Abstract
Background
We tested the hypothesis that HIV-related immunosuppression alters the host-hepatitis C virus (HCV) interaction, resulting in fewer amino acid changing substitutions in HCV viral variants. Higher HCV RNA levels in persons coinfected with HIV compared with HCV infection alone suggests increased viral replication. If this increase is dependent on decreased immune selective pressure, then a reduced rate of nucleotide changes resulting in amino acid replacements (nonsynonymous changes, dN), would be expected.
Methods
We investigated HCV envelope sequences over time in 79 persons with chronic HCV infection that were HIV negative (group 1), or HIV positive without (group 2) or with (group 3) severe immunodeficiency. We amplified a 1026 nucleotide region of the HCV genome, which encodes a portion of the envelope glycoproteins E1 and E2, including HVR-1 for direct sequence analysis.
Results
The overall divergence between paired sequences, dS, dN, and dN/dS all showed no significant differences among the three groups.
Conclusions
By measuring nucleotide substitutions in HCV sequences over time, we found no significant differences in the genetic divergence between HCV monoinfected control subjects and HIV/HCV coinfected subjects with various levels of immunodeficiency as measured by CD4+ T cell counts.
INTRODUCTION
Hepatitis C virus (HCV) is a major etiological agent of liver disease in most regions of the world, with approximately 170 million infected individuals 1;2. Coinfection with Human immunodeficiency virus (HIV) is found in approximately one third of HCV infected persons 3. This is important because HIV coinfection increases HCV-related liver diseases 4.
HCV is a single-stranded RNA virus, which encodes a polyprotein of approximately 3000 amino acids, which is cleaved into three structural (Core, E1, and E2) and several nonstructural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins 5. HCV, like HIV, exists in each infected person as a quasispecies, or “swarm” of closely related but divergent genetic sequences 6;7. These divergent sequences are generated by an error-prone polymerase (the NS5B protein), driven by the highly dynamic nature of HCV replication8. HCV quasispecies arise by nucleotide substitutions that either preserve protein structure (synonymous substitutions) or substitutions that change the amino acid composition (nonsynonymous substitutions). Whereas both types of substitutions arise from error prone viral replication, their relative proportions may be informative due to different selective forces. Synonymous changes are generally well-tolerated by the virus except in regions of RNA secondary structure,9 and thus are thought to be nearly neutral in the envelope gene region. In contrast, nonsynonymous substitutions may be deleterious due to effects on protein structure and function, or may be advantageous when they mediate immune escape. Thus, a reduced ratio of nonsynonymous to synonymous changes (dN/dS) is an indication of decreased immunologic pressure 10;11.
There is conflicting information on the effect of HIV on the course of HCV evolution. It has been shown that HCV RNA levels rise approximately 0.5 log10 in HIV coinfected subjects, suggestive of increased viral replication12–15. If this increased replication is due to reduced immunologic pressure, then a decrease in dN/dS would be anticipated. Therefore, we hypothesized that HIV infection would alter the host-HCV interaction resulting in lower dN/dS. To test this hypothesis, we examined HCV envelope sequences over time in persons with chronic HCV infection that were either HIV negative or HIV positive, with or without severe immunodeficiency.
MATERIALS AND METHODS
Subjects
We studied subjects participating in a longitudinal study of the natural history of HCV infection among drug users that were enrolled between November 2, 1999 and December 13, 2000 in the Bronx, New York 14;16. Subjects underwent semiannual standardized interviews and phlebotomy for several tests including HIV-1 viral loads, HIV antibody, T-lymphocyte subset assays and HCV RNA assays as previously described 14;16. In order to assess the possible relationship of HIV infection and immunodeficiency to changes in HCV sequences over time, we chose a nested sample of 115 individuals having chronic HCV viremia who could be classified at the time of their last study phlebotomy into one of the following three groups: Group 1 consisted of HIV-negative, HCV RNA positive subjects; Group 2 consisted of subjects with a persistently high CD4+ T cell count of >350 cells/mm3, and Group 3 consisted of subjects with a persistently low CD4+ lymphocyte count of <200 cells/mm3.
