214.34 Hence T-cell ALL is also a heterogeneous disease. Thus far, mutations of NOTCH1 and FBXW7 (observed in 50% of T-ALL) have generally been associated with a favorable prognosis, and NUP214-ABL1 fusion has been associated with responsiveness to tyrosine kinase inhibition.43,44,49
Genetic Determinants of Relapse in ALL
To explore the genetic basis of relapse, genome-wide studies were conducted using matched diagnosis and relapse samples from the same patients.54,55 Although 90% of the cases exhibited differential CNAs (gaining or losing genetic lesions) from diagnosis to relapse, most relapse samples are clonally related to diagnosis samples, and backtracking studies showed that the relapse clones were often present as minor populations at diagnosis, suggesting that they were selected during treatment. Notably, many of the genetic alterations that emerge in the predominant clone at relapse involve genes that have been implicated in treatment resistance (eg, CDKN2A/B, IKZF1).54,55 Interestingly, some cases had focal deletion of MSH6, reduced expression of which was associated with resistance to mercaptopurine and prednisone, providing another plausible mechanism of drug resistance at relapse.54 Parallel gene expression studies, which have identified a proliferative gene signature that emerges at relapse and consistent upregulation of genes such as survivin, provide attractive targets for novel therapeutic intervention.56
Inherited Susceptibility to ALL
Although candidate gene approaches have implicated inherited polymorphisms of several genes in leukemogenesis, the findings have not been consistent. Two recent genome-wide studies on patients of European ancestry failed to confirm these previously reported gene associations but independently identified germline polymorphisms of the IKZF1 gene and ARID5B gene to be associated with an increased risk of childhood ALL.57,58 The risk alleles of ARID5B, a gene that belongs to a family of transcription factors important in embryonic development, cell type–specific gene expression, and cell growth regulation, were specifically enriched in patients with hyperdiploid ALL and were also associated with greater methotrexate polyglutamate accumulation.57,58 Thus the same genetic variation of ARID5B that predisposes to the development of hyperdiploid ALL may also underlie the superior response of this subtype of ALL to chemotherapy. A subsequent study was performed in patients of African ancestry, showing that ARID5B germline polymorphisms were also associated with the risk of developing hyperdiploid ALL in black patients.59 The lower frequency of this risk allele in the control populations of African ancestry compared with that of European ancestry might also partly explain the lower incidence of hyperdiploid B-cell precursor ALL in the black population.
CELLULAR AND MOLECULAR ORIGINS OF AML
Despite the extremely heterogeneous nature of AML, the various subtypes seem to share some common pathways leading to leukemogenesis, and the hierarchical nature of the disease is generally well established.60 Several lines of evidence have led to a model that AML arises from the cooperation between two classes of genetic alterations that regulate self-renewal and differentiation.61 For example, mutations or epigenetic alterations involving CBF or MLL genes, termed type II alterations, play key roles in modifying the ability of precursor cells to differentiate, but are usually insufficient by themselves to generate AML. In support of this concept is the finding that the AML1-ETO fusion transcript resulting from the t(8;21) translocation can be detected in neonatal blood from teenagers with AML characterized by this translocation.62 Such fusion transcripts have also been identified to persist in the bone marrow of some patients in long-term morphologic remission.63 When a cooperative mutation or type I mutation (such as FLT/ITD or c-KIT point mutations) occurs, a proliferative signal is provided, leading to overt AML. That type II mutations such as AML-ETO can be detected far more frequently in neonatal samples than the incidence of leukemia further supports the concept of cooperating mutations in leukemogenesis.64 Genome-wide studies have identified additional genetic or epigenetic changes that lead to type I and II mutations. Currently, more than 90% of pediatric AML cases are identified to have at least one known genomic alteration (Fig 1B).
Altered Transcription and Chromatin-Modifying Factors
Similar to ALL, many of the chromosomal abnormalities in AML result in alteration of the function of transcription factors critical for normal hematopoiesis and have diagnostic and therapeutic implications (as discussed in Risk-Adapted Treatment of AML). The t(8;21)(q22;q22) and the inv(16)(p13q22) involving CBFα and β subunits respectively account for about a quarter of all cases. These translocations impair CBF transcriptional activation potential. Less common translocations include the t(15;17), 11q23/MLL rearrangements and the t(1;22). The t(15;17) generated PML-RARA fusion protein is resistant to physiologic concentrations of retinoic acid but sensitive to pharmacologic levels of all-trans-retinoic acid (ATRA), which destabilize the repressor complex, allowing for the expression of genes permissive for differentiation and making this subtype the first successfully treated acute leukemia by molecular targeting. The incidences of chromosomal rearrangements involving the MLL gene are age-dependent, being highest in children younger than 2 years and subsequently decreasing to less than 5% in adults.65 The t(1;22) translocation results in the fusion of the OTT gene on chromosome 1 to the Megakaryocytic Acute Leukemia gene on chromosome 22. The translocation is associated closely if not exclusively with acute megakaryoblastic leukemia (AMKL) and has been detected in up to one third of such childhood cases. Monosomy 7, monosomy 5, or 5q deletions are present in only approximately 2% to 4% of cases compared with more than 10% in adults.
Recent genome-wide microarray studies and direct-sequencing approaches have revealed important sub-karyotypic abnormalities. One important finding has been the high percentage of acquired uniparental disomy, resulting in homozygosity of chromosomal regions that occurs after the acquisition of a mutation in one allele.66,67 Examples of this have included FLT3-ITD and CEBPA mutations.68,69
Gene Mutations
Biallelic mutations in CEBPA, a key leucine zipper containing transcription factor regulating differentiation of several cell types, including myeloid precursors, have been identified in approximately 4% of the cases.70 The end result of such biallelic mutations is often that of a null phenotype or loss of function.71
WT1, originally described as a key oncogene in Wilms tumor, functions in non-nephrogenic tissues, including hematopoietic stem cells. Increased expression of WT1 has been used as a surrogate marker for minimal residual disease (MRD) in AML. Inactivating WT1 mutations were reported in 8% to 12% of patients with AML, and their prognostic significance remains uncertain and is likely treatment-dependent.
GATA1 is critical in regulating the differentiation of hematopoiesis, particularly for the erythroid and megakaryocyte lineages. Mutations leading to truncated forms of GATA1 are primarily observed in the AMKL or the megakaryoblastic transient myeloproliferative disease that occur in children with Down syndrome.72 Although not sufficient by themselves in causing leukemia, these GATA1 mutations have been established as a first genetic hit in these disorders and have even been detected prenatally.
Activating mutations of several cytokine receptors have been shown to result in altered signal transduction and increased leukemia cell proliferation, survival, and chemotherapeutic resistance. FLT3-ITD, which involves a duplication of the internal juxtamembrane domain leading to constitutive receptor activation, is rare in infants but is present in 5% to 10% of 5- to 10-year-old patients, 20% of young adult patients, and more than 35% of patients older than 55 years of age with AML.73 An increased FLT3-ITD mutant to wild-type allelic ratio portends a poor prognosis.74,75 Children's Oncology Group (COG) trials are using a mutant to wild-type allelic ratio of ≥ 0.4 as a criterion for allogeneic hematopoietic stem-cell transplantation (HSCT) in first remission. Point mutations in the activating loop of the receptor (FLT3/ALM) also result in constitutively activated FLT3 but do not confer a poor prognosis.73 This finding may be in part due to differences in signaling pathway activation by the different forms of FLT3. FLT3/ALM occurs in only 6% to 8% of children with AML and is mutually exclusive of FLT3-ITD.73
Activating mutations of the c-KIT receptor also result in cytokine-independent proliferation, survival, and drug resistance. Such mutations preferentially activate STAT3 signaling and appear to be clustered with CBF abnormalities, occurring in up to 25% of CBF AML cases but in only ≤ 5% of overall childhood AML.74–77 In contrast to initial reports,74,75 recent studies failed to demonstrate c-KIT mutations to have adverse prognostic impact.78 Mutations involving FMS and PDGFR, class III type of tyrosine kinases, have been reported in adults but not in children with hematologic malignancies.
