Abstract
Sperm chromatin is organized in a protamine-based, highly condensed form, which protects the paternal chromosome complement in transit, facilitates fertilization, and supports correct gene expression in the early embryo. Very few histones remain selectively associated with genes and defined regulatory sequences essential to embryonic development, while most of the genome becomes bound to protamine during spermiogenesis. Chromatin remodeling processes resulting in the dramatically different nuclear structure of sperm are poorly understood. This study shows that perturbation of poly(ADP-ribose) (PAR) metabolism, which is mediated by PAR polymerases and PAR glycohydrolase in response to naturally occurring endogenous DNA strand breaks during spermatogenesis, results in the abnormal retention of core histones and histone linker HIST1H1T (H1t) and H1-like linker protein HILS1 in mature sperm. Moreover, genetic or pharmacological alteration of PAR metabolism caused poor sperm chromatin quality and an abnormal nuclear structure in mice, thus reducing male fertility.
Keywords: chromatin remodeling, condensation, histone H1 linker, PARG, PARP, poly(ADP)ribose, poly(ADP-ribose) glycohydrolase, poly(ADP-ribose) polymerase, sperm, spermatid, spermatogenesis, spermiogenesis, testis, TOP2A, TOP2B, topoisomerase II beta, transition protein
Genetic and pharmacological perturbation of poly(ADP-ribose) (PAR) metabolism during spermiogenesis results in abnormal retention of core and linker histones in mature sperm.
INTRODUCTION
Nuclear DNA integrity and composition of chromatin proteins are important to normal sperm function [1]. Spermatid development is characterized by significant changes in cell physiology and nuclear organization, such as a dramatic decrease in nuclear size that is associated with a complete reorganization of the haploid genome from histone-bound nucleosomal DNA to a very condensed and mostly protamine-bound form (Fig. 1). During this process, histones are removed from the DNA and first replaced by transition proteins TP1 and TP2 and then by protamines P1 and P2. Only a small, but apparently well-defined fraction of the sperm genome remains histone-associated in mature sperm. The distribution of residual histones in sperm nuclei is not random but appears associated with promoters of master gene regulators such as Hox and pluripotency genes as well as CTCF binding sites [2–4]. Abnormally increased amounts of histones in sperm are associated with decreased fertility and increased risk of embryonic failure after fertilization [5]. Therefore, histone retention and protamine deficiency in sperm are hallmarks of certain forms of idiopathic infertility [6–8], but the genetic and mechanistic causes underlying this defect remain enigmatic.
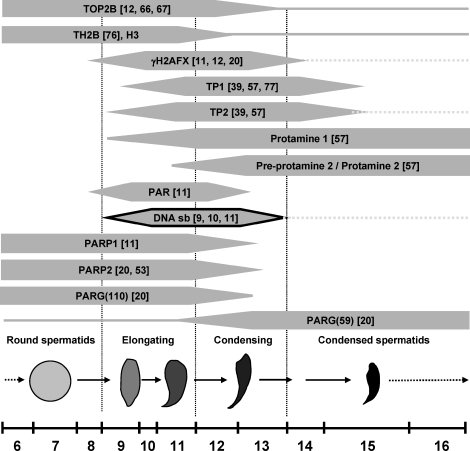
FIG. 1.
Spermatid nuclear condensation encompasses dramatic changes in nuclear protein composition. Schematic depiction of spermatid developmental steps 6–16 when nuclear remodeling is most evident. Spermatids begin the elongation process at step 9, and the changes in shape are indicated by the drawings that are not to scale. DNA strand breaks (black outline) occur at stages 9–13 with a distinct peak frequency during stages 10 and 11. Protein transitions are shown along with some, but not all, relevant references. Uninterrupted lines indicate where low-level presence of a given protein has been documented, and broken lines indicate where proteins or DNA strand breaks are retained abnormally in mouse mutants with disruption of the PAR pathway.
The transition from histone- to protamine-based chromatin during spermiogenesis is associated with a transient occurrence of physiological DNA strand breaks (Fig. 1) [9–11]. These DNA strand breaks, which likely permit topological changes associated with DNA relaxation during nucleoprotein exchange in spermiogenesis, have recently been attributed to the activity of topoisomerase II beta (TOP2B) [12]. Notably, these DNA strand breaks induce DNA damage signaling involving H2AFX phosphorylation and formation of poly(ADP-ribose) (PAR) [11]. Poly(ADP-ribosyl)ation (PARsylation) is involved in many different fundamental cellular functions, including recruitment of XRCC1 and DNA-PK for DNA repair as well as modulation of chromatin structure during transcription initiation [13–17]. A large body of data obtained from exogenous induction of DNA strand breaks in somatic tissues or cells using ionizing radiation or alkylating substances shows that DNA strand breaks are efficiently recognized by the DNA-binding domains of PAR polymerases 1 and 2 (PARP1 and PARP2, respectively) [18]. Binding of PARP1 and PARP2 to DNA strand breaks activates the enzymes, which cleave NAD+ to generate an ADP-ribose polymer that is subsequently attached to target proteins as a posttranslational modification. PARP enzymes themselves also poly(ADP-ribosyl)ate themselves; such automodification inactivates these enzymes and allows them to dissociate from the break. PARsylation marks are very short-lived as a result of a quick and specific degradation of PAR by poly(ADP-ribose) glycohydrolase (PARG), an essential enzyme that is ubiquitously expressed. The coordinated actions of PARPs and PARG therefore result in rapid and transient formation of PAR in response to detection of DNA strand breaks. Hence, reduction of PARG activity in the hypomorphic Parg(110)−/− mouse causes extended and prolonged automodification of PARPs with PAR, which inhibits these enzymes [19, 20].
PARP1 and PARP2 appear to have partially overlapping functions, and the double knock out of both genes in mice is embryonic lethal [21]. Similarly, complete ablation of Parg gene expression is lethal to the embryo at an early stage as a result of toxic accumulation of PAR [22]. The Parg gene encodes four enzymatically active isoforms [23], and mice with a partial Parg gene knockout (Parg(110)−/−) are viable because of at least one remaining, functional shorter PARG isoform. PARG activity in those mice is reduced to ∼25% of wild-type activity, resulting in an impaired PAR turnover [19, 24].
In MCF-7 breast cancer cells, endogenous DNA strand breaks created by TOP2B activate PARP1. The resulting PARsylation mediates a nucleosome-specific exchange of histone linker H1 for high-mobility group B proteins, and this alteration of chromatin structure to a more open state facilitates nuclear receptor-dependent transcription [25].
In earlier studies, we observed that spermatid steps undergoing chromatin remodeling contain endogenous DNA strand breaks and form PAR [11]. These observations led us to test the hypothesis that PAR metabolism may facilitate histone H1 removal and nuclear condensation in spermiogenesis. We report here that impairment of PAR metabolism by Parg gene disruption or by pharmacological PARP inhibition during spermiogenesis results in abnormal chromatin condensation and histone retention in mature sperm. Taken together, these results suggest that PAR metabolism mediated by PARP1, PARP2, and PARG is a newly identified component of chromatin condensation in spermiogenesis that assists in H1 linker-like protein removal, which further facilitates chromatin remodeling. Consequentially, genetic and biochemical parameters of PAR metabolism emerge as candidate culprits in patients diagnosed with idiopathic infertility characterized by histone retention and immature sperm chromatin.
MATERIALS AND METHODS
Mouse Models
Parp1 gene-disrupted mice (Parp1tm1Zqw [26]; Parp1−/−) as well as Parg gene-disrupted mice (Parg(110)−/− [24]) were maintained according to the guidelines of the University of Pennsylvania Institutional Animal Care and Use Committee. Mouse strains were maintained both as heterozygous Parg(110)−/+ and Parg(110)−/− homozygous lines in a 129SVE background. Crossbreeds of Parg(110)−/− and Parp1−/− mice (Parp1−/−/Parg(110)−/− double gene-disrupted mice) were reported previously [20]. Wild-type controls used in the described studies were littermates of knockout mice from heterozygous parents. Parg(110)−/− mice have a targeted deletion of exons 2 and 3 in the Parg gene, which leads to ablation of the three large PARG protein isoforms of 110, 102, and 98 kDa but the two smaller ones of 63 kDa (ubiquitous) and 58 kDa (mitochondrial) [24, 27, 28] are still expressed. Mice homozygous for complete knock out of Parg show an early embryonic lethal phenotype [22]. Wild-type 129SVE males (129S6/SvEvTac) used for the inhibitor studies were purchased from Taconics Inc. (Hudson, NY).