HCV RNA analyses
To characterize the HCV RNA sequence we amplified a 1026 nucleotide (nt) region of the HCV genome, which encodes the envelope glycoproteins E1 and E2, including hypervariable region 1 (HVR1), as previously described 17;18. Prior work has indicated that the consensus sequence is an excellent approximation of the master sequence, or most-commonly-observed sequence.19 Therefore, RT-PCR PCR products were purified and directly sequenced using the reverse primer 1868a21 (positions 1868–1848 relative to HCV-H77, Genbank accession number AF009606) 5′-GAAGCAATAYACYGGRCCACA-3′17. Sequences generated in this study are available in GenBank, with accession numbers ____ through _____.
Phylogenetic analysis
Sequences were aligned from raw tracings using Aligner version 2.0.4 (CodonCode Corporation), and trimmed to 756 nucleotides (positions 1041–1796 relative to HCV-H77) encompassing approximately most of E1 and a third of E2 including HVR1, using BioEdit 21. Sites containing a gap in any sequence were stripped during analysis. Genetic distance (divergence) between paired sequences was calculated using the distance matrix calculation scripts provided with HyPhy version 0.99,22 with model selection and parameter estimation by maximum likelihood (inferred model parameters available on request from the authors). Synonymous and nonsynonymous genetic distances were determined using the MG94custom model as implemented in HyPhy.23 To determine the specific regions of the HCV genome where these changes were occurring we used the VarPlot program (available from S.C.R. at http://sray.med.som.jhmi.edu/SCRoftware),18 which uses the Nei and Gojobori method.24 Using this software we calculated values for dS, dN and the dN/dS ratio in a sliding window of 20 nucleotides 18. This process was then repeated for an overlapping segment of 20 bp, which was shifted by 1 bp (the step size), and continued across the alignment. At each step all pairwise comparisons for a subject were performed and values averaged. The mean values for all subjects were then averaged, ensuring that each subject was given equal weight. The Jukes-Cantor correction was used to correct for underestimation of distance due to multiple substitutions at the same site 25.
Statistical Analysis
The chi-square test was used to analyze categorical data and a Wilcoxon signed rank test was used to compare HIV viral loads and CD4+ T cell counts nonparametrically. The Kruskal-Wallis rank test was used for multiple comparisons among the 3 groups of subjects.
RESULTS
Initially 115 subjects were chosen for analysis at two time points. Twenty two subjects were excluded because HCV-specific RT-PCR failed at both time points, and 8 were excluded because only one of two specimens was successfully amplified. Two subjects were excluded because sequencing of RT-PCR products yielded sequence product length less than 80% of the mean length of other subjects and four subjects were excluded because of possible HCV reinfection. Among the excluded subjects twenty were from group 1, fourteen from group 2 and two from group 3. The primers used in this study were designed to amplify genotype 1, which is the most common genotype in the United States and as expected a majority of the pairs that we failed to amplify, were non-genotype 1. Therefore, a total of 79 subjects (74.7%) at two time points were evaluable (Table 1). The median time between visits was 933 days (range 360–1303). Thirty eight subjects were HIV negative, 21 had CD4+ T cell counts > 350 cells/mm3 at both time points and 20 had CD4+ counts < 200 cells/mm3 at both time points. No significant differences were observed in the gender or age between the 3 groups. The subjects were primarily Hispanic and a significant difference in race composition was detected between the three groups. By design, the median CD4+ T cell count was significantly higher in group 2 compared to group 3; in addition, a significantly higher HIV viral load was observed for group 3 compared to group 2. No significant differences were detected in the HCV viral load or the estimated duration of HCV infection among the three groups at either visit.
Table 1.
