Abstract
During the determination of DNA sequences, frameshift errors are not the most frequent but they are the most bothersome as they corrupt the amino acid sequence over several residues. Detection of such errors by sequence alignment is only possible when related sequences are found in the databases. To avoid this limitation, we have developed a new tool based on the distribution of non-overlapping 3-tuples or 6-tuples in the three frames of an ORF. The method relies upon the result of a correspondence analysis. It has been extensively tested on Bacillus subtilis and Saccharomyces cerevisiae sequences and has also been examined with human sequences. The results indicate that it can detect frameshift errors affecting as few as 20 bp with a low rate of false positives (no more than 1.0/1000 bp scanned). The proposed algorithm can be used to scan a large collection of data, but it is mainly intended for laboratory practice as a tool for checking the quality of the sequences produced during a sequencing project.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985 May 24;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Claverie J. M. Detecting frame shifts by amino acid sequence comparison. J Mol Biol. 1993 Dec 20;234(4):1140–1157. doi: 10.1006/jmbi.1993.1666. [DOI] [PubMed] [Google Scholar]
- Claverie J. M., Sauvaget I., Bougueleret L. K-tuple frequency analysis: from intron/exon discrimination to T-cell epitope mapping. Methods Enzymol. 1990;183:237–252. doi: 10.1016/0076-6879(90)83017-4. [DOI] [PubMed] [Google Scholar]
- Cosmina P., Rodriguez F., de Ferra F., Grandi G., Perego M., Venema G., van Sinderen D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol Microbiol. 1993 May;8(5):821–831. doi: 10.1111/j.1365-2958.1993.tb01629.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabret C., Quentin Y., Guiseppi A., Busuttil J., Haiech J., Denizot F. Analysis of errors in finished DNA sequences: the surfactin operon of Bacillus subtilis as an example. Microbiology. 1995 Feb;141(Pt 2):345–350. doi: 10.1099/13500872-141-2-345. [DOI] [PubMed] [Google Scholar]
- Fichant G., Gautier C. Statistical method for predicting protein coding regions in nucleic acid sequences. Comput Appl Biosci. 1987 Nov;3(4):287–295. doi: 10.1093/bioinformatics/3.4.287. [DOI] [PubMed] [Google Scholar]