PARP Inhibitor Studies
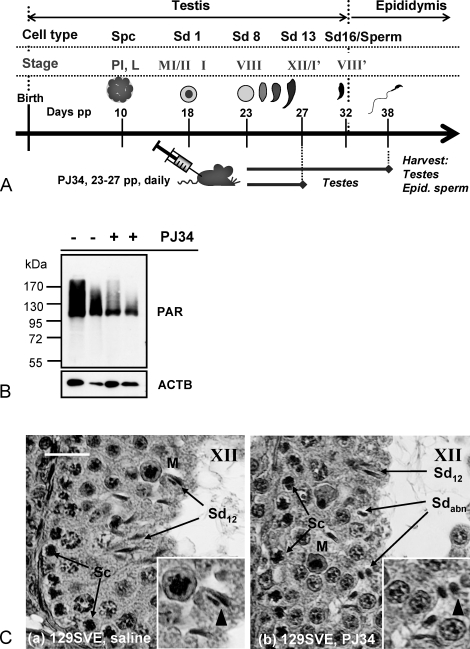
Groups of five wild-type 129SVE male mice were each treated with PJ34 (N-(6-oxo-5,6-dihydro-phenanthridin-2-yl)-N,N-dimethylacetamide HCl) [29–32], (Axxora, San Diego, CA) at a concentration of 10 mg/kg bodyweight by daily intraperitoneal injections on Days 23–27 pp (postpartum). Testes and epididymides were either harvested on Day 27 for SDS-PAGE protein analysis or fixed in Bouin solution (Sigma-Aldrich, St. Louis, MO) for histology, or on Day 40 for isolation of epididymal sperm.
Tissue Preparation
Male mice of the different genotypes, that is, Parg(110)−/−, Parp1−/−, Parp1−/−/Parg(110)−/− double knockout, and wild type, were closely age-matched for the studies. All animals were 79–81 days old except where noted. Tissues were either used fresh or immediately frozen in liquid nitrogen and stored at −80°C. Isolation of sonication-resistant spermatid nuclei (SRN), representing testicular sperm nuclei and spermatid steps 12 and later, and cauda epididymal sperm was performed as described previously [20] with minor modifications as described below. Epididymal sperm were isolated in phosphate buffered saline (PBS) with CaCl2 and MgCl2 (Invitrogen, Carlsbad, CA, catalog no. 14040-117). For chromomycin A3 staining, sperm smears were prepared on glass slides and air dried. For all the other investigations, sperm suspensions were treated with somatic cell lysis buffer (0.1% SDS, 0.5% Triton X-100 in diethylpyrocarbonate H2O) for 20 min on ice [4], then centrifuged and resuspended in PBS with CaCl2 and MgCl2, and finally counted using a hemacytometer chamber.
Protein Extracts and Immunoblotting
SDS extracts of whole testis were made from frozen testes by weighing unthawed decapsulated tissue in cooled, weighed 1.5 ml reaction tubes. Three volumes (v/w) of cooled 1× RIPA buffer (150 mM NaCl, 10 mM Tris-HCl, pH 7.4, 1% NP-40, 1% deoxycholic acid [DOC], 0.1% SDS), prepared with complete protease inhibitor cocktail (Roche, Indianapolis, IN) were added and the samples homogenized using 1.5 ml reaction tubes with tight-fitting micropestles (VWR, Radnor, PA). Six volumes (v/w) of 2× Laemmli sample buffer (Biorad, Hercules, CA, catalog no. 161-0737) were then added to give a final concentration of 100 ng/μl total testis fresh weight.
Extraction of proteins from SRN for either SDS-PAGE or acid-urea PAGE (AU-PAGE) was done by homogenization of four fresh testes (one per animal) per genotype as described previously [20, 33]. Isolated nuclei resistant to ultrasonic shear were diluted and counted using a hemacytometer. Samples were divided in half and either resuspended in boiling 2× SDS sample buffer to give a final concentration of 6.7 × 104 nuclei/μl or nucleoproteins were extracted with HCl using a published method [34] with the following modifications: pellets of purified SRN were resuspended in 720 μl of 5 mM MgCl2, 10 mM Tris-HCl, pH 7.5, 10 mM dithiothreitol (DTT), and 0.5 mM phenylmethylsulfonylfluoride per two testes starting material and incubated on ice for 10 min. Then, 10 M HCl was added to achieve a final HCl concentration of 0.5 M, and the samples were mixed and incubated on ice for 30 min. After centrifugation, acid-soluble proteins in the supernatant were precipitated with 25% trichloroacetic acid and the pellets washed with acidified acetone and then again with regular acetone. Semidry pellets were resuspended in 5% acetic acid according to the starting number of nuclei to give 1 × 105/μl. Before loading the samples on the AU gel, 2× loading buffer (5 M urea, 5% acetic acid, 5% beta-mercaptoethanol with methyl green dye) was added. Protein samples were separated by 15% AU-PAGE, 15% (w/v) urea, and electrotransferred to small pore polyvinylidene fluoride (PVDF) membranes (Immobilon-PSQ, Millipore, Billerica, MA) using a wet blot system with 0.1 M glycine/formic acid, pH 3.5, 10% methanol.
Testis lysates in Laemmli sample buffer were separated by 8% or 15% SDS-PAGE and transferred to PVDF membranes. All the protein blots were subjected to antibody detection by enhanced chemiluminescence. Quantification of bands was achieved using the ImageJ software package (Wayne Rasband, National Institutes of Health, http://rsb.info.nih.gov/ij/). For Western blot detection, the following primary antibodies were used: rabbit anti-PAR (LP96-10, 1:2000; BD Bioscience, San Diego, CA), mouse anti-ACTB (AC-15, 1:5000; Sigma, St. Louis, MO), rabbit anti-histone H3 (1:1000; Abcam, Cambridge, MA), rabbit anti-testis-specific H1t (HIST1H1T) (1:2000) [35], rabbit anti-HILS1 (1:1000) [36], and rabbit anti-macroH2A1 (1:1000; Millipore) [37], rabbit anti-TP1 (1:2000) [38], rabbit anti-TP2 (1:2000) [39], rabbit anti-MnSOD (1:2000; Stressgen, Ann Arbor, MI), rabbit anti-histone H3K4me3 (1:1000, Abcam), rabbit anti-histone H3K9me2 (1:1000; Millipore), rabbit anti-histone H3K9me3 (1:1000; Abcam), rabbit anti-histone H3K27me3 (1:1000 or 1:500 for immunofluorescence detection; Millipore), rabbit anti-hyperacetylated histone H4 (1:1000; Millipore, catalog no. 06-946), and horseradish peroxidase- or TRITC (tetramethylrhodamine isothiocyanate)-coupled donkey anti-rabbit donkey anti-mouse secondary antibodies (Jackson Immunolabs, West Grove, PA), according to standard procedures [20]. Testis-specific histone H2B (HIST2H2BA; Th2B) was detected using mouse anti-tyrosine hydroxylase (clone 2/40/15, 1:1000; Millipore). Signals were quantified using ImageJ software (Wayne Rasband).
Chromomycin A3 Staining of Sperm Nuclei
Chromomycin A3 (CMA3) assays were performed as described previously with minor modifications [7, 40]. Fluorescence microphotographs were performed, and monochrome images were evaluated as described below. CMA3 staining density was recorded for each nucleus (n > 500), and the values were grouped into fluorescence intensity categories and plotted as histograms for each individual mouse.
Immunofluorescence Staining of Histone H3K27 in Spermatozoa
Sperm smears were air dried and stored at −20°C until use when they were rehydrated with PBS and then partially decondensed for immunostaining as described previously [41]. Briefly, decondensation solution (10 mM DTT, 200 international units of heparin, 0.2% Triton-X/ml in PBS) were layered onto the slide for 8 min at room temperature; and then the slides were fixed with 4% neutral paraformaldehyde for 15 min at room temperature. Immunofluorescence staining was done as described [27] using H3K27me3-specific antiserum (see above) at a 1:500 dilution in PBS/3% bovine serum albumin overnight and using a secondary anti-rabbit TRITC antibody.