N-RAS has been reported to be mutated in approximately 10% of children and up to 30% of adults with AML.79 These activating mutations most frequently involve single-base changes of codons 12, 13, and 61, resulting in inhibition of RAS-GTPase. No clinical significance has been associated with mutations in RAS in pediatric AML, and RAS-directed therapies have not been successful thus far.80 However, the recent application of synthetic lethal screening has started to reveal some alternative pathways that could be therapeutically targeted.81
NPM1 mutations are associated with a favorable prognosis. They appear more frequently in AML with normal karyotypes and up to 20% of childhood and 50% of adult cases.82,83 NPM1 mutations seem to be present in leukemia-initiating cell populations and are usually maintained at relapse, suggesting they may represent a primary oncogenic event.84,85
Genome-Wide Analysis
The initial whole-genome sequencing of an adult with M1 AML and normal karyotype disclosed 10 somatic mutations and two leukemia-specific SNPs.86 Two of the mutations involved FLT3 and NPM1, whereas the other eight genes involved nonsense and missense mutations that had not previously been described in AML.86 These novel mutations were not identified in 200 additional AML samples, raising the question of whether they were merely passenger mutations. Analysis of the second case of cytogenetically normal AML revealed 12 mutations in annotated protein-coding genes or regulatory RNAs and 52 mutations in noncoding regions of the genome.87 Four genes, NPM1, NRAS, IDH1, and a conserved region on chromosome 10, were found in at least one additional sample of 180 adult AMLs. Although the IDH1 mutation was observed in 16% of 80 cytogenetically normal adult AML samples,87 it has not been found in childhood AML.88 Independent analyses of IDH1 and IDH2 mutations showed that they were more frequently associated with AML having a normal karyotype, but had no prognostic or therapeutic impact.89–92
Although RNA and microRNA expression signatures can accurately discriminate cases with specific chromosomal translocations and identify novel subsets of AML,93 two independent pediatric studies failed to identify an expression pattern with consistent prognostic significance.94,95 Some of the differences observed in gene expression studies are also beginning to be understood in terms of epigenetic regulation of chromatin function. The finding of a similar expression profile between an AML subset with the methylation and silencing of the CEBPA promoter and another characterized by CEBPA mutations illustrates the critical role that epigenetic changes play in AML.94 Of further interest, the distinctive myeloid/T-lymphoid characteristics of this subtype have been linked to the finding that decreased CEBPA expression leads to increased expression of T-lineage genes in hematopoietic precursors.94,95 On a genome-wide scale, altered CpG methylation profiles have been associated with distinct subsets of adult AML.96 A 15-gene methylation classifier has been reported to have prognostic significance.97
Inherited Susceptibility of AML
Several inherited syndromes associated with an increased incidence of AML have been instrumental in demonstrating the importance of key molecular pathways in the development of somatically acquired AML.98,99 Transient myeloproliferative disease and AMKL in patients with Down syndrome are characterized by specific mutations in the GATA1 gene, which have been documented even during prenatal development.72 Fanconi anemia (mutations in DNA repair genes), dyskeratosis congenita (X-linked mutations in dyskerin or autosomal-recessive forms with mutations in genes involved in telomere maintenance and RNA processing), Schwachman-Diamond's syndrome (SBDS gene involved in ribosome processing), and Kostmann's syndrome (defects in neutrophil elastase ELA2) are all associated with an increased incidence of AML. Mutations in the neurofibromin gene, a RAS-inactivating GTPase, are the cause of neurofibromatosis type I, along with its increased incidence of both juvenile myelomonocytic leukemia and AML. Similarly, mutations of the PTPN11 gene, which encodes the SHP-2 tyrosine phosphatase, cause Noonan's syndrome, which is also associated with the development of juvenile myelomonocytic leukemia and, less commonly, AML. This group of mutations are all linked to the activation of the RAS pathway. Familial platelet disorder with a propensity to develop AML, as well as congenital amegakaryocytic thrombocytopenia, are associated with an increased incidence of AML and caused by mutations in CFFA2 and the thrombopoietin receptor, c-mpl, respectively. Inherited mutations in the CEBPA have also been linked to familial AML.
RISK-ADAPTED TREATMENT OF ALL
The principal and strategies of contemporary risk-adapted treatment of ALL have been the subject of recent reviews.31,100 Therefore, only a few selected high-risk subgroups will be discussed here.
Philadelphia Chromosome–Positive ALL
The Philadelphia chromosome–positive ALL had historically been associated with a dismal outcome, even with allogeneic HSCT, especially in older patients who presented with a high leukocyte count or had a slow early response to initial therapy. In a recent COG study, the addition of continuous exposure of imatinib into an intensive chemotherapy regimen has yielded a 3-year event-free survival of 80%, more than twice that of the historical controls.37 Patients who were treated with intensive chemotherapy plus imatinib fared as least as well as those historical controls who underwent matched-related or matched-unrelated transplantation. Continuous exposure of imatinib seemed to abrogate the prognostic impact of age, leukocyte count, high level of MRD at the end of induction, and even induction failure. Additional follow-up is needed to determine whether the treatment improved the cure rate rather than merely prolonging the disease-free survival. Meanwhile, many pediatric oncologists have reserved transplantation for therapy after relapse in children with Philadelphia chromosome–positive ALL. Future studies will need to address whether the aggressive backbone therapy that was used in the COG study can be safely omitted or reduced in combination with a tyrosine kinase inhibitor and whether the new generation of tyrosine kinase inhibitors (eg, dasatinib, nilotinib) can further improve outcome. Because of its dual targeting of ABL and SRC, more potent suppression of the BCR-ABL1 signaling, and efficacy to most imatinib-resistant BCR-ABL1 mutants (except for T3151), as well as good tolerability in adult clinical trials,101 dasatinib is now being tested in children.
High-Risk T-Cell ALL
With the use of intensive treatment including asparaginase and dexamethasone, T-cell cases fared as well as B-cell precursor cases in some studies.6,10 Although high-dose methotrexate at 5 g/m2 has been suggested to improve outcome in T-cell ALL, methotrexate was given parenterally at only 30 mg/m2 in the DFCI 95-01 study, which featured intensive asparaginase and doxorubicin for consolidation therapy and has perhaps the best treatment result for this subtype of ALL, with a 5-year event-free survival rate of 85%.102 In the current COG study for T-cell ALL, the relative efficacy and safety of high-dose methotrexate at 5 g/m2 with leucovorin rescue are being compared with escalating medium doses of methotrexate without leucovorin rescue plus PEG-asparaginase. In the same study, patients are also randomly assigned to receive or not receive nelarabine (a prodrug of 9-β-D-arabinofuranosylguanine), which has considerable antileukemic effects for T-cell ALL in phase II studies.103
Early T-cell precursor ALL, a subset of T-cell ALL characterized by a gene expression profile similar to that of normal early thymic precursor cells with multilineage differentiation potential and a distinct immunophenotype (CD1a-negative, CD8-negative, CD5-weak, and the expression of stem-cell or myeloid markers), was recently identified.45 These cases have extremely poor outcome, despite the use of transplantation. Because of the significantly improved outcome of patients with T-cell ALL who were treated with high-dose dexamethasone (10 mg/m2 per day) during remission induction in the recently completed Associazione Italiana Ematologia ed Oncologia Pediatrica Berlin-Frankfurt-Muenster AIEOP-BFM-2000 study, albeit with increased toxicity,104 this dose of dexamethasone is being tested during remission induction for this subset of patients in the current St Jude Total Therapy Study XVI.
Infant ALL
Even with intensive therapy including high doses of cytarabine and methotrexate, treatment outcome for infants with ALL remains poor, and for those with MLL gene rearrangement, the event-free survival rates range between 30% and 40% only.100,105,106 Although several small studies have suggested that allogeneic HSCT improves outcome, results from large group studies failed to show any benefit of this treatment modality.105,106 The Interfant-06 Study Group is conducting a randomized trial to test whether the addition of two early intensification courses typically used for AML might improve outcomes for infants with medium-risk or high-risk ALL, including those with MLL rearrangement. The role of allogeneic HSCT in first remission is being investigated in very high-risk cases, as defined by MLL rearrangement, age less than 6 months, and WBCs more than 300 ×109/L. The current COG trial randomly assigns MLL-rearranged infant cases to receive the FLT3 inhibitor lestaurtinib, which has been shown to have schedule-dependent synergistic effects with chemotherapy for MLL-rearranged leukemia in preclinical studies.107 Recent findings suggested that aberrant DNA methylation occurs in the majority of infant ALL case with MLL rearrangement,108,109 raising the possibility of using DNA methyltransferase inhibitors in these patients.
Adolescent ALL
Older adolescents 15 to 21 years of age have had an inferior outcome as compared with that of children and younger adolescents with ALL, partly due to an increased incidence of Philadelphia chromosome– positive and T-cell ALL, lower incidence of ETV6–RUNX1 fusion and hyperdiploidy, and poorer compliance.31,110–112 The relatively poor outcome of older adolescents 16 to 21 years of age with ALL has prompted comparisons of outcome of patients in this age group treated in pediatric versus adult clinical trials in North America and Western Europe. Consistently, pediatric trials have yielded significantly better outcome than adult trials.112–114 This finding, in all likelihood, reflects differences in the more intensive use of nonmyelosuppressive agents such as dexamethasone, vincristine, and asparaginase; the incorporation of high-dose methotrexate; and early and frequent administration of intrathecal therapy in pediatric regimens.
The importance of these treatment components is supported by several other pediatric trials. In the Children's Cancer Group 1961 trial, which included 262 adolescents 16 to 21 years of age treated from 1996 to 2002,112 the 5-year event-free survival rate was 71.5%. This randomized trial demonstrated that augmented rather than standard postinduction intensification therapy with additional doses of vincristine and PEG-asparaginase in lieu of mercaptopurine during interim maintenance therapy and increased doses of intravenous methotrexate without leucovorin rescue improved outcome for patients with a rapid early response. The Dana-Farber Cancer Institute (DFCI) protocols (1991 to 2000)6 and St Jude Total Study XV (2000 to 2007),115 which yielded excellent results for adolescents, likewise featured early intensification therapy with vincristine and asparaginase. Although some contemporary adult ALL trials have recommended allogeneic transplantation as the best treatment option for good-risk adult ALL patients in first remission,116 the excellent results obtained in these pediatric trials do not support the routine use of transplantation in older adolescents with ALL.
There are three other important observations from the Children's Cancer Group 1961 study.112 First, there was no statistical difference in treatment outcome between adolescents with a rapid response who were randomly assigned to receive either one or two courses of postinduction intensification therapy. Second, there was an increased risk of toxic deaths among older adolescents, with 18% of the first events being nonrelapse deaths that occurred during induction or in first remission. Third, there was an increased risk of osteonecrosis. Thus it will be of interest to test whether prednisone pulses, which are commonly given during maintenance therapy, can be omitted in adolescents and young adults with ALL and rapid early response.
