Abstract
Ciliary dysfunction leads to a broad range of overlapping phenotypes, termed collectively as ciliopathies. This grouping is underscored by genetic overlap, where causal genes can also contribute modifying alleles to clinically distinct disorders. Here we show that mutations in TTC21B/IFT139, encoding a retrograde intraflagellar transport (IFT) protein, cause both isolated nephronophthisis (NPHP) and syndromic Jeune Asphyxiating Thoracic Dystrophy (JATD). Moreover, although systematic medical resequencing of a large, clinically diverse ciliopathy cohort and matched controls showed a similar frequency of rare changes, in vivo and in vitro evaluations unmasked a significant enrichment of pathogenic alleles in cases, suggesting that TTC21B contributes pathogenic alleles to ∼5% of ciliopathy patients. Our data illustrate how genetic lesions can be both causally associated with diverse ciliopathies, as well as interact in trans with other disease-causing genes, and highlight how saturated resequencing followed by functional analysis of all variants informs the genetic architecture of disorders.
Introduction
Genetic and functional studies have recognized that defects in genes encoding components of the ciliary apparatus lead to an overlapping set of clinical phenotypes that include retinal degeneration, renal cystic disease, polydactyly and other skeletal abnormalities, fibrosis of various organs, and a complex range of anatomical and functional defects of the central and peripheral nervous system. The recognition of the clinical overlap between discrete clinical entities attributable to ciliary dysfunction has led to the unification of such disorders under the ciliopathy module1,2. This integration has also been reflected in the genetic architecture of ciliopathies: although discrete groups of genes and proteins have been causally linked with specific ciliopathies, single-locus allelism has been insufficient to explain the variable penetrance and expressivity of such disorders, leading to the suggestion that genetic variation across multiple sites of the ciliary proteome influences clinical outcome. Consistent with this notion, mutations in NPHP1-4 have been shown to exacerbate extra-renal phenotypes in NPHP patients with primary lesions at one of the other NPHP genes3, whereas mutations in some of the genes that cause MKS can also contribute mutations in patients diagnosed with NPHP4,5, Joubert syndrome (JBTS)6-13 and Bardet-Biedl syndrome (BBS)14. Recently, epistatic mutations in RPGRIP1L, loss of function of which causes MKS and JBTS, have been shown to modify the penetrance of retinal degeneration across multiple ciliopathies15, while an allele in AHI1 can modify retinal phenotypes in individuals with NPHP16.
These observations have raised two possibilities. First, if the model outlined above were correct, one would predict that some genes critical to ciliary function would contribute alleles that will appear (genetically) to be necessary and sufficient to cause discrete ciliopathy phenotypes in humans. Second, the same loci should contribute epistatic alleles across the same phenotypic spectrum, some of which would be expected to contribute to the severity and/or pleiotropy of the disease.
To test these hypotheses, we have initiated a systematic screening of all genes known to be necessary for ciliary biogenesis and function, coupled to functional assessment of variants detected (irrespectively of their genetically-derived pathogenic potential under a strict Mendelian model). We chose to test this model on TTC21B for several reasons. First, it encodes a bone fide axonemal protein that is required for retrograde intraflagellar transport17. Second, homozygous null mutations at this locus are responsible for the embryonic lethal alien phenotype in the mouse, a mutant whose anatomical features overlap with some of the severe ciliopathies in humans18. Third, TTC21B contains several tetratricopeptide (TPR) domains that seem both important for ciliary function by virtue of their enrichment in the ciliary proteome and photoreceptor sensory cilium proteome19,20, and are also present in numerous ciliopathy-causing proteins in humans such as BBS421 and BBS822.
Results
Saturated resequencing of TTC21B identified numerous novel coding variants
To comprehensively assess the potential contribution of this transcript, we sequenced all 29 coding exons of TTC21B in a clinically diverse cohort of 753 patients, without preselection for known ciliopathy gene mutations or genetic linkage data. The cohort spanned the clinical severity spectrum, including NPHP (with and without extra-renal phenotypes), JBTS, BBS, JATD, and MKS (Supplementary Table 1). In parallel, to assess the extent of coding variation at this locus, we also sequenced 398 normal individuals (Suppl. Table 1). We identified 48 different novel variants that affected the protein sequence, including two frameshifts, two nonsense, four splice junction, and 40 missense. The majority of changes were present in the heterozygous state and their rarity necessitated functional characterization so that their genetic contribution to ciliopathies could be modeled in detail. We therefore proceeded to test all missense variants, including all nonsynonymous alleles present in SNP databases (Supplementary Table 2).
Functional assays to test the pathogenicity of TTC21B variants
In vivo complementation assays
We have shown previously that in vivo rescue of zebrafish ciliary phenotypes with wild-type (wt) or mutant human mRNA represents a robust, physiologically relevant tool to assay allele neutrality14,15,23. Upon reciprocal BLAST, we identified the single ortholog of TTC21B in the zebrafish genome (68% identity, 84% similarity), and we were able to detect endogenous message by RT-PCR as early as zebrafish embryonic shield-stage (data not shown). Next, we designed a translation-blocking morpholino (tb-MO) targeting ttc21b, which we injected into two-cell stage embryos (n=50-100/injection dose; 2ng, 4ng, 6ng, 8ng and 10ng). Masked scoring of live embryos at the eight-to-nine somite stage reproduced previously reported gastrulation phenotypes in a dose-dependent manner (Supplementary Fig. 1). These defects included shortening of the embryonic axis, widening and kinking of the notochord, and broadening and thinning of the somites, each of which enabled grouping into phenotypic classes according to previously-described objective phenotypic criteria14 (Normal, Class I, Class II; Fig. 1a,b). Importantly, we were able to rescue morphant phenotypes; co-injection of wt human TTC21B mRNA producing significant improvement (Fig. 1b, Supplementary Table 3). Finally, to ensure the specificity of the ttc21b tb-MO further, we repeated the in vivo rescue assay with an ttc21b splice-blocking (sb) MO, which reduced wt ttc21b by >80%; we observed dose-dependent gastrulation phenotypes similar to the tb-MO (Supplementary Fig. 1), which could also be rescued with a similar efficiency as the tb-MO (data not shown).