- Fickett J. W., Tung C. S. Assessment of protein coding measures. Nucleic Acids Res. 1992 Dec 25;20(24):6441–6450. doi: 10.1093/nar/20.24.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuma S., Fujishima Y., Corbell N., D'Souza C., Nakano M. M., Zuber P., Yamane K. Nucleotide sequence of 5' portion of srfA that contains the region required for competence establishment in Bacillus subtilus. Nucleic Acids Res. 1993 Jan 11;21(1):93–97. doi: 10.1093/nar/21.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Milleret F., Mugnier C., Jacobzone M., Gautier C. ACNUC: a nucleic acid sequence data base and analysis system. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):121–127. doi: 10.1093/nar/12.1part1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigó R., Knudsen S., Drake N., Smith T. Prediction of gene structure. J Mol Biol. 1992 Jul 5;226(1):141–157. doi: 10.1016/0022-2836(92)90130-c. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Bork P., Sander C. Yeast chromosome III: new gene functions. EMBO J. 1994 Feb 1;13(3):493–503. doi: 10.1002/j.1460-2075.1994.tb06287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossé M. O., Linder P., Lazowska J., Slonimski P. P. A comprehensive compilation of 1001 nucleotide sequences coding for proteins from the yeast Saccharomyces cerevisiae (= ListA2) Curr Genet. 1993 Jan;23(1):66–91. doi: 10.1007/BF00336752. [DOI] [PubMed] [Google Scholar]
- Mouchiroud D., Fichant G., Bernardi G. Compositional compartmentalization and gene composition in the genome of vertebrates. J Mol Evol. 1987;26(3):198–204. doi: 10.1007/BF02099852. [DOI] [PubMed] [Google Scholar]
- Perrière G., Gouy M., Gojobori T. NRSub: a non-redundant data base for the Bacillus subtilis genome. Nucleic Acids Res. 1994 Dec 25;22(25):5525–5529. doi: 10.1093/nar/22.25.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
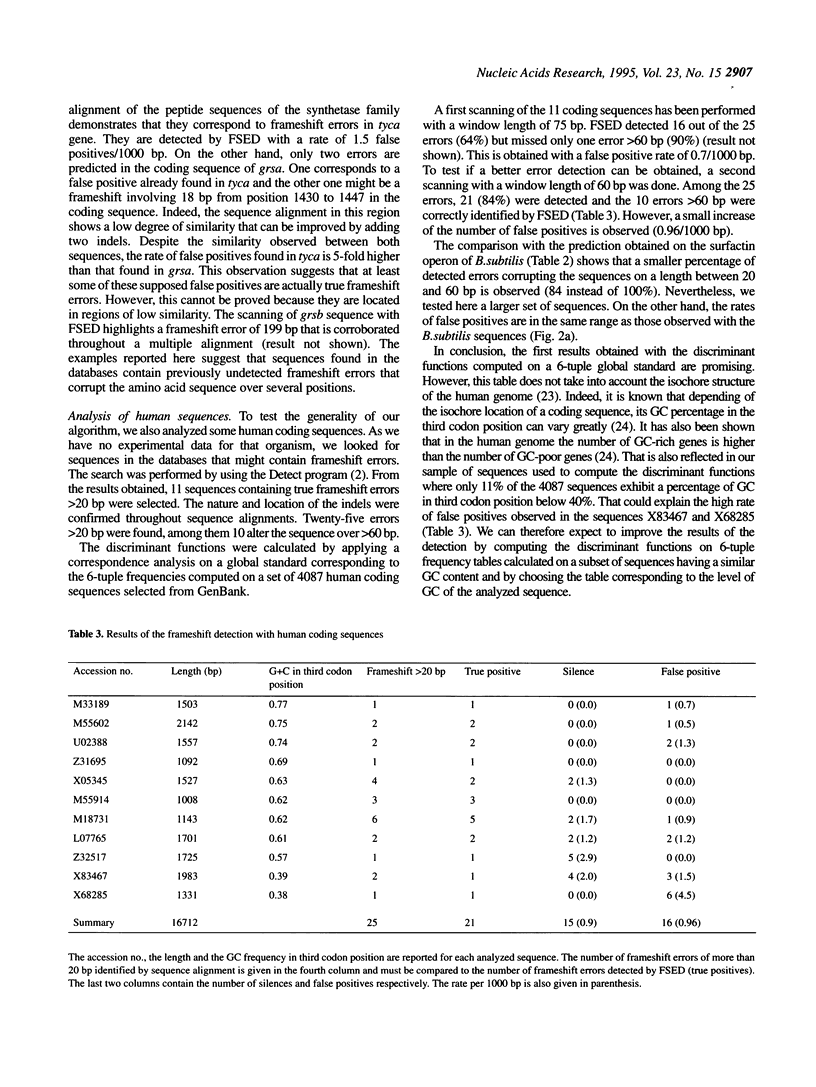
- Posfai J., Roberts R. J. Finding errors in DNA sequences. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4698–4702. doi: 10.1073/pnas.89.10.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y., Fichant G. A. Fast identification of repetitive elements in biological sequences. J Theor Biol. 1994 Jan 7;166(1):51–61. doi: 10.1006/jtbi.1994.1004. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder E. E., Stormo G. D. Identification of coding regions in genomic DNA sequences: an application of dynamic programming and neural networks. Nucleic Acids Res. 1993 Feb 11;21(3):607–613. doi: 10.1093/nar/21.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Searching for patterns in protein and nucleic acid sequences. Methods Enzymol. 1990;183:193–211. doi: 10.1016/0076-6879(90)83014-z. [DOI] [PubMed] [Google Scholar]
- Uberbacher E. C., Mural R. J. Locating protein-coding regions in human DNA sequences by a multiple sensor-neural network approach. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11261–11265. doi: 10.1073/pnas.88.24.11261. [DOI] [PMC free article] [PubMed] [Google Scholar]