Fluorescence Microscopy
Fluorescence microscopy was performed using a Nikon TE2000-U, equipped with monochrome CCD camera (Photometrics Coolsnap, Photometrics, Tucson, AZ) connected to a computer work station using ImagePro Plus version 5.1.2 software (Media Cybernetics, Bethesda, MD) for documentation. Automated analysis of large numbers of pictures was performed to measure areas of stained nuclei and to quantify fluorescence signal intensity (i.e., the density) in CMA3 assays.
Real-Time PCR
Total RNA was isolated from testes using a kit (miRNA isolation kit, Quiagen, Valencia, CA) and subsequently reverse transcribed using the transcriptor high-fidelity complementary DNA synthesis kit (Roche) with oligo-dT primers. Complementary DNA was amplified using the Lightcycler FastStart DNA Master SYBR Green I kit according to the manufacturer's instructions (Roche) with the LightCycler detection system. A melting curve was always determined afterward to control for primer amplification events (65°C to 95°C, slope 0.1 deg C/sec) with continuous measurement. Gapdh served as a reference gene, and samples without complementary DNA or without oligonucleotides were used as negative controls. PCR cycling conditions and primer sequences for Prm1, Prm2, Tnp1, and Tnp2 were used as published [42]. The control Gapdh fragment was amplified using as primers Gapdh-for, 5′-CCTGGAGAAACCTGCCAAGTATGATGAC-3′, and Gapdh-rev, 5′-GAGGTCCACCACCCTGTTGCTGTAG-3′. All the primers were designed to include an intron wherever possible, and the generation of single PCR bands using these primers was confirmed by gel electrophoresis (Supplemental Fig. S1, available online at www.biolreprod.org). All the measurements were performed at least twice.
RESULTS
Reduced Chromatin Compaction in Parg(110)−/− Sperm Nuclei
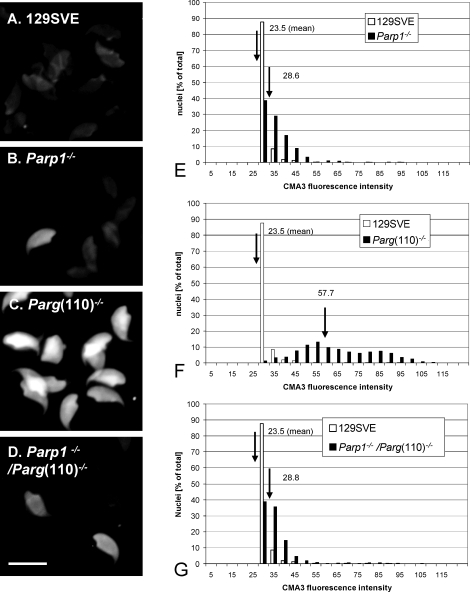
We previously showed that Parg(110)−/− mice have higher testicular steady-state levels of PAR and automodified PARP enzymes and that these animals are subfertile with reduced litter sizes [20]. Sperm counts and sperm motility are not different in Parg(110)−/− mice compared to the wild type; however, epididymal sperm of Parg(110)−/− and Parp1−/−/Parg(110)−/−, but not wild-type or Parp1−/−, mice are insufficiently elongated [20], indicative of an incomplete chromatin maturation process. Therefore, in the present study, epididymal sperm were stained with CMA3, which allows for assessment of sperm DNA protamine association [40, 43–45]. CMA3 is a fluorescent dye that intercalates into DNA, but because of its large size, it can do so in sperm only if the normal degree of chromatin compaction is compromised, indicating protamine deficiency.
CMA3-stained sperm nuclei from Parg(110)−/− but not from wild-type mice were intensely fluorescent and, when compared to wild-type sperm, the difference in fluorescence intensity was quite dramatic (P < 0.0001), indicating general protamine insufficiency in these mice (Fig. 2, C and F). In contrast, there were only modest increases in frequency and intensity of fluorescent sperm nuclei in Parp1−/− and Parp1−/−/Parg(110)−/− (Fig. 2, B and D) mice, resulting in a significant (P < 0.0001), but smaller shift in mean fluorescence values. This finding indicates that abnormally low chromatin condensation is most pronounced in sperm from animals with reduced PAR degradation. Unexpectedly, deletion of the Parp1 gene in the Parp1−/−/Parg(110)−/− animals provided a partial protective effect from the sperm chromatin defects caused by the Parg gene disruption. This observation indicated that in the Parp1−/−/Parg(110)−/−mouse residual PARG activity in the Parg gene-disrupted animals was sufficient to counteract automodification, that is, self-inactivation, of the remaining PARP2, which has overlappingfunctions with PARP1.
FIG. 2.
Low sperm chromatin quality in Parg(110)−/− mice. A–D) Chromomycin A3 (CMA3) intercalation into the DNA of epididymal spermatozoa was used as an indicator of incomplete chromatin maturation, as through insufficient DNA protamination or the presence of residual histone proteins in sperm. Bar = 10 μm. E–G) Quantitative measurement of CMA3 fluorescence revealed significantly (P < 0.001, ANOVA test) higher numbers of highly positive sperm nuclei in Parg(110)−/− mice (mean arbitrary fluorescence intensity F = 57.7, n = 12 animals) than in the wild-type 129 SVE mice (mean arbitrary intensity E–G = 23.5, n = 8 animals), Parp1−/− mice (mean arbitrary intensity E = 28.6, n = 4), or Parp1−/−/Parg(110) −/− animals (mean arbitrary intensity G = 28.6, n = 3 mice). Histogram analysis of data from a representative experiment (left panel, A–D) is shown in which labeled nuclei were categorized according to their measured fluorescence intensity and calculated as the percentage of that category relative to the total number of analyzed nuclei (n = 698 ± 77 nuclei/genotype). Statistical analyses showed that sperm from Parg(110)−/− mice were highly significantly different from all the other populations (P < 0.0001) whereas Parp1−/− and Parp1−/−/Parg(110)−/− populations were different from the wild type (P < 0.001, ANOVA test) but not from each other (P = 0.67).
Mice with Disrupted PAR Metabolism Show Increased Retention of Nonprotamine Nucleoproteins in Epididymal Sperm
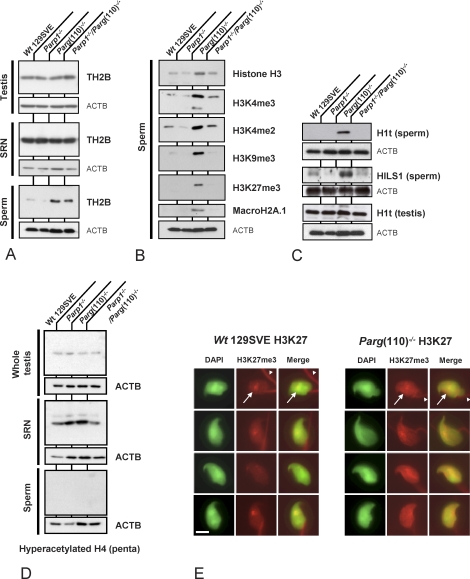
Next, we investigated whether the observed maturation defect involves incomplete nucleoprotein exchange in spermiogenesis with retention of histone proteins in sperm (Fig. 3). Sperm of Parg(110)−/− and, to a lesser extent, of Parp1−/−/Parg(110)−/− double knockout animals retained increased levels of the noncanonical testis-specific core histone variant TH2B (Fig. 3A). Because elevated levels of TH2B were not detected in whole testis lysates or SRN representing spermatid steps 12 and later (Fig. 3A, top two panels), it is unlikely that TH2B expression was affected. Rather, the exchange of TH2B for other DNA binding proteins such as transition proteins TP1 or TP2 or protamines P1 or P2 appeared reduced in Parg gene-disrupted animals.
FIG. 3.