The excellent results recently achieved in older adolescents with ALL have prompted many centers and cooperative groups to start treating young adults with pediatric-like regimens. A recent report showed 6-year event-free survival rates of 60% for the 35 adolescents 15 to 18 years old and 63% for the 46 young adults 19 to 30 years old.117 The DFCI Consortium attained a 2-year rate of 72.5% among 75 adults 18 to 50 years of age.118 Investigators at Princess Margaret Hospital in Toronto have also adapted the DFCI treatment regimen and achieved a 3-year relapse-free survival rate of 77% among 64 adults with BCR-ABL–negative ALL who did not undergo transplantation.119 To this end, a US adult intergroup study is testing a pediatric-like regimen in adolescents and young adults up to 39 years of age.
Other High-Risk Subgroups
Hypodiploidy (< 44 chromosomes), t(17;19)(q22;p13.3) [TCF3-HLF], remission induction failure, and the presence of MRD more than 1% at the end of remission induction are also associated with dismal outcome and collectively occur in approximately 5% of children with ALL.31 Although allogeneic transplantation is frequently used to treat these patients, there is no strong evidence to document the efficacy of this approach. Among many novel therapeutics under investigation (Table 4),101 killer-cell immunoglobulin-like receptor–mismatch natural cell therapy120,121 and immunotherapy with a T-cell–engaging CD19-/CD3-bispecific antibody construct (blinatumomab) seem to be particularly promising.122
Table 4.
Novel Therapeutics Under Investigation in Childhood ALL and AML
| Category and Agent | Properties |
|---|---|
| New formulations | |
| Pegylated asparaginase | Long half-life, reduced immunogenicity (ALL) |
| Sphingosomal vincristine | Decreased neuropathy, higher tissue concentration, non-vesicant (ALL) |
| Liposomal annamycin | Decreased cardiotoxicity (ALL/AML) |
| Liposomal doxorubicin | Decreased cardiotoxicity (ALL/AML) |
| Liposomal cytarabine | Long half-life, potential neurotoxicity when administered intrathecally (ALL/AML) |
| Nucleoside analogues | |
| Clofarabine | Effective for ALL and AML |
| Nelarabine | Selective for T-cell ALL, potential neurotoxicity |
| Forodesine (BCX-1777) | Oral phosphorylase inhibitor (ALL) |
| Monoclonal antibodies | |
| Rituximab (anti-CD20) | Potentiate chemotherapy for CD20+ B-lineage ALL |
| Alemtuzumab (anti-CD52) | Potentiate chemotherapy for CD52+ ALL |
| Epratuzumab (anti-CD22) | May have synergistic effect with anti-CD20 antibody |
| CAT-8015; HA22; inotuzumab ozogamicin (anti-CD22) | Cytotoxic for CD22+ ALL |
| Gemtuzumab ozogamicin (anti-CD33) | Cytotoxic for CD33+ leukemia (AML) |
| Blinatumomab | Bispecific antibodies that direct CD3+ T-cell against CD19+ ALL |
| Molecularly targeted agents | |
| Tyrosine kinase inhibitors (imatinib, nilotinib, dasatinib, MK-O457, bosutinib, AP24534, DCC2036, sorafenib); Aurora kinase inhibitor (danusertib); Src-family kinase inhibitor; VEGF inhibitors (bosutinib); bevacizumab, SU5416, AZD2171 | Potentiate chemotherapy for Ph+ ALL; targeting c-KIT, VEGF in AML |
| Fms-like tyrosine kinase −3 (Lestaurtinib, Midostaurin, Tandutinib) | Phase I studies on MLL-rearranged leukemia/FLT3-ITD positive AML |
| NOTCH1 inhibitors (γ–secretase inhibitor, antagonists targeting NOTCH transactivation complex, NOTCH1 receptor inhibitors) | Preclinical studies for T-cell ALL |
| mTOR inhibitors (rapamycin, temsirolimus, everolimus) | Testing in post-transplantation use to decrease graft-versus-host disease and to suppress leukemia (ALL); in combination with chemotherapy in AML |
| Demethylating agents (decitabine, azacytidine) | Potentiate chemotherapy by epigenetic modulation of leukemia cells with MLL rearrangement and in AML/MDS |
| Histone deacetylase inhibitor (vorinostat, valproic acid, depsipeptide) | Potentiate chemotherapy by epigenetic moderation of leukemia cells in ALL and AML |
| Heat shock protein inhibitor (17-allylaminogeldanamycin) | Potentiate the cytotoxicity of histone deacetylase inhibitor |
| Proteasome inhibitor (bortezomib, carfilzomib, ONX 0912) | Potentiate mTOR inhibitors and anthracyclines; clinical trial in AML |
| Farnesylation (tipifarnib) | Directed at RAS inhibition in AML |
| JAK2 inhibitor (lestaurtinib) | Phase I trial for ALL and AML with JAK mutations |
| Microenvironment | |
| Integrin and cell adhesion inhibition (AMD3100, plerixafor) | CXCR4 inhibition; phase I trials in AML planned |
| Immunomodulation | |
| IL-2, IL-6, GM-CSF, WT1, RHAMM-R3 and PR1 peptides, GVAX, dendritic cells | Immunostimulatory approaches to increase T-lymphocyte mediated antileukemic responses in AML |
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; VEGF, vascular endothelial growth factor; Ph+, Philadelphia chromosome positive; mTOR, mammalian target of rapamycin; MDS, myelodysplastic syndrome; IL, interleukin; GM-CSF, granulocyte-macrophage colony-stimulating factor.
Cases at High Risk of CNS Relapse
Two recent studies showed that with effective chemotherapy, prophylactic cranial irradiation can be safely omitted altogether in the treatment of childhood ALL,10,123 showing isolated CNS relapse rates of 2.7% and 2.6%. The complete omission of prophylactic cranial irradiation in these studies allowed the clear identification of risk factors for CNS relapse, which were any CNS involvement at diagnosis, t(1;19)[TCF3-PBX1], and T-cell ALL.10,123 In the current St Jude Total Therapy Study XVI, triple intrathecal therapy is further intensified for these patients with twice weekly administration in the first 2 weeks of remission induction.
RISK-ADAPTED TREATMENT OF AML
Similar to ALL, risk-adapted therapy has also become an important approach in childhood AML. Many traditional prognostic factors have been replaced by cytogenetic and molecular features, as well as flow cytometric assessment of MRD (Table 5).
Table 5.
Characteristics and Clinical Outcomes of Selected Subtypes of Childhood AML
| Subtype | Affected Genes | Gene Functions | Frequency (%) | Clinical Characteristics | 5-Year EFS (%) | 5-Year OS (%) |
|---|---|---|---|---|---|---|
| Rearrangements | ||||||
| t(8;21)(q22;q22) | ETO-AML1 | Transcription factors | 12 | Associated with chloromas | 55–71 | 75–85 |
| inv(16)(p13;q22) | MYH11-CBF | Muscle protein/transcription factor | 8 | Eosinophilia with dysplastic basophilic granules | 72–88 | 75–85 |
| t(8;16) | MOZ-CBP | Transcription factors | 1 | High WBCs, chloromas, etoposide-related secondary AML | ID*† | ID |
| t(15;17)(q22;q12) | PML-RAR | Transcription factors/retinoid receptor | 12 | Associated with FAB M3, Auer rods common; FAB M3; ATRA-sensitive | 71† | 90† |
| t(11;17)(q23;q12) | PLZF-RARA | Transcription factors/retinoid receptor | Rare | Associated with FAB M3, Auer rods common; FAB M3; ATRA-resistant | ID | ID |
| t(1;22) | RBM15-MKL1 | RNA binding protein, DNA binding protein | 2–3 | Associated with FAB M7 in Down syndrome and non–Down syndrome | ID | ID |
| t(6;9)(p23;q34) | DEK-NUP214(CAN) | Transcription factor/nuclear transport | Rare | Basophilia and multilineage dysplasia; associated with FLT3-ITD and TdT+ | ID | ID |
| MLL | MLL (partner genes) | Histone methyltransferase | 18 | |||
| t(1;11)(q21;q23) | AF1q (MLLT11) | Function unknown, causes short half-life | 3 | 76% < age 2 years; 20% and 48% FAB M4 and M5, respectively | 92 | 100 |
| t(4;11)(q21;q23) | AF4 (MLLT2) | Associated with EAP10 | 2 | 61% < age 2 years; 17% and 42% FAB M4 and M5, respectively | 29 | 27 |
| t(6;11)(q27;q23) | AF6 (MLLT4) | Functions as dimerization domain | 5 | 9% < age 2 years; 57% ≥ 10 years; 35% and 41% FAB M4 and M5, respectively | 11 | 22 |
| t(9;11)(p22;q23) | AF9 (MLLT3) | ENL homolog, associated with EAP | 43 | 42% < age 2 years; 81% is FAB M5 | 50 | 63 |
| t(10;11)(p11.2;q23) | AF10 (MLLT10) | Interacts with DOT1L | 2 | 75% < age 2 years; 27% and 55% FAB M4 and M5, respectively | 17 | 27 |
| t(10;11)(p12;q23) | AF10 (MLLT10) | Interacts with DOT1L | 13 | 62% < age 2 years; 72% is FAB M5 | 31 | 45 |
| t(11;19)(q23;p13) | ELL or ENL (MLLT1) | Binds histone H3, Assembles EAP | 4 | 58% < age 2 years; 42% and 45% FAB M4 and M5, respectively | 49 | 49 |
| t(11;19)(q23;p13.1) | ELL or ENL (MLLT1) | Binds histone H3, Assembles EAP | 4 | 41% < age 2 years; 30% and 33% FAB M4 and M5 respectively | 46 | 61 |
| t(11;19)(q23;p13.3) | ELL or ENL (MLLT1) | Binds histone H3, Assembles EAP | 3 | 36% < age 2 years; 44% ≥ age 10 years; 20% and 40% FAB M4 and M5 | 46 | 47 |
| t(11;17)(q23;q21) | AF17 (MLLT6), LASP1 | F-actin rick cytoskeletal activity | 2 | 33% < age 2 years; 42% ≥ age 10 years; 33% and 50% FAB M4 and M5 | 11 | 22 |
| Other | 19 | 50% < age 2 years; 29% and 50% FAB M4 and M5, respectively | 39 | 54 | ||
| Normal karyotype | 20 | |||||
| Gene mutations | ||||||
| NPM | Nucleophosmin | Nuclear transporter RNA processing | 23 | 8%-10% of childhood AML | 65–80 | 75–85 |
| CEBPα | CCAAT/enhancer binding protein α | Transcription factor | 14 | 4%-6% of all childhood AML; more common in older patients, FAB M1 or M2 | 70 | 83 |
| FLT3/ALM | Fms-like tyrosine kinase 3 activation loop domain | Receptor for FLT3 | 3 | 6%-7% of all childhood AML | 50–60 | 60–70 |
| FLT3-ITD | Fms-like tyrosine kinase 3 internal tandem duplication | Receptor for FLT3 | 18 | 10%-15% of all childhood AML | < 35 | < 35 |
| WT1 | Wilms tumor 1 | Transcription factor | 13 | 8%-10% of all childhood AML | 22–35 | 35–56 |
| RAS | Rat sarcoma gene | Signal transduction | 3 | 5% of all childhood AML | ID | ID |
| PTPN11 | Protein tyrosine phosphatase, non-receptor type 11 | Tyrosine phosphatase | 2 | Most commonly associated with JMML | ID | ID |
| No known mutations | 24 | 40 | 50 | |||
| Poor-risk cytogenetics | < 15 | < 40 | < 40 | |||
| Del 5q/E5 | 1 | |||||
| −7 | 2 |
Abbreviations: AML, acute myeloid leukemia; EFS, event-free survival; ID, insufficient data; FAB, French-American-British; ATRA, all-trans-retinoic acid; JMML, juvenile myelomonocytic leukemia.