Demographic, immunologic and virologic characteristics
| Characteristic | Group 1 (n=38) | Group 2 (n=21) | Group 3 (n= 20) | P |
|---|---|---|---|---|
| HIV status | Negative | Positive | Positive | NAa |
| Age | ||||
| median (IQRb) | 47 (43–50) | 45 (41–50) | 44 (39–52) | 0.315c |
| Estimated years of HCV infectiond | 27g | 25g | 29g | 0.633c |
| Gender (% male) | 74 | 67 | 85 | 0.395e |
| Race (%) | 0.037e | |||
| Hispanic | 71 | 52 | 75 | |
| African American | 16 | 48 | 15 | |
| Caucasian | 13 | 0 | 10 | |
| CD4+ T cell count, median (IQR), cells/mm3 | ||||
| Time point (TP) 1 | ND | 680 (476–754) | 84 (28–138) | <0.0001f |
| TP 2 | ND | 681 (510–804) | 54(34–83) | <0.0001f |
| HCV RNA, median (IQR), log10 IU/mL | ||||
| TP 1 | 5.7 (5.4–5.9)h | 5.6 (5.4–5.9)h | 5.8 (5.6–5.9)h | 0.404c |
| TP 2 | 6.3 (5.7–6.6) | 6.6 (6.0–6.7)h | 6.7 (5.9–6.9)h | 0.112c |
| HIV RNA median (IQR), log10 copies/mL | ||||
| TP 1 | NA | 2.3 (1.7–3.3) | 3.9 (2.6–4.3) | 0.009c |
| TP 2 | NA | 3.2 (1.9–3.9) | 4.6 (4.0–4.6) | 0.003c |
NA, not applicable
IQR, interquartile range
Kruskal-Wallis test
Estimated from the time of first injection drug use
Chi-Square test
Wilcoxon Signed-rank test
Time of first injection drug use unknown for n=5 (group1), n=2 (group 2), n = 2 (group 3)
HCV RNA unknown for n=1 (group 1, TP1), n=1 (group 2, TP1), n=1 (group 3, TP1), n=3 (group 2, TP2), n=2 (group 3, TP2)
Effective antiretroviral therapy (ART) has been shown to alter the complexity and diversity of HCV quasispecies. Among the 41 HIV infected individuals in our study 20 had reported ART. Seven of the 21 subjects in Group 2 reported ART, 3 at both time points and 4 at one time point. Thirteen of the 20 subjects in Group 3 reported ART, 2 at both time points and 11 at a single time point. Over the time course of this study only 4/20 subjects on ART maintained HIV RNA levels less than 2.6 log10 copies/mL, which precluded an analysis of the effect of ART on HCV sequence divergence.
Overall, divergence between paired sequences in the three groups was similar (Table 2). In addition, no significant differences in divergence were detected between groups when the four excluded subjects (possible reinfection) were included in the analysis. Because a significant difference in racial composition was detected between groups we analyzed divergence controlling for race. However, since our study only had 7 Caucasian subjects, and group 2 did not contain Caucasian subjects they were not included in this analysis. No significant differences in divergence stratified by race were detected (P > 0.50, Kruskal-Wallis Test).
Table 2.
Phylogenetic analysis of paired sequence data
| Characteristic | Group 1 (HIV -) | Group 2 (HIV+) | Group 3 (HIV+) | Pa |
|---|---|---|---|---|
| Divergence (range) | ||||
| Median (range) | 0.0054 (0 – 0.094) | 0.0044 (0 – 0.049) | 0.0053 (0 – 0.071) | 0.903 |
| Interval between specimens | ||||
| Median months (range) | 36 (13–40) | 31 (12–43) | 27(13–43) | 0.036b |
| Rate of divergence/year | 0.0018 (0 – 0.050) | 0.0022 (0 – 0.033) | 0.0024 (0 – 0.022) | 0.470 |
| Nonsynonymous (dS) | ||||
| Distance Median (range) | 0.0014 (0 – 0.065) | 0.0026 (0 – 0.024) | 0.0027 (0 – 0.031) | 0.916 |
| Rate Median (range) | 0.00067 (0 – 0.034) | 0.00092 (0 – 0.016) | 0.0010 (0 – 0.0096) | 0.647 |
| Synonymous (dN) | ||||
| Distance Median (range) | 0.0016 (0 – 0.034) | 0.0031 (0 – 0.024) | 0.0035 (0 – 0.044) | 0.533 |
| Rate Median (range) | 0.00055 (0 – 0.024) | 0.0014 (0 – 0.016) | 0.0013 (0 – 0.014) | 0.288 |
| dN/ dS ratio | ||||
| Median (range) | 0.94 (0 – 13) | 0.79 (0 – 15) | 0.69 (0 – 23) | 0.379 |
Sequence results compared using the Kruskal-Wallis Test
Group 1 significantly longer interval between visits compared to group 3 using Dunn’s multiple comparison test.
The median number of days between specimens was significantly different among the three groups (P = 0.036, Kruskal-Wallis Test), with group 1 having a significantly longer interval between the specimens compared to group 3 (Dunn’s pairwise multiple comparison). The rate of divergence/year was also calculated and was not significantly different among the groups (Table 2). Divergence rate was also stratified by race but no statistical differences were detected (P >0.50 Kruskal-Wallis Test).