Disruption of Parg but not Parp1 results in increased retention of core histones and H1 linker proteins in sperm. Immunoblot analyses of whole testis (A, upper row), sonication-resistant testicular spermatid nuclei (SRN) (A, middle row), and sperm (A, lower row) show partial retention of TH2B in sperm from Parg(110)−/− and Parp1−/−/Parg(110)−/−, but not Parp1−/− or wild-type 129SV, mice. Whole testis lysates showed no significant differences between the mouse strains. B) Core histones H3 and macroH2A1.1 as well as histone H3 tail modifications such as H3K4me3, H3K4me2, H3K9me3, andH3K27me3 were found to be elevated in Parg(110)−/− sperm compared to the wild type, but not strongly elevated in Parp1−/−/Parg(110)−/− sperm. C) Histone H1-like linker proteins H1t and HILS1 are retained in Parg(110)−/− sperm, which may contribute to positive CMA3 staining and overall poor chromatin quality of these mice. However, overall testicular expression of H1t as an example of a histone linker protein was not altered. D) The amount of histone H4 hyperacetylation is not altered in whole testis lysates (upper row) and SRN (middle row) of the different genotypes, and no hyperacetylated histone H4 is detectable in sperm lysates (lower row) of any of the tested genotypes. E) Histones retained in Parg(110)−/− sperm are located in the nucleus as exemplified by immunodetection using an H3K27me3-specific, antibody. The red staining of the midpiece (white arrowheads in upper row) is due to cross-reaction of the secondary TRITC-coupled antibody and was also always visible in secondary antibody controls in the absence of the primary antibody. The nuclear H3K27me3 signals (white arrows in upper row) are specific. The DNA-specific 4′,6-diamidino-2-phenylindole (DAPI) stain is shown in green pseudocolor. Bar = 10 μm.
Elevated histone retention in sperm of Parg gene-disrupted animals was further confirmed by analyses of two other histone proteins, histone H3 and macroH2A1 (Fig. 3B). Epididymal sperm histone H3 content was elevated in Parg(110)−/− mice compared to the other genotypes (Fig. 3B). Retained sperm histones carried epigenetically relevant methylation marks such as H3K4me2, H3K4me3, H3K9me3, and H3K27me3 were readily detectable in Parg(110)−/−, whereas they were much lower or below the detection limit in the other genotypes.
Strikingly, retention of testis-specific histone linker proteins HIST1H1T, hereafter referred to as H1t and HILS1, was particularly pronounced in the sperm of Parg(110)−/− mice but not in Parp1−/− or double gene-disrupted mice (Fig. 3C) where H1t always remained undetectable. H1t is a testis-specific H1 linker protein that is expressed in late spermatocytes and spermatids but is not normally found in sperm. H1t levels in Parg(110)−/− whole testis lysates were indistinguishable from the wild type, indicating that overall expression of the protein was not affected. The presence of H1t in cauda epididymal sperm of Parg(110)−/− mice therefore clearly suggests that Parg(110)−/− mice are unable to fully eliminate H1t during spermiogenesis as it normally occurs in elongating spermatids [46].
One of the known prerequisites for the removal of nucleosomes from spermatid chromatin is hyperacetylation of histone H4 (HIST4H4) [12, 47, 48]. Therefore, lysates of whole testis, SRN, and epididymal sperm were analyzed for this posttranslational modification but no differences between gene-disrupted mice and the wild type could be detected (Fig. 3D). To exclude the possibility that the retained histones detected by immunoblotting were simply a result of elevated levels of residual cytoplasmic droplets or other extranuclear material, these experiments were repeated after sonicating the sperm samples and checking for purity of the sperm nuclear fraction prior to preparation of the SDS protein samples. Alternatively, a somatic cell lysis buffer to remove potential contaminating somatic cells was used, but all the experimental setups yielded the same results (data not shown). In order to analyze the presence of retained histones in situ, sperm from Parg(110)−/− and wild-type mice was stained using an antibody specific to H3K27me3. Immunofluorescence detection showed that the retained histones were indeed located in the sperm nucleus (Fig. 3E).
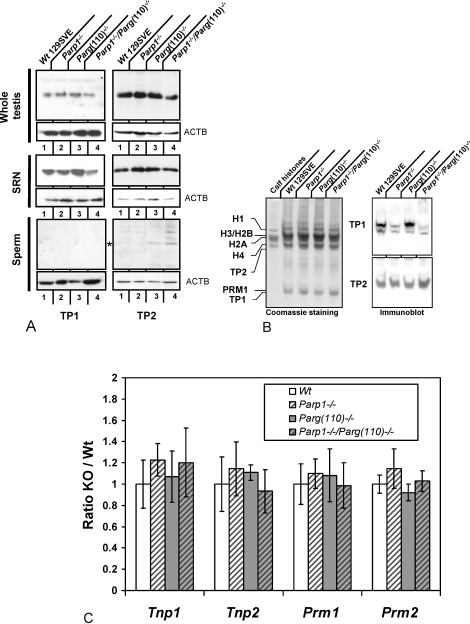
The transient presence of transition proteins TP1 and TP2 during spermiogenesis is essential for chromatin remodeling and genetic integrity of mouse sperm [33, 49, 50]. Therefore, their expression levels were determined in SDS lysates of whole testis as well as SRN, representing spermatid nuclei of step 12 and later, and epididymal sperm nuclei (Fig. 4A). Immunoblot analyses with specific antisera (kind gift of Dr. Steve Kistler) showed that TP1 was slightly decreased in Parp1−/− and Parp1−/−/Parg(110)−/−, but not Parg(110)−/−, SRN relative to wild type. This result, which suggests a role of PARP1 in TP1 deposition on the DNA or removal from the spermatid nucleus, was confirmed by biochemical extraction of the acid-soluble protein fraction of SRN followed by acid-urea PAGE and immunoblotting (Fig. 4B). TP1 was not detected in epididymal sperm. TP2 levels did not vary among genotypes in whole testis or SRN but remained detectable in sperm of Parg(110)−/− and Parp1−/−/Parg(110)−/− males, albeit at low levels. Because appropriate expression levels of transition proteins and protamines are critical for protamination of the sperm nucleus [33, 42, 49, 51, 52], quantitative RT-PCR analyses of testicular Tnp1, Tnp2, Prm1, and Prm2 were performed to analyze if alteration of the PAR pathway influences their expression at the transcriptional level. However, mRNA levels in the four different mice genotypes did not show significant alterations of transition protein or protamine expression (Fig. 4C). Taken together, these results suggest that the incomplete nucleoprotein exchange observed in spermatid nuclei in the Parg(110)−/− genotype is neither due to altered expression of transition proteins or protamines nor to reduced histone H4 hyperacetylation.
FIG. 4.
Disruption of Parp1 but not Parg results in reduced levels of TP1 in nuclei resistant to ultrasound disruption. Immunoblot analyses of whole testis (A, upper row), sonication-resistant testicular spermatid nuclei (SRN) (A, middle row), and sperm (A, lower row) show moderate but reproducible retention of residual TP2, but not TP1, in sperm of Parg(110)−/− and Parp1−/−/Parg(110)−/−, but not Parp1−/− or wild-type 129SV, mice. In contrast, TP1 but not TP2 levels were reduced in Parp1−/− and Parp1−/−/Parg(110)−/−, but not Parg(110)−/−, SRN using SDS-PAGE and Western blot analysis. Whole testis lysates showed no significant differences between the mouse strains. B) Acid/urea gel electrophoresis of basic proteins extracted from SRN followed by immunoblot analysis confirmed the notion that TP1 but not TP2 was underrepresented in SRN of Parp1−/− and Parp1−/−/Parg(110)−/−, but not Parg(110)−/−, testes. The left panel shows a Coomassie-stained gel; the right two panels are results of the respective immunoblot analysis. (Loading equiv.: 5 × 104 SRN/lane.) C) Real-time RT-PCR analysis of whole testis total RNA shows that disruption of the Parp1 and Parg genes had no significant effect on Tnp1, Tnp2, Prm1, or Prm2 mRNA levels relative to Gapdh in the four mouse genotypes tested. All the animals (n = 5 in wild type, n = 4 in Parp1−/−, n = 9 in Parg(110)−/−, and n = 5 in Parp1−/−/Parg(110)−/−) showed some individual variance in expression of Tnp1, Tnp2, Prm1, and Prm2. KO = knockout mice; Wt = wild-type mice.