AML Characterized by Chimeric Transcription Factors: “Mostly Low-Risk” AML
With the introduction of intensively dosed and/or timed chemotherapy regimens, AML characterized by alterations of core-binding and transcription factors have emerged as a favorable prognostic group. Both t(8;21) and inv(16) AML have an approximately 80% overall survival rate when treated with three to four courses of intensive chemotherapy.15–28 Although patients with t(8;21) AML may have a higher relapse rate than those with inv(16), they are still able to be cured with allogeneic transplantation after attaining a second remission.127
Similar to adults, children with acute promyelocytic leukemia (APL) have an excellent overall survival in the 75% to 85% range, with some reports as high as 90%.124,128 This leukemia is particularly sensitive to ATRA and arsenic trioxide. Children with APL also benefit from maintenance therapy with ATRA and antimetabolites.129,130 Allogeneic transplantation is not recommended for these children in first remission. Survival after relapse is approximately 70% after reinduction, usually with arsenic trioxide, and then autologous or allogeneic transplantation.131,132
MLL-rearranged AML represents a diverse group with quite variable outcomes.65 In an international study, survival was 100%, 63%, 27%, and 22% for patients with the t(1;11), the t(9;11), the t(4;11), and the t(6;11), respectively.65 In part because of the variability in outcomes, small numbers of patients, and lack of definitive data demonstrating the superiority of HSCT, most cooperative trials do not include patients with MLL-rearranged AML in the high-risk group requiring allogeneic HSCT in first remission.133 Once associated with a poor prognosis,134 AMKL with the t(1;22) has improved outcome with contemporary treatment, and HSCT may not be indicated in these patients.26,135
AML Characterized by Mutation Altered Genes: The “Good” and the “Bad”
FLT3-ITD mutations, particularly when associated with a high mutant to wild-type allelic ratio, are associated with an overall survival rate of usually less than 30%.136 Although controversial, there are some indications that transplantation might improve outcome of children with this type of AML.137 Some clinical trials are testing the efficacy of various FLT3 inhibitors with different degrees of specificity plus allogeneic transplantation in these children. In the next COG trial for newly diagnosed AML, patients with a high FLT3-ITD to wild-type allelic ratio will be offered an allogeneic HSCT in first remission. Of note, patients with point mutations of FLT3 (eg, FLT3/ALM) do not have poor outcome and should be treated with chemotherapy only.
The c-KIT mutations, most commonly observed in CBF AML, has been reported to portend a poor prognosis in some studies, but not in a recent COG study.138 Similarly, RAS mutations have not been definitively associated with a poor prognosis. Thus patients with these mutations are usually treated with chemotherapy only.
NPM mutations convey both increased chemosensitivity and improved outcome.83 However, whether or not the presence of NPM mutations can partially abrogate the adverse affect of coexpressed FLT3-ITD is controversial.82 CEBPA mutations are often associated with normal karyotype AML and improved overall survival.70 Thus HSCT in first remission is not recommended for pediatric patients with isolated NPM or CEBPA mutations. WT1 mutations had an independent, poor prognostic impact in one study,125 but not in another (except in patients with coexpression of FLT3-ITD).126 Most clinical trials do not recommend allogeneic transplantation based on isolated WT1 mutations.
The Importance of MRD
Other than a Berlin-Frankfurt-Munster study, the detection of MRD has been associated with adverse prognostic significance.26,139 COG and St Jude trials are using this factor to stratify patients for risk-directed treatment.
Combined Approaches to Risk Stratification
In COG, the combination of cytogenetic, molecular, and MRD information is being used to stratify patients into two groups for risk-directed therapy. Low-risk AML includes patients with mutations involving CBF, CEBPA, and NPM and those with no MRD at the end of induction therapy. This group represents approximately 73% of patients, with a predicted survival close to 75%. The high-risk group (the remaining 27% of patients with survival < 35%) includes patients with adverse cytogenetic abnormalities (monosomy 7, del(5q)–, −5), high FLT3-ITD to wild-type allelic ratio, or MRD at the end of induction and will be offered HSCT in first remission with the most suitable donor.
Molecularly targeted drugs can be tested in both low- and high-risk groups with the intention of reducing toxic therapy in the former group while improving survival of the latter group (Table 4). For example, the use of bortezomib to augment chemotherapy effects on AML stem cells will be randomized in the next COG phase III trial. In addition, FLT3-ITD inhibitors (ie, sorafenib) will be tested in the high-risk group with a high FLT3-ITD to wild-type allelic ratio.
Special Subtypes
Down syndrome.
In view of superior prognosis of Down syndrome patients with AML,140 current trials are testing strategies to reduce overall drug exposure, especially cardiotoxic drugs, because of special concerns in this population.141
APL.
Since the demonstration that ATRA significantly improved the outcome of patients with APL, these patients have been treated separately from other patients with AML, resulting in 5-year overall survival rates as high as 87%.128 A major, adverse prognostic factor is presenting WBC more than 10 × 109/L, associated with an event-free survival of approximately 60%.128 Approximately 3% of patients died during induction from hemorrhagic complications, accounting for half of the induction failures. The microgranular variant (M3v), a bcr3 PML breakpoint, and the presence of FLT3-ITD had also been associated with a poor prognosis,142 partly due to increased presenting WBCs. In the current pediatric studies, patients are considered low risk or high risk on the basis of WBCs ≤ or more than 10 × 109/L, respectively.
Although intensive anthracycline treatment (cumulative doses ranging from 400 to 750 mg/m2) has been attributed to improve outcome,130 one of the key issues for pediatric patients has been to reduce exposure to these cardiotoxic drugs. The efficacy of arsenic in relapsed and newly diagnosed patients has made it an attractive alternative to anthracyclines.143–145 Thus the current COG trial replaces a course of anthracycline-containing chemotherapy with arsenic, reducing anthracycline exposure to 355 mg/m2 of daunorubicin equivalents for standard-risk patients with negative MRD and to 455 mg/m2 for high-risk patients and standard-risk patients with positive MRD after the third treatment course. This trial also includes high-dose cytarabine based on the survival advantage observed in a European trial.145 Maintenance therapy with ATRA plus antimetabolites will be given to all patients for approximately 2 years. This trial is similar to the European trial, with one primary difference being the introduction of arsenic during consolidation. Future studies may include other targeted agents, such as anti-CD33 monoclonal antibody therapy.146,147
AML in neonates and infants.
Because spontaneous remissions have been reported more frequently in the neonate age group, it is commonly recommended to initially observe such patients and, if cytoreduction is necessary, to perform an exchange transfusion. However, AML-directed therapy with dosing adjustments is usually required. Outcome in this age group is worse than that in older children, despite similar therapy, as a result of treatment toxicities and resistant disease.148 Novel approaches to directly target MLL149 and bcl-2150 are being developed (Table 4). In contrast to neonates, infants more than 1 month old seem to do as well as older children when treated with current, intensive regimens.15–30 The role of transplantation in infants is controversial because it has not been shown to definitely improve outcome and has significant adverse sequelae.
In conclusion, cure rates for childhood leukemias have improved largely through more intensive use of conventional cytotoxic agents evaluated in the context of large, randomized clinical trials. Clinical factors, genetic features of the leukemia, and initial response to therapy are now used in concert to personalize treatment for all patients. Advances in high-throughput genomics have led to the discovery of additional recurrent somatic lesions in the leukemic cell that offers not only additional prognostic information, but also opportunities for the application of novel targeted treatment. Finally, the recognition of host factors that are associated with the risk of leukemic transformation and the response to therapy will likely lead to more sophisticated treatment strategies in the near future.151,152
Acknowledgment
We thank many colleagues for their discussions and work on childhood leukemia. Our apologies go to colleagues whose work we were not able to reference because of space limitations.