To specifically address our hypothesis that increased immunosuppression would alter the host-HCV interaction by decreasing amino acid changing mutations, we determined synonymous and nonsynonymous substitution frequencies. The median dS, dN substitution frequencies and rates, and dN/dS ratiosare listed in Table 2. Analysis of these measures for the three groups showed no significant differences (Kruskal-Wallis Test).
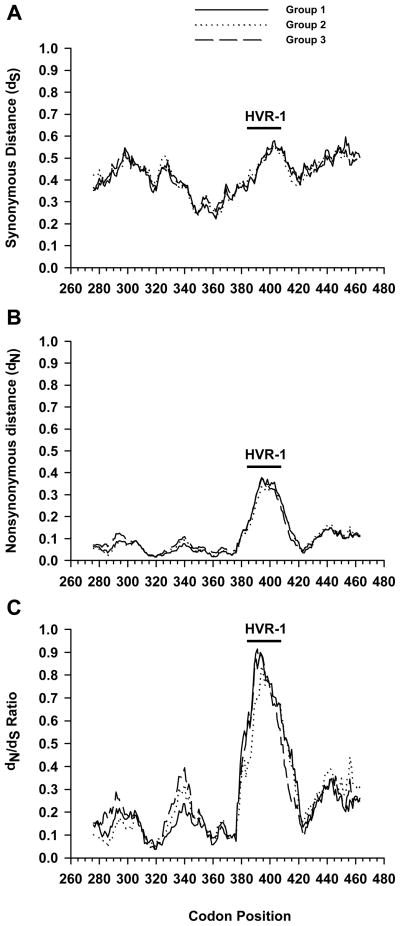
Since we found no global differences between the three groups in selective pressure, a high-resolution analysis was performed to determine if specific regions within the genome segment we analyzed were under greater selective pressures (Figure 1). Using the program VarPlot we calculated values for dS, dN and the dN/dS ratio in a sliding window. Comparison of the three groups using this method revealed little difference within dS or dN (Figure 1A and B). However, in general synonymous distances were greater than non-synonymous distance with median values for the three groups being 0.436 versus 0.081, respectively. Interestingly, synonymous changes were suppressed in the region centered on codon 362. The highest rate of nonsynonymous mutations was observed for the HVR-1 region centered on codon 398 in the E2 protein, as has been described previously18.
Figure 1.
Variability plots of HCV envelope region. For each subject the intersample dS (A), dN (B) or dN/dS ratio (C) variability was calculated for each position along the alignment, for a sliding window of 20 codons. The graph shows the mean values for each group (HIV negative, group 1; HIV positive absolute CD4+ T cell count >350 cells/mm3, group 2 or HIV positive CD4+ T cell count <200 cells/mm3, group 3). The indicated amino acid positions in the HCV polyprotein are according to HCV-H77 Genbank accession number AF009606. The position of hypervariable region 1 (HVR-1) is indicated.
Based on our initial assumptions we would expect that increasing immunosuppression as a result of HIV infection would tend to decrease immune selection (dN) and thus decrease the dN/dS ratio particularly in the group with the lowest CD4 counts. The HVR-1 dN/dS ratios in this study reached a maximum of approximately 0.9. However, the three groups essentially had overlapping dN/dS curves with the highest ratio centered on HVR-1.
DISCUSSION
In this study we examined the hypothesis that immunosuppression resulting from HIV infection would modify the host-HCV interaction resulting in decreased HCV sequence evolution over time. By measuring genetic divergence, synonymous and nonsynonymous nucleotide substitutions in HCV sequences over time, we found no significant differences in the master sequence between HCV monoinfected control subjects and HIV/HCV coinfected subjects with various levels of immunodeficiency as measured by CD4+ T cell counts.
Most previous studies have primarily analyzed complexity and diversity of the quasispecies. These analyses are efficient in determining the temporal composition of the HCV population and have sensitivity to allow determination of both minor and major variants. We decided to examine evolution (divergence) by directly sequencing RT-PCR products at two time points. This approach gives a more global or average weight to the quasispecies, but is insensitive for detection of minor variants of the population. Based on our approach, our data suggest that the shift in the quasispecies master sequence was similar among the three groups. However, a shift in minor variants could be occurring, but would not be detectable by our approach.