The complete absence of PARP1 in the Parp1−/− mouse, where PARP2 is still expressed [20, 53], does not lead to increased histone retention in sperm, and furthermore, histones are much less retained in the Parp1−/−/Parg(110)−/− genotype than in the single Parg(110) knockout. This finding suggests an amelioration of the phenotype from the Parg gene disruption compared to the double gene-disrupted mouse model. We conclude that PARP2 is sufficient to allow for normal nucleoprotein exchange in spermatids, but failure to degrade PAR produced by PARP1 leads to histone retention in sperm in Parg(110)−/− mice.
PARP Enzymatic Activity Is Necessary for Proper Spermatid Development
The aforementioned results suggest that inhibition of PARP1, and likely PARP2, by means of excessive automodification in the Parg-disrupted mouse has an inhibitory effect on chromatin reorganization. Accordingly, we tested whether pharmacological PARP inhibition has a similar effect on sperm chromatin composition and condensation.
Pubertal wild-type mice undergoing the first wave of spermatogenesis were treated with PJ34, a potent, highly specific, and well-characterized PARP1 and PARP2 inhibitor during the short time window in which the first spermatids undergo chromatin remodeling and condensation (see Fig. 5A). Spermiogenesis was then allowed to commence until the first wave of sperm appeared in the epididymis at Day 38. At this time, germ cells that had been subject to PARP inhibition as elongating spermatids during the nuclear-remodeling process had developed into sperm, and tissues were harvested and sperm isolated from the epididymides.
FIG. 5.
Pharmacological inhibition of PAR formation during spermatid elongation results in abnormal sperm nuclear structure and immature chromatin composition. A) Male mice were injected daily with 10mg/kg of PJ34 between Day 23–27 pp to inhibit PAR metabolism during spermatid developmental steps 8–12 (tubule stages VIII–XII) of the first wave of spermatogenesis. Control animals were injected with the relevant drug solvent (saline) only. Animals were killed, and testes as well as epididymides were harvested either at Day 27 or 38 pp, that is, after the relevant cells arrived in the epididymides. B) Immunoblot analyses of Day 27 whole testis extracts using PAR-specific antibodies showed that testicular PARP activity, measured as PAR accumulation, was significantly (P < 0.0001) but not completely inhibited 3 h after the last PJ34 injection in all the animals tested (n = 6). C) Testicular histology indicated abnormal (Sdabn) and normal (Sd12) subpopulations of spermatid nuclei in stage XII tubules of Day 27 pp mice previously treated with PJ34 (b) compared to saline-injected controls (a). Bar = 25 μm. Sc, spermatocyte; M, meiotic figure.
In order to verify the inhibition of testicular PAR formation by PJ34 treatment, tissues from several animals were collected immediately after completion of the treatment regimen and levels of testicular PAR formation as well as testis histology were analyzed (Fig. 5, B and C, respectively). Treatment with PJ34 reduced PARsylation of testicular proteins to about one-third of control levels (Fig. 5B). Testicular histology at the end of the PJ34 treatment period demonstrated abnormal spermatid development with an apparent failure to undergo normal nuclear elongation (Fig. 5C, arrows highlighting abnormal spermatids, Sdabn).
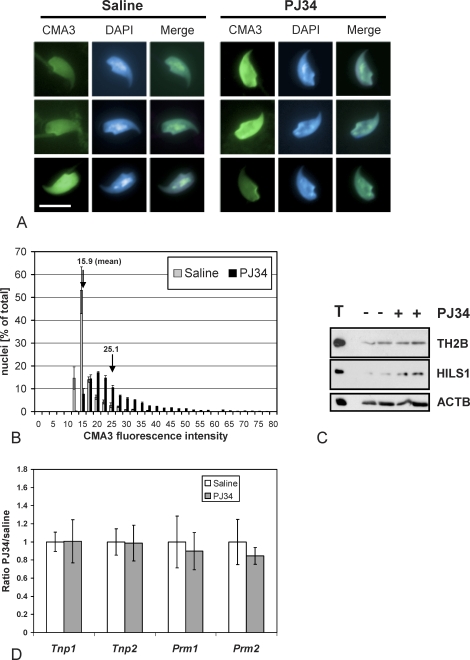
In comparison to saline-injected control animals, PJ34-treated males had epididymal sperm with significantly higher average percentages of CMA3-positive nuclei (P < 0.0001, Fig. 6, A and B). Histogram analysis revealed that the mean fluorescence intensity of cell populations from PJ34-treated animals, broken down into categories of comparable fluorescence intensity, was strongly shifted, indicating that a large fraction of the sperm cells exhibited abnormally low chromatin compaction (Fig. 6B). Furthermore, immunoblot analysis of nuclear sperm proteins demonstrated an approximately 2-fold increase of retained HILS1 in epididymal sperm from PJ34-injected mice and a mild but reproducible increase in TH2B of 1.2- to 1.3-fold (Fig. 6C). Quantitative gene expression analysis of whole testis from RNA extracted 3 h after the last injection of PJ34 or saline did not indicate any significant alteration of the spermatid-transcribed genes Tnp1, Tnp2, Prm1, and Prm2 (Fig. 6D). These findings demonstrate that PARP activity is necessary for proper nuclear condensation and maturation with adequate removal of histones, and particularly of H1-like linker protein HILS1.
FIG. 6.
Compromised sperm chromatin integrity induced by pharmacological inhibition of PARsylation during spermatid development. A) CMA3 staining of epididymal sperm nuclei retrieved at Day 39 pp showed that PJ34-mediated inhibition of PARP enzymes during the spermatid elongation phase results in poor chromatin integrity in the majority of sperm cells (right panel). Representative normal (left panel, top and middle rows) sperm nuclei as well as an example of rare abnormal CMA3-positive nuclei (left panel, bottom row) in saline-injected animals are shown. Bar = 10 μm. B) CMA3 fluorescence is significantly shifted in cauda epididymal sperm populations from PJ34-treated males (n ≥ 900 nuclei, data pooled from two experiments, each with four individuals/group) compared to saline-injected control animals (ANOVA test, P < 0.0001). C) Immunoblot analyses of two representative wild-type 129SVE animals treated with PJ34 (+) according to the regimen shown in Figure 5A and two corresponding control saline-injected mice (−) reveals elevated retention of HILS1 in sperm of the PJ34-treated animals. An equivalent of 5 × 104 sperm was loaded per lane. D) Real-time RT-PCR of whole testis RNA did not show significant alteration of Tnp1, Tnp2, Prm1, or Prm2 expression by inhibition of PARP activities using 10 mg/kg PJ34. The slight trend of decreasing of Prm2 expression in the presence of PJ34 was not statistically significant (six animals in each treatment group in three independent experiments).
DISCUSSION
During mammalian spermiogenesis, packaging of the paternal chromosome complement into the highly condensed transport form is not a well-understood process, despite its central role for the survival of the species and despite increasing evidence for associations between low sperm chromatin quality and infertility, increased miscarriage rates, and likely also embryonic failure [5, 54–56]. One of the hallmarks of adequate sperm chromatin condensation is the removal of most of the histones and their replacement first with transition proteins and then protamines (Fig. 1). Another hallmark is the relaxation of DNA torsion in the elongating spermatid genome that involves the transient formation of natural, endogenous DNA strand breaks. The results presented in this report focus on a novel role of PARsylation, a short-lived posttranslational modification of proteins mediated by PARP enzymes in response to DNA strand breaks, in this essential exchange of spermatid nucleoproteins.
We reported earlier that male Parg(110)−/− mice are subfertile with abnormally shaped sperm nuclei that are rounder and larger than wild-type control nuclei and have reduced sperm chromatin integrity with DNA strand breaks leading to reduced litter sizes [20]. Here we show that decreased PARG activity in the Parg(110)−/− mice [24] as well as pharmacological inhibition of PARPs in PJ34-treated wild-type mice both lead to nuclear condensation defects in differentiating spermatids as a result of their failure to adequately remove core histones, histone H1 linker-like nucleoproteins, and TP2 prior to sperm maturation. Alteration of the nuclear shape that we described previously likely reflects the lesser degree of chromatin condensation and may be a direct consequence of the histone retention reported here. Histone retention in sperm results in a higher proportion of sperm chromatin being in a nucleosomal, more bulky form instead of a protaminated, very condensed form and therefore is expected to reduce the degree of condensation that can be achieved, ultimately resulting in larger nuclei.