Footnotes
Supported by the National Cancer Institute (Grant No. CA21765), CureSearch, and American Lebanese Syrian Associated Charities. R.J.A. is supported in part through an endowed King Fahd Professorship in Pediatric Oncology.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Ching-Hon Pui, Genzyme (C) Stock Ownership: None Honoraria: Ching-Hon Pui, Enzon Pharmaceuticals, sanofi-aventis Research Funding: None Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Ching-Hon Pui, Robert J. Arceci
Financial support: Ching-Hon Pui
Administrative support: Ching-Hon Pui
Provision of study materials or patients: Ching-Hon Pui, Robert J. Arceci
Collection and assembly of data: Ching-Hon Pui, Robert J. Arceci
Data analysis and interpretation: Ching-Hon Pui, Robert J. Arceci
Manuscript writing: Ching-Hon Pui, William L. Carroll, Soheil Meshinchi, Robert J. Arceci
Final approval of manuscript: Ching-Hon Pui, William L. Carroll, Soheil Meshinchi, Robert J. Arceci
REFERENCES
- 1.Conter V, Aricò M, Basso G, et al. Long-term results of the Italian Association of Pediatric Hematology and Oncology (AIEOP) Studies 82, 87, 88, 91 and 95 for childhood acute lymphoblastic leukemia. Leukemia. 2010;24:255–264. doi: 10.1038/leu.2009.250. [DOI] [PubMed] [Google Scholar]
- 2.Möricke A, Zimmermann M, Reiter A, et al. Long-term results of five consecutive trials in childhood acute lymphoblastic leukemia performed by the ALL-BFM study group from 1981 to 2000. Leukemia. 2010;24:265–284. doi: 10.1038/leu.2009.257. [DOI] [PubMed] [Google Scholar]
- 3.Gaynon PS, Angiolillo AL, Carroll WL, et al. Long-term results of the Children's Cancer Group studies for childhood acute lymphoblastic leukemia 1983-2002: A Children's Oncology Group Report. Leukemia. 2010;24:285–297. doi: 10.1038/leu.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escherich G, Horstmann MA, Zimmermann M, et al. Cooperative study group for childhood acute lymphoblastic leukaemia (COALL): Long-term results of trials 82,85,89,92 and 97. Leukemia. 2010;24:298–308. doi: 10.1038/leu.2009.249. [DOI] [PubMed] [Google Scholar]
- 5.Stary J, Jabali Y, Trka J, et al. Long-term results of treatment of childhood acute lymphoblastic leukemia in the Czech Republic. Leukemia. 2010;24:425–428. doi: 10.1038/leu.2009.255. [DOI] [PubMed] [Google Scholar]
- 6.Silverman LB, Stevenson KE, O'Brien JE, et al. Long-term results of Dana-Farber Cancer Institute ALL Consortium protocols for children with newly diagnosed acute lymphoblastic leukemia (1985–2000) Leukemia. 2010;24:320–334. doi: 10.1038/leu.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stark B, Nirel R, Avrahami G, et al. Long-term results of the Israeli National Studies in childhood acute lymphoblastic leukemia: INS 84, 89 and 98. Leukemia. 2010;24:419–424. doi: 10.1038/leu.2009.254. [DOI] [PubMed] [Google Scholar]
- 8.Schmiegelow K, Forestier E, Hellebostad M, et al. Long-term results of NOPHO ALL-92 and ALL-2000 studies of childhood acute lymphoblastic leukemia. Leukemia. 2010;24:345–354. doi: 10.1038/leu.2009.251. [DOI] [PubMed] [Google Scholar]
- 9.Pui CH, Pei D, Sandlund JT, et al. Long-term results of St Jude Total Therapy Studies 11, 12, 13A, 13B, and 14 for childhood acute lymphoblastic leukemia. Leukemia. 2010;24:371–382. doi: 10.1038/leu.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pui CH, Campana D, Pei D, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med. 2009;360:2730–2741. doi: 10.1056/NEJMoa0900386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuchida M, Ohara A, Manabe A, et al. Long-term results of Tokyo Children's Cancer Study Group trials for childhood acute lymphoblastic leukemia, 1984-1999. Leukemia. 2010;24:383–396. doi: 10.1038/leu.2009.260. [DOI] [PubMed] [Google Scholar]
- 12.Liang DC, Yang CP, Lin DT, et al. Long-term results of Taiwan Pediatric Oncology Group studies 1997 and 2002 for childhood acute lymphoblastic leukemia. Leukemia. 2010;24:397–405. doi: 10.1038/leu.2009.248. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell C, Richards S, Harrison CJ, et al. Long-term follow-up of the United Kingdom medical research council protocols for childhood acute lymphoblastic leukaemia, 1980-2001. Leukemia. 2010;24:406–418. doi: 10.1038/leu.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamps WA, van der Pal-de Bruin KM, Veerman AJ, et al. Long-term results of Dutch Childhood Oncology Group studies for children with acute lymphoblastic leukemia from 1984 to 2004. Leukemia. 2010;24:309–319. doi: 10.1038/leu.2009.258. [DOI] [PubMed] [Google Scholar]
- 15.Lange BJ, Smith FO, Feusner J, et al. Outcomes in CCG-2961, a Children's Oncology Group phase 3 trial for untreated pediatric acute myeloid leukemia: A report from the Children's Oncology Group. Blood. 2008;111:1044–1053. doi: 10.1182/blood-2007-04-084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burnett AK, Hills RK, Milligan DW, et al. Attempts to optimize induction and consolidation treatment in acute myeloid leukemia: Results of the MRC AML12 trial. J Clin Oncol. 2010;28:586–595. doi: 10.1200/JCO.2009.22.9088. [DOI] [PubMed] [Google Scholar]
- 17.Tomizawa D, Tabuchi K, Kinoshita A, et al. Repetitive cycles of high-dose cytarabine are effective for childhood acute myeloid leukemia: Long-term outcome of the children with AML treated on two consecutive trials of Tokyo Children's Cancer Study Group. Pediatr Blood Cancer. 2007;49:127–132. doi: 10.1002/pbc.20944. [DOI] [PubMed] [Google Scholar]
- 18.Armendariz H, Barbieri MA, Freigeiro D, et al. Treatment strategy and long-term results in pediatric patients treated in two consecutive AML-GATLA trials. Leukemia. 2005;19:2139–2142. doi: 10.1038/sj.leu.2403854. [DOI] [PubMed] [Google Scholar]
- 19.Lie SO, Abrahamsson J, Clausen N, et al. Long-term results in children with AML: NOPHO-AML Study Group—Report of three consecutive trials. Leukemia. 2005;19:2090–2100. doi: 10.1038/sj.leu.2403962. [DOI] [PubMed] [Google Scholar]
- 20.Creutzig U, Zimmermann M, Ritter J, et al. Treatment strategies and long-term results in paediatric patients treated in four consecutive AML-BFM trials. Leukemia. 2005;19:2030–2042. doi: 10.1038/sj.leu.2403920. [DOI] [PubMed] [Google Scholar]
- 21.Smith FO, Alonzo TA, Gerbing RB, et al. Long-term results of children with acute myeloid leukemia: A report of three consecutive phase III trials by the Children's Cancer Group: CCG 251, CCG 213 and CCG 2891. Leukemia. 2005;19:2054–2062. doi: 10.1038/sj.leu.2403925. [DOI] [PubMed] [Google Scholar]
- 22.Pession A, Barbieri E. Treatment and prevention of tumor lysis syndrome in children. Contrib Nephrol. 2005;147:80–92. doi: 10.1159/000082546. [DOI] [PubMed] [Google Scholar]
- 23.Entz-Werle N, Suciu S, van der Werff ten Bosch J, et al. Results of 58872 and 58921 trials in acute myeloblastic leukemia and relative value of chemotherapy vs allogeneic bone marrow transplantation in first complete remission: The EORTC Children Leukemia Group report. Leukemia. 2005;19:2072–2081. doi: 10.1038/sj.leu.2403932. [DOI] [PubMed] [Google Scholar]
- 24.Perel Y, Auvrignon A, Leblanc T, et al. Treatment of childhood acute myeloblastic leukemia: Dose intensification improves outcome and maintenance therapy is of no benefit—Multicenter studies of the French LAME (Leucemie Aigue Myeloblastique Enfant) Cooperative Group. Leukemia. 2005;19:2082–2089. doi: 10.1038/sj.leu.2403867. [DOI] [PubMed] [Google Scholar]
- 25.Ravindranath Y, Chang M, Steuber CP, et al. Pediatric Oncology Group (POG) studies of acute myeloid leukemia (AML): A review of four consecutive childhood AML trials conducted between 1981 and 2000. Leukemia. 2005;19:2101–2116. doi: 10.1038/sj.leu.2403927. [DOI] [PubMed] [Google Scholar]
- 26.Rubnitz JE, Inaba H, Dahl G, et al. Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: Results of the AML02 multicentre trial. Lancet Oncol. 2010;11:543–552. doi: 10.1016/S1470-2045(10)70090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson BE, Wheatley K, Hann IM, et al. Treatment strategy and long-term results in paediatric patients treated in consecutive UK AML trials. Leukemia. 2005;19:2130–2138. doi: 10.1038/sj.leu.2403924. [DOI] [PubMed] [Google Scholar]
- 28.Tsukimoto I, Tawa A, Horibe K, et al. Risk-stratified therapy and the intensive use of cytarabine improves the outcome in childhood acute myeloid leukemia: The AML99 trial from the Japanese Childhood AML Cooperative Study Group. J Clin Oncol. 2009;27:4007–4013. doi: 10.1200/JCO.2008.18.7948. [DOI] [PubMed] [Google Scholar]
- 29.Quintana J, Advis P, Becker A, et al. Acute myelogenous leukemia in Chile PINDA protocols 87 and 92 results. Leukemia. 2005;19:2143–2146. doi: 10.1038/sj.leu.2403959. [DOI] [PubMed] [Google Scholar]
- 30.Franklin J, Alonzo T, Hurwitz CA, et al. COG AAML03P1: Efficacy and safety in a pilot study of intensive chemotherapy including gemtuzumab in children newly diagnosed with acute myeloid leukemia (AML) ASH Annual Meeting Abstracts. 2008;112(abstr 13):56. [Google Scholar]
- 31.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 32.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 33.Reaman GH, Smith FO. Childhood Leukemias. Heidelberg, Germany: Springer Verlag; 2010. (in press) [Google Scholar]