In our study we analyzed HCV sequence variability over a 2–3 year interval in essentially two groups, HIV positive and HIV negative subjects. One possible explanation for our inability to detect differences among these groups is that our assay was insensitive. However, the median dS value observed in our subjects was similar to synonymous distances found when analyzing full length genomes in this region and the HVR-1 dN/dS ratios, which reached a maximum of approximately 0.9 were also similar to another study 26. The synonymous and nonsynonymous substitution rates that were observed in our study were also consistent with previous reports of HCV sequence evolution 27. Furthermore, the highest rate of nonsynonymous mutations in our subjects was observed for the HVR-1 region centered on codon 398 in the E2 protein as expected. In addition, synonymous changes in our subjects were suppressed in the region centered on codon 362, which has been previously described and may represent RNA secondary structure 26. Therefore, our results are consistent with previous reports and validates that our methodology was sufficient to detect earlier recognized HCV sequence variability.
In our analysis we looked at other factors that could influence HCV sequence evolution such as race, age, duration of HCV infection, sampling interval and HCV viral load. Keenan and colleagues examining HCV infected subjects found significantly higher frequencies of dN and dN/dS ratios in the HVR-1 region for Caucasian versus African American subjects28. However, no significant differences were detected in complexity and diversity among these groups. A limitation of our study was that the group of HIV seropositive persons with CD4+ counts >350 cells/mm3 had no Caucasian subjects and we therefore could not examine our data for differences between this group and African Americans. However, our HIV negative group contained 6 African Americans and 5 Caucasian but no significant differences were detected in divergence, dS, dN or the dN/dS ratio (data not shown). We therefore stratified our results based on race and omitted the data from Caucasian subjects in all groups based on the small sample size, and found no significant differences between Hispanics and African Americans.
Previous HCV quasispecies analyses of HIV/HCV coinfected subjects have produced conflicting results. In one report examining HCV HVR-1 diversity in HIV negative versus HIV/HCV positive subjects no differences were detected until the CD4+ T cell count decreased below 50/μL 29. Still others have detected either an increase 30;31 or decrease in HCV sequence variability 17;32;33. The role of HAART in this setting has also been examined with some groups finding increased HCV quasispecies diversity after immune restoration 33;34, and some finding no difference 35. It is difficult to reconcile the differences in these studies because they used different study designs (cross-sectional versus longitudinal), and variable duration of HCV infection existed among study subjects. In addition to the above mentioned factors the HCV sequence analysis methodology also differed. A majority of these studies utilized gel shift patterns to determine the diversity and complexity of the quasispecies while some utilized direct sequencing of PCR products. The gel shift analyses are used to examine viral heterogeneity (eg. complexity and diversity among clones) which has been shown to be influenced by the factors listed above. Since we did not examine viral heterogeneity we could have missed changes in minor quasispecies variants on which evolution could be acting. While we agree these changes in minor variants are likely occurring, we found in our study the major or most fit variant seems to be stable over the time period studied. Since the interaction of HIV and concomitant immunosuppression on HCV is likely a multifactorial process, small differences in experimental designs with small and diverse study populations make it difficult to precisely model this interaction.
There were limitations to our study. Antiretroviral treatment has been shown to influence complexity and diversity of the quasispecies 34,33. Since only 4 subjects in our study had effective ART we could not assess its role in HCV sequence divergence. Similarly, we did observe higher median rates of divergence in subjects with HIV and CD4 depletion compared with those without HIV infection that were not statistically significant, suggesting that future studies using similar methods may need to be larger than the one described here to determine whether there may be small significantly different divergence rates. Another potential limitation is that we only examined approximately 600 nucleotides in the E1 and E2 regions of the HCV genome and could have missed changes in other B and T cell epitopes outside of this region. Data on liver histology and aminotransferase levels were not available, precluding an examination of the effect of liver disease activity on HCV sequence divergence.
A possible explanation for our results could be that amino acid substitutions in the envelope glycoproteins, especially HVR-1, have saturated. By studying HCV sequence evolution in a cohort of Irish women infected by contaminated anti-D immunoglobulin, McAllister et.al. found that amino acid substitutions become saturated over short durations of divergence. Therefore, it is plausible with the greater than 20 years of estimated HCV infection that the subjects in the current study have reached their divergence or evolutionary plateau.
Acknowledgments
This study was supported by grant number R01 DA13248 from the National Institute on Drug Abuse and by a Center for AIDS Research Grant AI-051519 from the National Institute of Allergy and Infectious Diseases.