Alterations of mature sperm nucleoprotein composition involved in the subfertility of mouse models have mostly been reported to be caused by transcriptional deregulation or genetic deletion of transition proteins or protamines [42, 51, 52, 57, 58]. In contrast to these studies, the results presented here show that none of the transition proteins or protamines was abnormally expressed under conditions of PARP inhibition, supporting the view that PAR metabolism exerts its impact on chromatin condensation at the posttranslational level. Indeed, recent findings of interactions of PARP2, and probably PARP1, with the TP2-specific chaperone HSPA2 and TP2 suggest a possible role of PARsylation in TP2 DNA shuttling [59], which may explain the abnormal presence of TP2 in Parg(110)−/− sperm (Fig. 4A). A possible involvement of PAR metabolism in the regulation of transition protein function is further supported by the finding that TP1 was underrepresented in PARP1-deficient SRN (Fig. 4, A and B).
The apparently more severe—and intriguing—defect observed in Parg(110)−/− and PJ34-treated mice, however, is retention of H1 linker proteins such as H1t and HILS1 (Fig. 3C). Studies using somatic cells have established that PAR metabolism acts as a biochemical mechanism facilitating removal of histones and histone H1-like proteins to promote opening of condensed, inaccessible chromatin domains [60–62], which is in line with earlier reports proposing PARsylation as a DNA strand break-dependent shuttling mechanism removing histones from the DNA to allow access of other DNA-binding proteins, such as DNA repair proteins, DNA helicase A and topoisomerase I [63–65]. Nuclear PAR formation by PARP1 and PARP2 is generally dependent on the presence of DNA strand breaks, and the level of PAR formation reflects the amount of DNA strand breaks that are present at a given point. The simultaneous occurrence of significant amounts of TOP2B-mediated DNA strand breaks [12, 66, 67] with PAR formation in condensing spermatids [11] indicates that PARsylation may be involved in the temporal and spatial coordination of DNA relaxation with the removal of histones, particularly H1, afforded by PAR-mediated histone shuttling.
Core histones and histone H1t are known testicular acceptor proteins for PAR [68–71]. It is therefore not surprising that inhibition of PARP enzymatic activity using PJ34 leads to retention of H1 linker proteins in sperm, as observed in this study (Fig. 6C). Retention of core histones such as TH2B (HIST1H2BA) or H3 may be a secondary effect that is due to inaccessibility of chromatin domains that are stabilized by the untimely presence of linker H1 proteins. As a result, overall chromatin compaction and likely protamination of the DNA appear to be significantly impaired by PARP inhibition because of a lack of transient chromatin decondensation allowing for exchange of DNA-binding proteins and the resulting lack of histone removal from the condensing nucleus.
Murine sperm nuclei normally retain only a small amount of histones (with estimates in the range of 1%–5% of total nucleoproteins), and the largest proportion of nuclear proteins are protamines. In cases with increased histone retention, the decrease in protamines content in the sperm nuclei likely corresponds to the increase in histone content. While increases in histone content are readily detectable, even a several-fold increase of retained histones will likely only cause a relatively small reduction in total sperm protamine content, making it difficult to quantify the corresponding small alteration of total protamine content. There are cases in which a general deficiency of protamine gene expression causes failed spermatid condensation and histone retention [42, 72, 73]. We excluded this as the primary reason for the histone retention observed in this study because there were no obvious differences in levels of testicular protamine transcripts (Figs. 4C and 6D) or protamine protein content in SRN (e.g., Fig. 4B, left panel).
The fact that the overall phenotype observed here is mild can either be explained by the fact that neither mice with pharmacological PARP inhibition nor Parp1−/− or Parg(110)−/− mice have a completely abolished nuclear PAR metabolism, which is expected to be early embryonically lethal [21, 22]. The residual PARP activity might permit a sufficient degree of PARsylation to prevent complete failure of chromatin remodeling.
Adding to the anticipated multiplicity of PAR functions in histone removal during spermiogenesis is a recent report showing that PARP1 covalently ADP-ribosylates the amino-terminal histone tails of all core histones. ADP-ribose acceptor sites include lysine residues K27 and K37 for histone H3, and K16 for histone H4 [71]. The authors also show in the same report that previous acetylation of H4K16 inhibits ADP-ribosylation by PARP1, which suggests the existence of considerable cross talk between histone tail modifications mediated by different pathways of yet unknown extent. Recently, Lu et al. described that a lack of histone ubiquitination, and subsequent failure of histone H4 acetylation in Rnf8 knockout mice, caused spermatid condensation defects characterized by histone retention [48], drawing attention to the possibility that improper or missing posttranslational modifications can directly disturb proper nucleoprotein exchange during spermiogenesis. Although our data show no interference of PARP inhibition with histone H4 hyperacetylation, an involvement of PARP in the histone ubiquitination process cannot be completely ruled out. Taken together, it is, however, likely that PARP itself is directly involved in (ADP-ribosyl)ation of N-terminal histone tails and H1 linker proteins.
The notion that both up- and down-regulation of PAR levels, as in the Parg(110)−/− or the PJ34-treated mouse model, respectively, cause similar effects may at first seem puzzling. However, a plausible explanation is that in the Parg(110)−/− mouse inhibition of PARP1 and PARP2 activity occurs primarily as a consequence of delayed PAR removal where the reduced PARG activity prolongs the automodified, hence inactive, state of both PARPs. In these mice, the catabolic arm of the pathway appears to be overwhelmed, which may not be the case in the Parp1/Parg(110) double-gene-disrupted mouse, in which PARP1, the major PAR synthesizing enzyme, is absent. The residual PARG activity in the Parp1/Parg double gene-disrupted mice is apparently sufficient to counteract the activity of PARP2, which only accounts for a small fraction of the total DNA strand break-dependent PAR synthesis in elongated spermatids [74]. Consequently, Parg(110)−/− animals suffer from reduced PARP activity in that they lack sufficient amounts of non-PARsylated PARP1 and PARP2 available to initiate posttranslational modification of other target proteins. This lack of activity then leads to insufficient PARsylation, and consequently removal of H1 linker proteins and core histones, while at the same time accumulating high levels of PAR, partly explaining the complex phenotype of these mice. PARP1 and PARP2 have largely overlapping functions, but it is possible that PARP2 also has specific additional functions in spermatid differentiation [53, 75], which would be particularly impaired in the Parg(110)−/− mouse.
In PJ34-treated animals, PARP activity was directly inhibited with strongly reduced PAR formation (by ∼75%, Fig. 5B). Pharmacological inhibition of PARPs within the defined time window in stages IX–XII of spermiogenesis, which is characterized by DNA strand breaks and dramatic chromatin remodeling events (Fig. 1), severely impaired spermatid development in terms of nuclear structure and chromatin condensation (Figs. 5C and 6A). Because our preliminary data also show reduced fertility of PJ34-treated wild-type males, similar to what we observed in the Parg(110)−/− mice (data not shown), results from pharmacological and genetic disruption of PAR metabolism consistently support the view that PARsylation is important to male fertility in mice.
Sperm histones transmit epigenetic information in the form of diverse methylation marks with possible roles in gene regulation events in the early developing embryo as well as in the proper establishment of genetic imprinting patterns. Notably, in the present study, sperm histone content and consequently histone H3 tail modification were significantly increased by the Parg gene disruption.
The results presented here demonstrate that manipulation of the PAR metabolism pathways can be utilized to alter sperm chromatin composition and may offer the opportunity to study the relevance of epigenetics marks in the sperm genome for proper embryo development.
Supplementary Material
Acknowledgments
We are grateful for the following generous gifts: TP1 antiserum, TP2 antiserum, and H1t antiserum were provided by Dr. W. Steve Kistler (University of South Carolina, Columbia, SC); Hils1 antiserum was a generous gift from Dr. Hiromitsu Tanaka (Nagasaki International University, Nagasaki, Japan) and Dr. Yoshitake Nishimune (RCC-ERI, Bangkok, Thailand); and macroH2A1.2 antiserum was provided by Dr. John Pehrson (University of Pennsylvania, Philadelphia, PA). We thank Dr. Richard Schultz for critical reading of this manuscript.
