Abstract
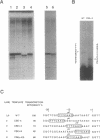
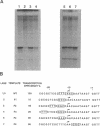
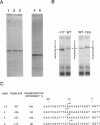
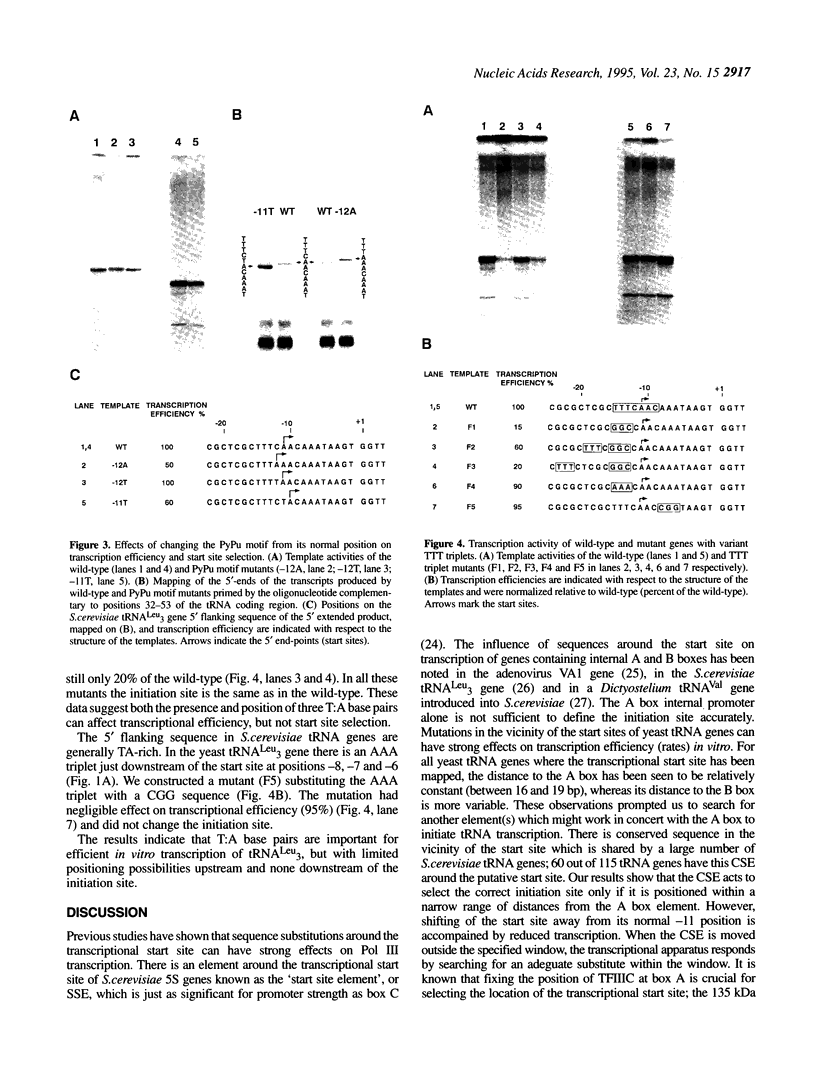
In addition to the well-known internal promoter elements of tRNA genes, 5' flanking sequences can also influence the efficiency of transcription by Saccharomyces cerevisiae extracts in vitro. A consensus sequence of yeast tRNA genes in the vicinity of the transcriptional start site can be derived. To determine whether the activity of this region can be attributed to particular sequence features we studied in vitro mutants of the start site region. We found that the start site can be shifted, but only to a limited extent, by moving the conserved sequence element. We found that both a pyrimidine-purine motif (with transcription initiating at the purine) and a small T:A base pair block upstream are important for efficient transcription in vitro. Thus the sequence surrounding the start site of transcription of the yeast tRNA(Leu3) gene does play a role in determining transcription efficiency and fixing the precise site of initiation by RNA polymerase III.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold G. J., Schmutzler C., Gross H. J. Functional dissection of 5' and 3' extragenic control regions of human tRNA(Val) genes reveals two different regulatory effects. DNA. 1988 Mar;7(2):87–97. doi: 10.1089/dna.1988.7.87. [DOI] [PubMed] [Google Scholar]
- Baker R. E., Camier S., Sentenac A., Hall B. D. Gene size differentially affects the binding of yeast transcription factor tau to two intragenic regions. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8768–8772. doi: 10.1073/pnas.84.24.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B., Kassavetis G. A., Geiduschek E. P. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991 Oct;11(10):5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker D. L., Sandmeyer S. B. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 1992 Jan;6(1):117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- Challice J. M., Segall J. Transcription of the 5 S rRNA gene of Saccharomyces cerevisiae requires a promoter element at +1 and a 14-base pair internal control region. J Biol Chem. 1989 Nov 25;264(33):20060–20067. [PubMed] [Google Scholar]
- Comai L., Tanese N., Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992 Mar 6;68(5):965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992 May 15;69(4):685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Coppo A., Fruscoloni P., Benedetti P., Di Segni G., Tocchini-Valentini G. P. Comparative mutational analysis of wild-type and stretched tRNA3(Leu) gene promoters. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8763–8767. doi: 10.1073/pnas.84.24.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furter-Graves E. M., Hall B. D. DNA sequence elements required for transcription initiation of the Schizosaccharomyces pombe ADH gene in Saccharomyces cerevisiae. Mol Gen Genet. 1990 Sep;223(3):407–416. doi: 10.1007/BF00264447. [DOI] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Hipskind R. A., Clarkson S. G. 5'-flanking sequences that inhibit in vitro transcription of a xenopus laevis tRNA gene. Cell. 1983 Oct;34(3):881–890. doi: 10.1016/0092-8674(83)90545-7. [DOI] [PubMed] [Google Scholar]
- Johnson J. D., Raymond G. J. Three regions of a yeast tRNALeu3 gene promote RNA polymerase III transcription. J Biol Chem. 1984 May 10;259(9):5990–5994. [PubMed] [Google Scholar]