Footnotes
The information was presented in part at the 12th Conference on Retroviruses and Opportunistic Infections. February 22–25, 2005, Boston, MA. Abstract #911
Reference List
- 1.World Health Organization. Weekly Epidemiological Record. 1997. Hepatitis C: global prevalence; pp. 341–48. [Google Scholar]
- 2.Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA, Kaslow RA, Margolis HS. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556–62. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- 3.Sherman KE, Rouster SD, Chung RT, Rajicic N. Hepatitis C Virus prevalence among patients infected with Human Immunodeficiency Virus: a cross-sectional analysis of the US adult AIDS Clinical Trials Group. Clin Infect Dis. 2002;34:831–37. doi: 10.1086/339042. [DOI] [PubMed] [Google Scholar]
- 4.Graham CS, Baden LR, Yu E, Mrus JM, Carnie J, Heeren T, Koziel MJ. Influence of human immunodeficiency virus infection on the course of hepatitis c virus infection: a meta-analysis. Clin Infect Dis. 2001;33:562–69. doi: 10.1086/321909. [DOI] [PubMed] [Google Scholar]
- 5.Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos C, Coit, Medina-Selby R, Barr PJ. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–55. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martell M, Esteban JI, Quer J, Genesca J, Weiner A, Esteban R, Guardia, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–29. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato N, Ootsuyama Y, Tanaka T, Nakagawa M, Nakazawa T, Muraiso K, Ohkoshi S, Hijikata M, Shimotohno K. Marked sequence diversity in the putative envelope proteins of hepatitis C viruses. Virus Res. 1992;22:107–23. doi: 10.1016/0168-1702(92)90038-b. [DOI] [PubMed] [Google Scholar]
- 8.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–7. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 9.Tuplin A, Wood J, Evans DJ, Patel AH, Simmonds P. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA. 2002;8:824–41. doi: 10.1017/s1355838202554066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans DT, O’Connor DH, Jing P, Dzuris JL, Sidney J, da Silva J, Allen TM, Horton H, Venham JE, Rudersdorf RA, Vogel T, Pauza CD, Bontrop RE, DeMars R, Sette A, Hughes AL, Watkins DI. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat Med. 1999;5:1270–1276. doi: 10.1038/15224. [DOI] [PubMed] [Google Scholar]
- 11.Ray SC, Mao Q, Lanford RE, Bassett S, Laeyendecker O, Wang YM, Thomas DL. Hypervariable region 1 sequence stability during hepatitis C virus replication in chimpanzees. J Virol. 2000;74:3058–66. doi: 10.1128/jvi.74.7.3058-3066.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherman KE, O’Brien J, Gutierrez AG, Harrison S, Urdea M, Neuwald P, Wilber J. Quantitative evaluation of hepatitis C virus RNA in patients with concurrent human immunodeficiency virus infections. J Clin Microbiol. 1993;31:2679–82. doi: 10.1128/jcm.31.10.2679-2682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas DL, Shih JW, Alter HJ, Vlahov D, Cohn S, Hoover DR, Cheung L, Nelson KE. Effect of human immunodeficiency virus on hepatitis C virus infection among injecting drug users. J Infect Dis. 1996;174:690–695. doi: 10.1093/infdis/174.4.690. [DOI] [PubMed] [Google Scholar]
- 14.Fishbein DA, Lo Y, Netski D, Thomas DL, Klein RS. Predictors of hepatitis C virus RNA levels in a prospective cohort study of drug users. J Acquir Immune Defic Syndr. 2006;41:471–76. doi: 10.1097/01.qai.0000218360.28712.f3. [DOI] [PubMed] [Google Scholar]
- 15.Thomas DL, Astemborski J, Vlahov D, Strathdee SA, Ray SC, Nelson KE, Galai N, Nolt KR, Laeyendecker O, Todd JA. Determinants of the quantity of hepatitis C virus RNA. J Infect Dis. 2000;181:844–51. doi: 10.1086/315314. [DOI] [PubMed] [Google Scholar]
- 16.Strasfeld L, Lo Y, Netski D, Thomas DL, Klein RS. The association of hepatitis C prevalence, activity, and genotype with HIV infection in a cohort of New York City drug users. J Acquir Immune Defic Syndr. 2003;33:356–64. doi: 10.1097/00126334-200307010-00010. [DOI] [PubMed] [Google Scholar]
- 17.Mao Q, Ray SC, Laeyendecker O, Ticehurst JR, Strathdee SA, Vlahov D, Thomas DL. Human immunodeficiency virus seroconversion and evolution of the hepatitis C virus quasispecies. J Virol. 2001;75:3259–67. doi: 10.1128/JVI.75.7.3259-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray SC, Wang YM, Laeyendecker O, Ticehurst J, Villano SA, Thomas DL. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as decoy. J Virol. 1998;73:2938–46. doi: 10.1128/jvi.73.4.2938-2946.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabot B, Esteban JI, Martell M, Genescà J, Vargas V, Esteban R, Guardia J, Gómez J. Structure of replicating hepatitis C virus (HCV) quasispecies in the liver may not be reflected by analysis of circulating HCV virions. J Virol. 1997;71:1732–34. doi: 10.1128/jvi.71.2.1732-1734.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–5. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 21.Hall TA. [accessed 20 Feb 2005];BioEdit: Biological sequence alignment editor for Windows 95/98/NT version 7.0.4. software. 2001 Distributed by author: http://www.mbio.ncsu.edu/RNaseP/info/programs/BIOEDIT/bioedit.html.