Footnotes
Supported by grants from the National Institutes of Health (NIH R01 HD48837 to R.G.M.) and the Mari Lowe Center for Comparative Oncology at the University of Pennsylvania.
REFERENCES
- Delbes G, Hales BF, Robaire B. Toxicants and human sperm chromatin integrity. Mol Hum Reprod 2010; 16: 14 22 [DOI] [PubMed] [Google Scholar]
- Li Y, Lalancette C, Miller D, Krawetz SA. Characterization of nucleohistone and nucleoprotamine components in the mature human sperm nucleus. Asian J Androl 2008; 10: 535 541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res 2009; 19: 1338 1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009; 460: 473 478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell DT, Hammoud SS. The human sperm epigenome and its potential role in embryonic development. Mol Hum Reprod 2010; 16: 37 47 [DOI] [PubMed] [Google Scholar]
- Zhang X, San Gabriel M, Zini A. Sperm nuclear histone to protamine ratio in fertile and infertile men: evidence of heterogeneous subpopulations of spermatozoa in the ejaculate. J Androl 2006; 27: 414 420 [DOI] [PubMed] [Google Scholar]
- Singleton S, Zalensky A, Doncel GF, Morshedi M, Zalenskaya IA. Testis/sperm-specific histone 2B in the sperm of donors and subfertile patients: variability and relation to chromatin packaging. Hum Reprod 2007; 22: 743 750 [DOI] [PubMed] [Google Scholar]
- Zini A, Gabriel MS, Zhang X. The histone to protamine ratio in human spermatozoa: comparative study of whole and processed semen. Fertil Steril 2007; 87: 217 219 [DOI] [PubMed] [Google Scholar]
- Smith A, Haaf T. DNA nicks and increased sensitivity of DNA to fluorescence in situ end labeling during functional spermiogenesis. Biotechniques 1998; 25: 496 502 [DOI] [PubMed] [Google Scholar]
- Marcon L, Boissonneault G. Transient DNA strand breaks during mouse and human spermiogenesis: new insights in stage specificity and link to chromatin remodeling. Biol Reprod 2004; 70: 910 918 [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Scherthan H, Burkle A, Meyer RG. Poly(ADP-ribosyl)ation during chromatin remodeling steps in rat spermiogenesis. Chromosoma 2005; 114: 67 74 [DOI] [PubMed] [Google Scholar]
- Leduc F, Maquennehan V, Nkoma GB, Boissonneault G. DNA damage response during chromatin remodeling in elongating spermatids of mice. Biol Reprod 2008; 78: 324 332 [DOI] [PubMed] [Google Scholar]
- Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol 2008; 20: 294 302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci 2008; 13: 3046 3082 [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Meyer RG, Jacobson EL, Jacobson MK. Poly(ADP-ribose) polymerases: managing genome stability. Int J Biochem Cell Biol 2005; 37: 920 926 [DOI] [PubMed] [Google Scholar]
- Caiafa P, Guastafierro T, Zampieri M. Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J 2009; 23: 672 678 [DOI] [PubMed] [Google Scholar]
- Gagne JP, Hendzel MJ, Droit A, Poirier GG. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr Opin Cell Biol 2006; 18: 145 151 [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev 2006; 70: 789 829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Coyle DL, Meyer-Ficca ML, Meyer RG, Jacobson EL, Wang ZQ, Jacobson MK. Altered poly(ADP-ribose) metabolism impairs cellular responses to genotoxic stress in a hypomorphic mutant of poly(ADP-ribose) glycohydrolase. Exp Cell Res 2007; 313: 984 996 [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Lonchar J, Credidio C, Ihara M, Li Y, Wang ZQ, Meyer RG. Disruption of poly(ADP-ribose) homeostasis affects spermiogenesis and sperm chromatin integrity in mice. Biol Reprod 2009; 81: 46 55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J 2003; 22: 2255 2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stoger T, Poirier GG, Dawson VL, Dawson TM. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A 2004; 101: 17699 17704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res 2004; 297: 521 532 [DOI] [PubMed] [Google Scholar]
- Cortes U, Tong WM, Coyle DL, Meyer-Ficca ML, Meyer RG, Petrilli V, Herceg Z, Jacobson EL, Jacobson MK, Wang ZQ. Depletion of the 110-kilodalton isoform of poly(ADP-ribose) glycohydrolase increases sensitivity to genotoxic and endotoxic stress in mice. Mol Cell Biol 2004; 24: 7163 7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 2006; 312: 1798 1802 [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev 1995; 9: 509 520 [DOI] [PubMed] [Google Scholar]
- Meyer RG, Meyer-Ficca ML, Whatcott CJ, Jacobson EL, Jacobson MK. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp Cell Res 2007; 313: 2920 2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatcott CJ, Meyer-Ficca ML, Meyer RG, Jacobson MK. A specific isoform of poly(ADP-ribose) glycohydrolase is targeted to the mitochondrial matrix by a N-terminal mitochondrial targeting sequence. Exp Cell Res 2009; 315: 3477 3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelkarim GE, Gertz K, Harms C, Katchanov J, Dirnagl U, Szabo C, Endres M. Protective effects of PJ34, a novel, potent inhibitor of poly(ADP-ribose) polymerase (PARP) in in vitro and in vivo models of stroke. Int J Mol Med 2001; 7: 255 260 [PubMed] [Google Scholar]
- Mabley JG, Jagtap P, Perretti M, Getting SJ, Salzman AL, Virag L, Szabo E, Soriano FG, Liaudet L, Abdelkarim GE, Hasko G, Marton A, et al. Anti-inflammatory effects of a novel, potent inhibitor of poly (ADP-ribose) polymerase. Inflamm Res 2001; 50: 561 569 [DOI] [PubMed] [Google Scholar]
- Fonfria E, Marshall IC, Benham CD, Boyfield I, Brown JD, Hill K, Hughes JP, Skaper SD, McNulty S. TRPM2 channel opening in response to oxidative stress is dependent on activation of poly(ADP-ribose) polymerase. Br J Pharmacol 2004; 143: 186 192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambi N, Tramontano F, Quesada P. Poly(ADPR)polymerase inhibition and apoptosis induction in cDDP-treated human carcinoma cell lines. Biochem Pharmacol 2008; 75: 2356 2363 [DOI] [PubMed] [Google Scholar]
- Yu YE, Zhang Y, Unni E, Shirley CR, Deng JM, Russell LD, Weil MM, Behringer RR, Meistrich ML. Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc Natl Acad Sci U S A 2000; 97: 4683 4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler WS, Henriksen K, Mali P, Parvinen M. Sequential expression of nucleoproteins during rat spermiogenesis. Exp Cell Res 1996; 225: 374 381 [DOI] [PubMed] [Google Scholar]
- Drabent B, Bode C, Bramlage B, Doenecke D. Expression of the mouse testicular histone gene H1t during spermatogenesis. Histochem Cell Biol 1996; 106: 247 251 [DOI] [PubMed] [Google Scholar]
- Iguchi N, Tanaka H, Yomogida K, Nishimune Y. Isolation and characterization of a novel cDNA encoding a DNA-binding protein (Hils1) specifically expressed in testicular haploid germ cells. Int J Androl 2003; 26: 354 365 [DOI] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 1998; 393: 599 601 [DOI] [PubMed] [Google Scholar]
- Heidaran MA, Showman RM, Kistler WS. A cytochemical study of the transcriptional and translational regulation of nuclear transition protein 1 (TP1), a major chromosomal protein of mammalian spermatids. J Cell Biol 1988; 106: 1427 1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso PJ, Kistler WS. Immunohistochemical localization of spermatid nuclear transition protein 2 in the testes of rats and mice. Biol Reprod 1993; 48: 522 529 [DOI] [PubMed] [Google Scholar]