- 34.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 35.Salzer WL, Devidas M, Carroll WL, et al. Long-term results of the pediatric oncology group studies for childhood acute lymphoblastic leukemia 1984-2001: A report from the Children's Oncology Group. Leukemia. 2010;24:355–370. doi: 10.1038/leu.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Attarbaschi A, Mann G, Panzer-Grümayer R, et al. Minimal residual disease values discriminate between low and high relapse risk in children with B-cell precursor acute lymphoblastic leukemia and an intrachromosomal amplification of chromosome 21: The Austrian and German acute lymphoblastic leukemia Berlin-Frankfurt-Munster (ALL-BFM) trials. J Clin Oncol. 2008;26:3046–3050. doi: 10.1200/JCO.2008.16.1117. [DOI] [PubMed] [Google Scholar]
- 37.Schultz KR, Bowman WP, Aledo A, et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: A Children's Oncology Group study. J Clin Oncol. 2009;27:5175–5181. doi: 10.1200/JCO.2008.21.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 39.Nachman JB, Heerema NA, Sather H, et al. Outcome of treatment in children with hypodiploid acute lymphoblastic leukemia. Blood. 2007;110:1112–1115. doi: 10.1182/blood-2006-07-038299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mullighan CG, Collins-Underwood JR, Phillips LA, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41:1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cario G, Zimmermann M, Romey R, et al. Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood. 2010;115:5393–5397. doi: 10.1182/blood-2009-11-256131. [DOI] [PubMed] [Google Scholar]
- 42.Harvey RC, Mullighan CG, Chen IM, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115:5312–5321. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meijerink JP, den Boer ML, Pieters R. New genetic abnormalities and treatment response in acute lymphoblastic leukemia. Semin Hematol. 2009;46:16–23. doi: 10.1053/j.seminhematol.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Graux C, Stevens-Kroef M, Lafage M, et al. Heterogeneous patterns of amplification of the NUP214-ABL1 fusion gene in T-cell acute lymphoblastic leukemia. Leukemia. 2009;23:125–133. doi: 10.1038/leu.2008.278. [DOI] [PubMed] [Google Scholar]
- 45.Coustan-Smith E, Mullighan CG, Onciu M, et al. Early T-cell precursor leukaemia: A subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mullighan CG, Su X, Zhang J, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Den Boer ML, van Slegtenhorst M, De Menezes RX, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: A genome-wide classification study. Lancet Oncol. 2009;10:125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mullighan CG, Zhang J, Harvey RC, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2009;106:9414–9418. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breit S, Stanulla M, Flohr T, et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108:1151–1157. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- 50.Gutierrez A, Sanda T, Grebliunaite R, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114:647–650. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mullighan CG, Miller CB, Radtke I, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 52.Hertzberg L, Vendramini E, Ganmore I, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: A report from the International BFM Study Group. Blood. 2010;115:1006–1017. doi: 10.1182/blood-2009-08-235408. [DOI] [PubMed] [Google Scholar]
- 53.van Grotel M, Meijerink JP, van Wering ER, et al. Prognostic significance of molecular-cytogenetic abnormalities in pediatric T-ALL is not explained by immunophenotypic differences. Leukemia. 2008;22:124–131. doi: 10.1038/sj.leu.2404957. [DOI] [PubMed] [Google Scholar]
- 54.Yang JJ, Bhojwani D, Yang W, et al. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood. 2008;112:4178–4183. doi: 10.1182/blood-2008-06-165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mullighan CG, Phillips LA, Su X, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhojwani D, Kang H, Moskowitz NP, et al. Biologic pathways associated with relapse in childhood acute lymphoblastic leukemia: A Children's Oncology Group study. Blood. 2006;108:711–717. doi: 10.1182/blood-2006-02-002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Treviño LR, Yang W, French D, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet 2009. 2009;41:1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang W, Trevino LR, Yang JJ, et al. ARID5B SNP rs10821936 is associated with risk of childhood acute lymphoblastic leukemia in blacks and contributes to racial differences in leukemia incidence. Leukemia. 2010;24:894–896. doi: 10.1038/leu.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lane SW, Scadden DT, Gilliland DG. The leukemic stem cell niche: Current concepts and therapeutic opportunities. Blood. 2009;114:1150–1157. doi: 10.1182/blood-2009-01-202606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gilliland DG, Jordan CT, Felix CA. The molecular basis of leukemia. Am Soc Hematol Educ Program. 2004:80–97. doi: 10.1182/asheducation-2004.1.80. [DOI] [PubMed] [Google Scholar]
- 62.Wiemels J, Kang M, Greaves M. Backtracking of leukemic clones to birth. Methods Mol Biol. 2009;538:7–27. doi: 10.1007/978-1-59745-418-6_2. [DOI] [PubMed] [Google Scholar]
- 63.Weisser M, Haferlach C, Hiddemann W, et al. The quality of molecular response to chemotherapy is predictive for the outcome of AML1-ETO-positive AML and is independent of pretreatment risk factors. Leukemia. 2007;21:1177–1182. doi: 10.1038/sj.leu.2404659. [DOI] [PubMed] [Google Scholar]
- 64.Mori H, Colman SM, Xiao Z, et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Natl Acad Sci U S A. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balgobind BV, Raimondi SC, Harbott J, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: Results of an international retrospective study. Blood. 2009;114:2489–2496. doi: 10.1182/blood-2009-04-215152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gupta M, Raghavan M, Gale RE, et al. Novel regions of acquired uniparental disomy discovered in acute myeloid leukemia. Genes Chromosomes Cancer. 2008;47:729–739. doi: 10.1002/gcc.20573. [DOI] [PubMed] [Google Scholar]
- 67.Raghavan M, Smith LL, Lillington DM, et al. Segmental uniparental disomy is a commonly acquired genetic event in relapsed acute myeloid leukemia. Blood. 2008;112:814–821. doi: 10.1182/blood-2008-01-132431. [DOI] [PubMed] [Google Scholar]
- 68.Radtke I, Mullighan CG, Ishii M, et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci U S A. 2009;106:12944–12949. doi: 10.1073/pnas.0903142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walter MJ, Payton JE, Ries RE, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci U S A. 2009;106:12950–12955. doi: 10.1073/pnas.0903091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ho PA, Alonzo TA, Gerbing RB, et al. Prevalence and prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia (AML): A report from the Children's Oncology Group. Blood. 2009;113:6558–6566. doi: 10.1182/blood-2008-10-184747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kirstetter P, Schuster MB, Bereshchenko O, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13:299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 72.Crispino JD. GATA1 mutations in Down syndrome: Implications for biology and diagnosis of children with transient myeloproliferative disorder and acute megakaryoblastic leukemia. Pediatr Blood Cancer. 2005;44:40–44. doi: 10.1002/pbc.20066. [DOI] [PubMed] [Google Scholar]
- 73.Meshinchi S, Alonzo TA, Stirewalt DL, et al. Clinical implications of FLT3 mutations in pediatric AML. Blood. 2006;108:3654–3661. doi: 10.1182/blood-2006-03-009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schnittger S, Kohl TM, Haferlach T, et al. KIT-D816 mutations in AML1-ETO-positive AML are associated with impaired event-free and overall survival. Blood. 2006;107:1791–1799. doi: 10.1182/blood-2005-04-1466. [DOI] [PubMed] [Google Scholar]
- 75.Cairoli R, Beghini A, Grillo G, et al. Prognostic impact of c-KIT mutations in core binding factor leukemias: An Italian retrospective study. Blood. 2006;107:3463–3468. doi: 10.1182/blood-2005-09-3640. [DOI] [PubMed] [Google Scholar]
- 76.Nanri T, Matsuno N, Kawakita T, et al. Mutations in the receptor tyrosine kinase pathway are associated with clinical outcome in patients with acute myeloblastic leukemia harboring t(8;21)(q22;q22) Leukemia. 2005;19:1361–1366. doi: 10.1038/sj.leu.2403803. [DOI] [PubMed] [Google Scholar]
- 77.Paschka P, Marcucci G, Ruppert AS, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): A Cancer and Leukemia Group B Study. J Clin Oncol. 2006;24:3904–3911. doi: 10.1200/JCO.2006.06.9500. [DOI] [PubMed] [Google Scholar]
- 78.Paschka P, Du J, Schlenk RF, et al. Mutations in the Fms-related tyrosine kinase 3 (FLT3) gene independently predict poor outcome in acute myeloid leukemia (AML) with t(8;21): A study of the German-Austrian AML Study Group (AMLSG) Am Soc Hematol Annual Meeting Abstracts. 2009;114(abstr 825):340. [Google Scholar]