- Kassavetis G. A., Blanco J. A., Johnson T. E., Geiduschek E. P. Formation of open and elongating transcription complexes by RNA polymerase III. J Mol Biol. 1992 Jul 5;226(1):47–58. doi: 10.1016/0022-2836(92)90123-2. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Braun B. R., Nguyen L. H., Geiduschek E. P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990 Jan 26;60(2):235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Riggs D. L., Negri R., Nguyen L. H., Geiduschek E. P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989 Jun;9(6):2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure A., Carbon P., Krol A. The different positioning of the proximal sequence element in the Xenopus RNA polymerase II and III snRNA promoters is a key determinant which confers RNA polymerase III specificity. Nucleic Acids Res. 1991 Feb 11;19(3):435–441. doi: 10.1093/nar/19.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure A., Murgo S., Carbon P., Krol A. The proximal promoter and the start site cooperate to specify correct U1 snRNA transcription initiation by RNA polymerase II. Nucleic Acids Res. 1992 Apr 11;20(7):1573–1578. doi: 10.1093/nar/20.7.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisser S., Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993 Apr 11;21(7):1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofquist A. K., Garcia A. D., Sharp S. J. A discrete region centered 22 base pairs upstream of the initiation site modulates transcription of Drosophila tRNAAsn genes. Mol Cell Biol. 1988 Oct;8(10):4441–4449. doi: 10.1128/mcb.8.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschalek R., Dingermann T. Identification of a protein factor binding to the 5'-flanking region of a tRNA gene and being involved in modulation of tRNA gene transcription in vivo in Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 25;16(14B):6737–6752. doi: 10.1093/nar/16.14.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Palida F. A., Hale C., Sprague K. U. Transcription of a silkworm tRNA(cAla) gene is directed by two AT-rich upstream sequence elements. Nucleic Acids Res. 1993 Dec 25;21(25):5875–5881. doi: 10.1093/nar/21.25.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Railey J. F., 2nd, Wu G. J. Organization of multiple regulatory elements in the control region of the adenovirus type 2-specific VARNA1 gene: fine mapping with linker-scanning mutants. Mol Cell Biol. 1988 Mar;8(3):1147–1159. doi: 10.1128/mcb.8.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond K. C., Raymond G. J., Johnson J. D. In vivo modulation of yeast tRNA gene expression by 5'-flanking sequences. EMBO J. 1985 Oct;4(10):2649–2656. doi: 10.1002/j.1460-2075.1985.tb03983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. A synthetic substrate for tRNA splicing. Anal Biochem. 1987 Oct;166(1):90–106. doi: 10.1016/0003-2697(87)90551-3. [DOI] [PubMed] [Google Scholar]
- Schultz M. C., Choe S. Y., Reeder R. H. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M. C., Reeder R. H., Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992 May 15;69(4):697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- Werner M., Chaussivert N., Willis I. M., Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993 Oct 5;268(28):20721–20724. [PubMed] [Google Scholar]
- White R. J., Jackson S. P., Rigby P. W. A role for the TATA-box-binding protein component of the transcription factor IID complex as a general RNA polymerase III transcription factor. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1949–1953. doi: 10.1073/pnas.89.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L. S., Takahashi N., Sprague K. U. Upstream sequences confer distinctive transcriptional properties on genes encoding silkgland-specific tRNAAla. Proc Natl Acad Sci U S A. 1986 Jan;83(2):374–378. doi: 10.1073/pnas.83.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubenko G. S., Mitchell A. P., Jones E. W. Mapping of the proteinase b structural gene PRB1, in Saccharomyces cerevisiae and identification of nonsense alleles within the locus. Genetics. 1980 Sep;96(1):137–146. doi: 10.1093/genetics/96.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]