- Bianchi PG, Manicardi G, Bizzaro D, Campana A, Bianchi U, Sakkas D. Use of the guanine-cytosine (GC) specific fluorochrome, chromomycin A3, as an indicator of poor sperm morphology. J Assist Reprod Genet 1996; 13: 246 250 [DOI] [PubMed] [Google Scholar]
- Singleton S, Mudrak O, Morshedi M, Oehninger S, Zalenskaya I, Zalensky A. Characterisation of a human sperm cell subpopulation marked by the presence of the TSH2B histone. Reprod Fertil Dev 2007; 19: 392 397 [DOI] [PubMed] [Google Scholar]
- Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 2007; 450: 119 123 [DOI] [PubMed] [Google Scholar]
- Bianchi PG, Manicardi GC, Bizzaro D, Bianchi U, Sakkas D. Effect of deoxyribonucleic acid protamination on fluorochrome staining and in situ nick-translation of murine and human mature spermatozoa. Biol Reprod 1993; 49: 1083 1088 [DOI] [PubMed] [Google Scholar]
- Manicardi GC, Bianchi PG, Pantano S, Azzoni P, Bizzaro D, Bianchi U, Sakkas D. Presence of endogenous nicks in DNA of ejaculated human spermatozoa and its relationship to chromomycin A3 accessibility. Biol Reprod 1995; 52: 864 867 [DOI] [PubMed] [Google Scholar]
- Sakkas D, Manicardi G, Bianchi PG, Bizzaro D, Bianchi U. Relationship between the presence of endogenous nicks and sperm chromatin packaging in maturing and fertilizing mouse spermatozoa. Biol Reprod 1995; 52: 1149 1155 [DOI] [PubMed] [Google Scholar]
- Fantz DA, Hatfield WR, Horvath G, Kistler MK, Kistler WS. Mice with a targeted disruption of the H1t gene are fertile and undergo normal changes in structural chromosomal proteins during spermiogenesis. Biol Reprod 2001; 64: 425 431 [DOI] [PubMed] [Google Scholar]
- Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thevenon J, Catena R, Davidson I, Garin J, Khochbin S, Caron C. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J Cell Biol 2007; 176: 283 294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell 2010; 18: 371 384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Shirley CR, Yu YE, Mohapatra B, Zhang Y, Unni E, Deng JM, Arango NA, Terry NH, Weil MM, Russell LD, Behringer RR, Meistrich ML. Targeted disruption of the transition protein 2 gene affects sperm chromatin structure and reduces fertility in mice. Mol Cell Biol 2001; 21: 7243 7255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich ML, Mohapatra B, Shirley CR, Zhao M. Roles of transition nuclear proteins in spermiogenesis. Chromosoma 2003; 111: 483 488 [DOI] [PubMed] [Google Scholar]
- Shirley CR, Hayashi S, Mounsey S, Yanagimachi R, Meistrich ML. Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biol Reprod 2004; 71: 1220 1229 [DOI] [PubMed] [Google Scholar]
- Torregrosa N, Dominguez-Fandos D, Camejo MI, Shirley CR, Meistrich ML, Ballesca JL, Oliva R. Protamine 2 precursors, protamine 1/protamine 2 ratio, DNA integrity and other sperm parameters in infertile patients. Hum Reprod 2006; 21: 2084 2089 [DOI] [PubMed] [Google Scholar]
- Dantzer F, Mark M, Quenet D, Scherthan H, Huber A, Liebe B, Monaco L, Chicheportiche A, Sassone-Corsi P, de Murcia G, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proc Natl Acad Sci U S A 2006; 103: 14854 14859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken RJ, De Iuliis GN, McLachlan RI. Biological and clinical significance of DNA damage in the male germ line. Int J Androl 2009; 32: 46 56 [DOI] [PubMed] [Google Scholar]
- Miller D, Brinkworth M, Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction 2010; 139: 287 301 [DOI] [PubMed] [Google Scholar]
- Zini A, Zhang X, San Gabriel M. Sperm nuclear histone H2B: correlation with sperm DNA denaturation and DNA stainability. Asian J Androl 2008; 10: 865 871 [DOI] [PubMed] [Google Scholar]
- Zhao M, Shirley CR, Mounsey S, Meistrich ML. Nucleoprotein transitions during spermiogenesis in mice with transition nuclear protein Tnp1 and Tnp2 mutations. Biol Reprod 2004; 71: 1016 1025 [DOI] [PubMed] [Google Scholar]
- Nair M, Nagamori I, Sun P, Mishra DP, Rheaume C, Li B, Sassone-Corsi P, Dai X. Nuclear regulator Pygo2 controls spermiogenesis and histone H3 acetylation. Dev Biol 2008; 320: 446 455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenet D, Mark M, Govin J, van Dorsselear A, Schreiber V, Khochbin S, Dantzer F. Parp2 is required for the differentiation of post-meiotic germ cells: identification of a spermatid-specific complex containing Parp1, Parp2, TP2 and HSPA2. Exp Cell Res 2009; 315: 2824 2834 [DOI] [PubMed] [Google Scholar]
- Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 2003; 299: 560 562 [DOI] [PubMed] [Google Scholar]
- Tulin A, Chinenov Y, Spradling A. Regulation of chromatin structure and gene activity by poly(ADP-ribose) polymerases. Curr Top Dev Biol 2003; 56: 55 83 [DOI] [PubMed] [Google Scholar]
- Wacker DA, Frizzell KM, Zhang T, Kraus WL. Regulation of chromatin structure and chromatin-dependent transcription by poly(ADP-ribose) polymerase-1: possible targets for drug-based therapies. Subcell Biochem 2007; 41: 45 69 [DOI] [PubMed] [Google Scholar]
- Realini CA, Althaus FR. Histone shuttling by poly(ADP-ribosylation). J Biol Chem 1992; 267: 18858 18865 [PubMed] [Google Scholar]
- Althaus FR, Hofferer L, Kleczkowska HE, Malanga M, Naegeli H, Panzeter PL, Realini CA. Histone shuttling by poly ADP-ribosylation. Mol Cell Biochem 1994; 138: 53 59 [DOI] [PubMed] [Google Scholar]
- Althaus FR, Hofferer L, Kleczkowska HE, Malanga M, Naegeli H, Panzeter P, Realini C. Histone shuttle driven by the automodification cycle of poly(ADP-ribose)polymerase. Environ Mol Mutagen 1993; 22: 278 282 [DOI] [PubMed] [Google Scholar]
- Laberge RM, Boissonneault G. On the nature and origin of DNA strand breaks in elongating spermatids. Biol Reprod 2005; 73: 289 296 [DOI] [PubMed] [Google Scholar]
- McPherson SM, Longo FJ. Nicking of rat spermatid and spermatozoa DNA: possible involvement of DNA topoisomerase II. Dev Biol 1993; 158: 122 130 [DOI] [PubMed] [Google Scholar]
- Quesada P, Farina B, Jones R. Poly(ADP-ribosylation) of nuclear proteins in rat testis correlates with active spermatogenesis. Biochim Biophys Acta 1989; 1007: 167 175 [DOI] [PubMed] [Google Scholar]
- Malanga M, Atorino L, Tramontano F, Farina B, Quesada P. Poly(ADP-ribose) binding properties of histone H1 variants. Biochim Biophys Acta 1998; 1399: 154 160 [DOI] [PubMed] [Google Scholar]
- Quesada P, d'Erme M, Parise G, Faraone-Mennella MR, Caiafa P, Farina B. Nuclear matrix-associated poly(ADPribosyl)ation system in rat testis chromatin. Exp Cell Res 1994; 214: 351 357 [DOI] [PubMed] [Google Scholar]
- Messner S, Altmeyer M, Zhao H, Pozivil A, Roschitzki B, Gehrig P, Rutishauser D, Huang D, Caflisch A, Hottiger MO. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res 2010; Jun 4 [Epub ahead of print] PMID: 20525793. [DOI] [PMC free article] [PubMed]
- Liu Z, Zhou S, Liao L, Chen X, Meistrich M, Xu J. Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J Biol Chem 2010; 285: 2758 2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, Eddy EM. Haploinsufficiency of protamine-1 or −2 causes infertility in mice. Nat Genet 2001; 28: 82 86 [DOI] [PubMed] [Google Scholar]
- Tramontano F, Malanga M, Quesada P. Differential contribution of poly(ADP-ribose)polymerase-1 and -2 (PARP-1 and -2) to the poly(ADP-ribosyl)ation reaction in rat primary spermatocytes. Mol Hum Reprod 2007; 13: 821 828 [DOI] [PubMed] [Google Scholar]
- Yelamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol Med 2008; 14: 169 178 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.