- 79.Shih LY, Liang DC, Huang CF, et al. Cooperating mutations of receptor tyrosine kinases and Ras genes in childhood core-binding factor acute myeloid leukemia and a comparative analysis on paired diagnosis and relapse samples. Leukemia. 2008;22:303–307. doi: 10.1038/sj.leu.2404995. [DOI] [PubMed] [Google Scholar]
- 80.Berman JN, Gerbing RB, Sung L, et al. Prevalence and clinical implications of N-RAS mutations in childhood AML: A report from the Children's Oncology Group. Am Soc Hematol Annual Meeting Abstracts. 2009;114(abstr 3115):1211. [Google Scholar]
- 81.Scholl S, Fricke HJ, Sayer HG, et al. Clinical implications of molecular genetic aberrations in acute myeloid leukemia. J Cancer Res Clin Oncol. 2009;135:491–505. doi: 10.1007/s00432-008-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brown P, McIntyre E, Rau R, et al. The incidence and clinical significance of nucleophosmin mutations in childhood AML. Blood. 2007;110:979–985. doi: 10.1182/blood-2007-02-076604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hollink IH, Zwaan CM, Zimmermann M, et al. Favorable prognostic impact of NPM1 gene mutations in childhood acute myeloid leukemia, with emphasis on cytogenetically normal AML. Leukemia. 2009;23:262–270. doi: 10.1038/leu.2008.313. [DOI] [PubMed] [Google Scholar]
- 84.Schnittger S, Kern W, Tschulik C, et al. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood. 2009;114:2220–2231. doi: 10.1182/blood-2009-03-213389. [DOI] [PubMed] [Google Scholar]
- 85.Haferlach C, Mecucci C, Schnittger S, et al. AML with mutated NPM1 carrying a normal or aberrant karyotype show overlapping biologic, pathologic, immunophenotypic, and prognostic features. Blood. 2009;114:3024–3032. doi: 10.1182/blood-2009-01-197871. [DOI] [PubMed] [Google Scholar]
- 86.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ho PA, Alonzo TA, Kopecky KJ, et al. Molecular alterations of the IDH1 gene in AML: A Children's Oncology Group and Southwest Oncology Group study. Leukemia. 2010;24:909–913. doi: 10.1038/leu.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thol F, Damm F, Wagner K, et al. Prognostic impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. Blood. 2010;116:614–616. doi: 10.1182/blood-2010-03-272146. [DOI] [PubMed] [Google Scholar]
- 90.Chou WC, Hou HA, Chen CY, et al. Distinct clinical and biologic characteristics in adult acute myeloid leukemia bearing the isocitrate dehydrogenase 1 mutation. Blood. 2010;115:2749–2754. doi: 10.1182/blood-2009-11-253070. [DOI] [PubMed] [Google Scholar]
- 91.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 Gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wagner K, Damm F, Gohring G, et al. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010;28:2356–2364. doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- 93.Wouters BJ, Löwenberg B, Delwel R. A decade of genome-wide gene expression profiling in acute myeloid leukemia: Flashback and prospects. Blood. 2009;113:291–298. doi: 10.1182/blood-2008-04-153239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wouters BJ, Jorda MA, Keeshan K, et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 2007;110:3706–3714. doi: 10.1182/blood-2007-02-073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Figueroa ME, Wouters BJ, Skrabanek L, et al. Genome-wide epigenetic analysis delineates a biologically distinct immature acute leukemia with myeloid/T-lymphoid features. Blood. 2009;113:2795–2804. doi: 10.1182/blood-2008-08-172387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen J, Odenike O, Rowley JD. Leukaemogenesis: More than mutant genes. Nat Rev Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Golub T, Arceci RJ. Acute myelogenous leukemia. In: Pizzo P, Poplack G, editors. Principles and Practice of Pediatric Oncology. Philadelphia, PA: Lippincott Williams & Wilkins; 2006. pp. 591–644. [Google Scholar]
- 99.Arceci RJ, Brown P. Myeloid leukemias and myelodysplastic syndromes, in Carroll W, Finlay J (eds): Cancer in Children: The Clinical Biology of Childhood Cancer. Boston, MA: Jones and Bartlett Publishers; 2010. pp. 185–215. [Google Scholar]
- 100.Stanulla M, Schrappe M. Treatment of childhood acute lymphoblastic leukemia. Semin Hematol. 2009;46:52–63. doi: 10.1053/j.seminhematol.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 101.Pui CH, Jeha S. New therapeutic strategies for the treatment of acute lymphoblastic leukaemia. Nat Rev Drug Discov. 2007;6:149–165. doi: 10.1038/nrd2240. [DOI] [PubMed] [Google Scholar]
- 102.Moghrabi A, Levy DE, Asselin B, et al. Results of the Dana-Farber Cancer Institute ALL Consortium Protocol 95-01 for children with acute lymphoblastic leukemia. Blood. 2007;109:896–904. doi: 10.1182/blood-2006-06-027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Berg SL, Blaney SM, Devidas M, et al. Phase II study of nelarabine (compound 506U78) in children and young adults with refractory T-cell malignancies: A report from the Children's Oncology Group. J Clin Oncol. 2005;23:3376–3382. doi: 10.1200/JCO.2005.03.426. [DOI] [PubMed] [Google Scholar]
- 104.Schrappe M, Zimmermann M, Möricke A, et al. Dexamethasone in induction can eliminate one third of all relapses in childhood acute lymphoblastic leukemia (ALL): Results of an international randomized trial in 3655 patients (Trial AEIOP-BFM ALL 2000) Am Soc Hematol Annual Meeting Abstracts. 2008;112(abstr 7):9. [Google Scholar]
- 105.Pieters R, Schrappe M, De Lorenzo P, et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): An observational study and a multicentre randomised trial. Lancet. 2007;370:240–250. doi: 10.1016/S0140-6736(07)61126-X. [DOI] [PubMed] [Google Scholar]
- 106.Pui CH, Gaynon PS, Boyett JM, et al. Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet. 2002;359:1909–1915. doi: 10.1016/S0140-6736(02)08782-2. [DOI] [PubMed] [Google Scholar]
- 107.Brown P, Levis M, McIntyre E, et al. Combinations of the FLT3 inhibitor CEP-701 and chemotherapy synergistically kill infant and childhood MLL-rearranged ALL cells in a sequence-dependent manner. Leukemia. 2006;20:1368–1376. doi: 10.1038/sj.leu.2404277. [DOI] [PubMed] [Google Scholar]
- 108.Stumpel DJ, Schneider P, van Roon EH, et al. Specific promoter methylation identifies different subgroups of MLL-rearranged infant acute lymphoblastic leukemia, influences clinical outcome, and provides therapeutic options. Blood. 2009;114:5490–5498. doi: 10.1182/blood-2009-06-227660. [DOI] [PubMed] [Google Scholar]
- 109.Schafer E, Irizarry R, Negi S, et al. Promoter hypermethylation in MLL-r infant acute lymphoblastic leukemia: Biology and therapeutic targeting. Blood. 2010;115:4798–4809. doi: 10.1182/blood-2009-09-243634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pulte D, Gondos A, Brenner H. Improvement in survival in younger patients with acute lymphoblastic leukemia from the 1980s to the early 21st century. Blood. 2009;113:1408–1411. doi: 10.1182/blood-2008-06-164863. [DOI] [PubMed] [Google Scholar]
- 111.Stock W, La M, Sanford B, et al. What determines the outcomes for adolescents and young adults with acute lymphoblastic leukemia treated on cooperative group protocols? A comparison of Children's Cancer Group and Cancer and Leukemia Group B studies. Blood. 2008;112:1646–1654. doi: 10.1182/blood-2008-01-130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nachman JB, La MK, Hunger SP, et al. Young adults with acute lymphoblastic leukemia have an excellent outcome with chemotherapy alone and benefit from intensive postinduction treatment: A report from the children's oncology group. J Clin Oncol. 2009;27:5189–5194. doi: 10.1200/JCO.2008.20.8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boissel N, Auclerc MF, Lheritier V, et al. Should adolescents with acute lymphoblastic leukemia be treated as old children or young adults? Comparison of the French FRALLE-93 and LALA-94 trials. J Clin Oncol. 2003;21:774–780. doi: 10.1200/JCO.2003.02.053. [DOI] [PubMed] [Google Scholar]
- 114.de Bont JM, Holt B, Dekker AW, et al. Significant difference in outcome for adolescents with acute lymphoblastic leukemia treated on pediatric vs adult protocols in the Netherlands. Leukemia. 2004;18:2032–2035. doi: 10.1038/sj.leu.2403538. [DOI] [PubMed] [Google Scholar]
- 115.Pui CH, Pei D, Campana D, et al. Improved prognosis for older adolescents with acute lymphoblastic leukemia. J Clin Oncol. doi: 10.1200/JCO.2010.32.0325. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goldstone AH, Richards SM, Lazarus HM, et al. In adults with standard-risk acute lymphoblastic leukemia, the greatest benefit is achieved from a matched sibling allogeneic transplantation in first complete remission, and an autologous transplantation is less effective than conventional consolidation/maintenance chemotherapy in all patients: Final results of the International ALL Trial (MRC UKALL XII/ECOG E2993) Blood. 2008;111:1827–1833. doi: 10.1182/blood-2007-10-116582. [DOI] [PubMed] [Google Scholar]
- 117.Ribera JM, Oriol A, Sanz MA, et al. Comparison of the results of the treatment of adolescents and young adults with standard-risk acute lymphoblastic leukemia with the Programa Espanol de Tratamiento en Hematologia pediatric-based protocol ALL-96. J Clin Oncol. 2008;26:1843–1849. doi: 10.1200/JCO.2007.13.7265. [DOI] [PubMed] [Google Scholar]