- 22.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–79. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 23.Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol. 1994;11:715–24. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- 24.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–26. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 25.Jukes TH, Cantor TR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 26.Smith DB, Simmonds P. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J Mol Evol. 1997;45:238–46. doi: 10.1007/pl00006226. [DOI] [PubMed] [Google Scholar]
- 27.Ina Y, Mizokami M, Ohba K, Gojobori T. Reduction of synonymous substitutions in the core protein gene of hepatitis C virus. J Mol Evol. 1994;38:50–56. doi: 10.1007/BF00175495. [DOI] [PubMed] [Google Scholar]
- 28.Keenan ED, Rouster SD, Shire NJ, Horn PS, Sherman KE. Complexity and diversity of hepatitis C virus RNA in african americans and whites: analysis of the envelope-coding domain. J Infect Dis. 2004;190:511–14. doi: 10.1086/421506. [DOI] [PubMed] [Google Scholar]
- 29.Toyoda H, Fukuda Y, Koyama Y, Takamatsu J, Saito H, Hayakawa T. Effect of immunosuppression on composition of quasispecies population of hepatitis C virus in patients with chronic hepatitis C coinfected with human immunodeficiency virus. J Hepatol. 1997;26:975–82. doi: 10.1016/s0168-8278(97)80105-5. [DOI] [PubMed] [Google Scholar]
- 30.Sherman KE, Andreatta C, O’Brien J, Gutierrez A, Harris R. Hepatitis C in human immunodeficiency virus-coinfected patients: Increased variability in the hypervariable envelope coding domain. Hepatology. 1996;23:688–94. doi: 10.1002/hep.510230405. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka Y, Hanada K, Hanabusa H, Kurbanov F, Gojobori T, Mizokami M. Increasing genetic diversity of hepatitis C virus in haemophiliacs with human immunodeficiency virus coinfection. J Gen Virol. 2007;88:2513–19. doi: 10.1099/vir.0.82974-0. [DOI] [PubMed] [Google Scholar]
- 32.Roque-Afonso AM, Robain M, Simoneau D, Rodriguez-Mathieu P, Gigou M, Meyer L, Dussaix E. Influence of CD4 cell counts on the genetic heterogeneity of hepatitis C virus in patients coinfected with human immunodeficiency virus. J Infect Dis. 2002;185:728–33. doi: 10.1086/339297. [DOI] [PubMed] [Google Scholar]
- 33.Shuhart MC, Sullivan DG, Bekele K, Harrington RD, Kitahata MM, Mathisen TL, Thomassen LV, Emerson SS, Gretch DR. HIV Infection and Antiretroviral Therapy: Effect on Hepatitis C Virus Quasispecies Variability. J Infect Dis. 2006;193:1211–18. doi: 10.1086/502974. [DOI] [PubMed] [Google Scholar]
- 34.Blackard JT, Yang Y, Bordoni P, Sherman KE, Chung RT. Hepatitis C virus (HCV) diversity in HIV-HCV-coinfected subjects initiating highly active antiretroviral therapy. J Infect Dis. 2004;189:1472–81. doi: 10.1086/382959. [DOI] [PubMed] [Google Scholar]
- 35.Babik JM, Holodniy M. Impact of highly active antiretroviral therapy and immunologic status on hepatitis C virus quasispecies diversity in human immunodeficiency virus/hepatitis C virus-coinfected patients. J Virol. 2003;77:1940–1950. doi: 10.1128/JVI.77.3.1940-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]