- 118.DeAngelo DJ, Dahlberg S, Silverman LB, et al. A multicenter phase II study using a dose intensified pediatric regimen in adults with untreated acute lymphoblastic leukemia. Am Soc Hematol Annual Meeting Abstracts. 2007;110(abstr 587):181a. [Google Scholar]
- 119.Storring JM, Minden MD, Kao S, et al. Treatment of adults with BCR-ABL negative acute lymphoblastic leukaemia with a modified paediatric regimen. Br J Haematol. 2009;146:76–85. doi: 10.1111/j.1365-2141.2009.07712.x. [DOI] [PubMed] [Google Scholar]
- 120.Imai C, Iwamoto S, Campana D. Genetic modification of primary natural killer cells overcomes inhibitory signals and induces specific killing of leukemic cells. Blood. 2005;106:376–383. doi: 10.1182/blood-2004-12-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Triplett B, Handgretinger R, Pui CH, et al. KIR-incompatible hematopoietic-cell transplantation for poor prognosis infant acute lymphoblastic leukemia. Blood. 2006;107:1238–1239. doi: 10.1182/blood-2005-07-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bargou R, Leo E, Zugmaier G, et al. Tumor regression in cancer patients by very low doses of a T cell-engaging antibody. Science. 2008;321:974–977. doi: 10.1126/science.1158545. [DOI] [PubMed] [Google Scholar]
- 123.Veerman AJ, Kamps WA, van den Berg H, et al. Dexamethasone-based therapy for childhood acute lymphoblastic leukaemia: Results of the prospective Dutch Childhood Oncology Group (DCOG) protocol ALL-9 (1997–2004) Lancet Oncol. 2009;10:957–966. doi: 10.1016/S1470-2045(09)70228-1. [DOI] [PubMed] [Google Scholar]
- 124.de Botton S, Coiteux V, Chevret S, et al. Outcome of childhood acute promyelocytic leukemia with all-trans-retinoic acid and chemotherapy. J Clin Oncol. 2004;22:1404–1412. doi: 10.1200/JCO.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 125.Hollink IH, van den Heuvel-Eibrink MM, Zimmermann M, et al. Clinical relevance of Wilms tumor 1 gene mutations in childhood acute myeloid leukemia. Blood. 2009;113:5951–5960. doi: 10.1182/blood-2008-09-177949. [DOI] [PubMed] [Google Scholar]
- 126.Ho PA, Zeng R, Alonzo TA, et al. Prevalence and prognostic implications of WT1 mutations in pediatric acute myeloid leukemia (AML): a report from the Children's Oncology Group. Blood. 2010;116:702–710. doi: 10.1182/blood-2010-02-268953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Abrahamsson J, Clausen N, Gustafsson G, et al. Improved outcome after relapse in children with acute myeloid leukaemia. Br J Haematol. 2007;136:229–236. doi: 10.1111/j.1365-2141.2006.06419.x. [DOI] [PubMed] [Google Scholar]
- 128.Ortega JJ, Madero L, Martin G, et al. Treatment with all-trans retinoic acid and anthracycline monochemotherapy for children with acute promyelocytic leukemia: A multicenter study by the PETHEMA Group. J Clin Oncol. 2005;23:7632–7640. doi: 10.1200/JCO.2005.01.3359. [DOI] [PubMed] [Google Scholar]
- 129.Gregory J, Kim H, Alonzo T, et al. Treatment of children with acute promyelocytic leukemia: Results of the first North American Intergroup trial INT0129. Pediatr Blood Cancer. 2009;53:1005–1010. doi: 10.1002/pbc.22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sanz MA, Grimwade D, Tallman MS, et al. Management of acute promyelocytic leukemia: Recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2009;113:1875–1891. doi: 10.1182/blood-2008-04-150250. [DOI] [PubMed] [Google Scholar]
- 131.de Botton S, Fawaz A, Chevret S, et al. Autologous and allogeneic stem-cell transplantation as salvage treatment of acute promyelocytic leukemia initially treated with all-trans-retinoic acid: A retrospective analysis of the European acute promyelocytic leukemia group. J Clin Oncol. 2005;23:120–126. doi: 10.1200/JCO.2005.03.127. [DOI] [PubMed] [Google Scholar]
- 132.Thirugnanam R, George B, Chendamarai E, et al. Comparison of clinical outcomes of patients with relapsed acute promyelocytic leukemia induced with arsenic trioxide and consolidated with either an autologous stem cell transplant or an arsenic trioxide-based regimen. Biol Blood Marrow Transplant. 2009;15:1479–1484. doi: 10.1016/j.bbmt.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 133.Oliansky DM, Rizzo JD, Aplan PD, et al. The role of cytotoxic therapy with hematopoietic stem cell transplantation in the therapy of acute myeloid leukemia in children: An evidence-based review. Biol Blood Marrow Transplant. 2007;13:1–25. doi: 10.1016/j.bbmt.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 134.Bernstein J, Dastugue N, Haas OA, et al. Nineteen cases of the t(1;22)(p13;q13) acute megakaryblastic leukaemia of infants/children and a review of 39 cases: Report from a t(1;22) study group. Leukemia. 2000;14:216–218. doi: 10.1038/sj.leu.2401639. [DOI] [PubMed] [Google Scholar]
- 135.Duchayne E, Fenneteau O, Pages MP, et al. Acute megakaryoblastic leukaemia: A national clinical and biological study of 53 adult and childhood cases by the Groupe Francais d'Hematologie Cellulaire (GFHC) Leuk Lymphoma. 2003;44:49–58. doi: 10.1080/1042819021000040279. [DOI] [PubMed] [Google Scholar]
- 136.Pratz KW, Sato T, Murphy KM, et al. FLT3-mutant allelic burden and clinical status are predictive of response to FLT3 inhibitors in AML. Blood. 2010;115:1425–1432. doi: 10.1182/blood-2009-09-242859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Meshinchi S, Arceci RJ, Sanders JE, et al. Role of allogeneic stem cell transplantation in FLT3/ITD-positive AML. Blood. 2006;108:400–401. doi: 10.1182/blood-2005-12-4938. [DOI] [PubMed] [Google Scholar]
- 138.Pollard JA, Alonzo TA, Gerbing RB, et al. Prevalence and prognostic significance of KIT mutations in pediatric patients with core binding factor AML enrolled on serial pediatric cooperative trials for de novo AML. Blood. 2010;115:2372–2379. doi: 10.1182/blood-2009-09-241075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sievers EL, Lange BJ, Alonzo TA, et al. Immunophenotypic evidence of leukemia after induction therapy predicts relapse: Results from a prospective Children's Cancer Group study of 252 acute myeloid leukemia patients. Blood. 2003;101:3398–3406. doi: 10.1182/blood-2002-10-3064. [DOI] [PubMed] [Google Scholar]
- 140.Gamis AS. Acute myeloid leukemia and Down syndrome evolution of modern therapy: State of the art review. Pediatr Blood Cancer. 2005;44:13–20. doi: 10.1002/pbc.20207. [DOI] [PubMed] [Google Scholar]
- 141.O'Brien MM, Taub JW, Chang MN, et al. Cardiomyopathy in children with Down syndrome treated for acute myeloid leukemia: A report from the Children's Oncology Group Study POG 9421. J Clin Oncol. 2008;26:414–420. doi: 10.1200/JCO.2007.13.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tallman MS. The expanding role of arsenic in acute promyelocytic leukemia. Semin Hematol. 2008;45:S25–S29. doi: 10.1053/j.seminhematol.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 143.Powell BL. Effect of consolidation with arsenic trioxide (As2O3) on event-free survival (EFS) and overall survival (OS) among patients with newly diagnosed acute promyelocytic leukemia (APL): North American Intergroup Protocol C9710. J Clin Oncol. 2007;25(abstr 2):1s. [Google Scholar]
- 144.Gore SD, Hermes-DeSantis ER. Enhancing survival outcomes in the management of patients with higher-risk myelodysplastic syndromes. Cancer Control. 2009;16(suppl):2–10. doi: 10.1177/107327480901604s03. [DOI] [PubMed] [Google Scholar]
- 145.Adès L, Chevret S, Raffoux E, et al. Is cytarabine useful in the treatment of acute promyelocytic leukemia? Results of a randomized trial from the European Acute Promyelocytic Leukemia Group. J Clin Oncol. 2006;24:5703–5710. doi: 10.1200/JCO.2006.08.1596. [DOI] [PubMed] [Google Scholar]
- 146.Estey EH, Giles FJ, Beran M, et al. Experience with gemtuzumab ozogamycin (“mylotarg”) and all-trans retinoic acid in untreated acute promyelocytic leukemia. Blood. 2002;99:4222–4224. doi: 10.1182/blood-2001-12-0174. [DOI] [PubMed] [Google Scholar]
- 147.Lo Coco F, Ammatuna E, Noguera N. Treatment of acute promyelocytic leukemia with gemtuzumab ozogamicin. Clin Adv Hematol Oncol 4:57- 2006;62:76–77. [PubMed] [Google Scholar]
- 148.Ishii E, Oda M, Kinugawa N, et al. Features and outcome of neonatal leukemia in Japan: Experience of the Japan infant leukemia study group. Pediatr Blood Cancer. 2006;47:268–272. doi: 10.1002/pbc.20599. [DOI] [PubMed] [Google Scholar]
- 149.Liedtke M, Cleary ML. Therapeutic targeting of MLL. Blood. 2009;113:6061–6068. doi: 10.1182/blood-2008-12-197061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Robinson BW, Behling KC, Gupta M, et al. Abundant anti-apoptotic BCL-2 is a molecular target in leukaemias with t(4;11) translocation. Br J Haematol. 2008;141:827–839. doi: 10.1111/j.1365-2141.2008.07100.x. [DOI] [PubMed] [Google Scholar]
- 151.Yang JJ, Cheng C, Yang W, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Treviño LR, Shimasaki N, Yang W, et al. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J Clin Oncol. 2009;27:5972–5978. doi: 10.1200/JCO.2008.20.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]