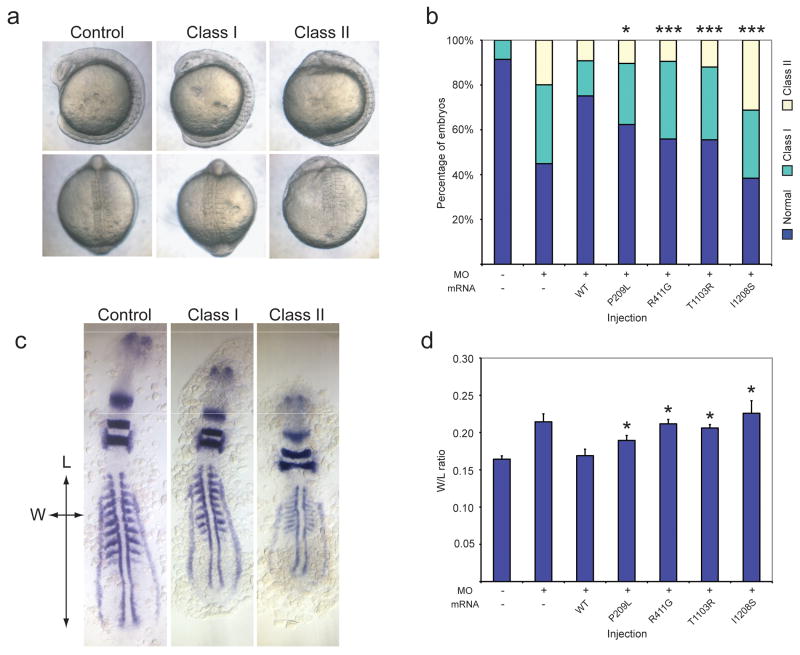
Figure 1. In vivo assay of TTC21B variants in mid-somitic zebrafish embryos.
a. Lateral and dorsal views of ttc21b morphants. Morpholino (MO)-based suppression of ttc21b results in gastrulation defects. Class I: shortened anterior-posterior body axis with small anterior structures and mild somite defects; Class II: severely shortened body axis, severely affected anterior structures, broadening and kinking of the notochord, thinning and widening of the somites, and tail extension defects.
b. in vivo rescue assay of ttc21b MO with human mRNA. Co-injection of wild-type (WT) human TTC21B with ttc21b translation-blocking (tb) MO results in significant rescue at the 10-somite stage, whereas mRNAs encoding missense alleles result in either partial rescue (P209L, R411G, T1103R), or are indistinguishable from MO alone (I1208S). χ2 values for rescue vs. WT are denoted as (*), p<0.05; or (***), p<0.0001.
c. Whole embryo flat mounts hybridized in situ with krox20, pax2, and myoD riboprobes. Measurements reflect phenotypes at two different axes; length of the notochord as indicated by adaxial cell labeling (L), and width spanning from the lateral ends of the fifth appreciable somites (W), expressed as a ratio (W/L).
d. Quantitative morphological data for age-matched embryos, measured as indicated in (c). The variants shown are significantly different from WT rescue. t-test values for rescue vs. WT are denoted as (*), p<0.05. Error bars show standard error of the mean (SEM).
We subsequently tested each of the 40 novel nonsynonymous TTC21B changes using an in vivo rescue assay. Injection cocktails containing tb-MO and each mutant TTC21B message were injected into zebrafish embryos (71-180 embryos/injection; masked scoring) and compared to the rescue efficiency of wt rescue or tb-MO alone (Fig. 1b, Supplementary Table 3). Using this approach we observed that, first, nearly half of TTC21B variants (n=18/40), were significantly worse than wt rescue, yet significantly better than tb-MO alone (p<0.05 for each comparison), indicating they were likely hypomorphic alleles. We also observed functionally null alleles (n=11; not significantly improved from tb-MO alone). The remaining alleles were scored as benign (n=11; not significantly exacerbated in comparison to wt rescue), which was consistent with our genetic prediction; 3/11 alleles were detected in similar frequencies in patients and controls, and 8/11 were detected exclusively in controls. We also tested four SNPs from HapMap encoding nonsynonymous TTC21B changes (rs1432273, rs7592429, rs16851307, and rs2163649) and found each to be benign, increasing our confidence in the specificity of the assay (Supplementary Table 2, Supplementary Table 3). Finally, to eliminate the possibility of mutant mRNA toxicity being the primary cause for gastrulation defects, we injected each of the 40 transcripts in the absence of tb-MO and found each to be similar to wt mRNA alone (χ2 for each comparison not significant; Supplementary Table 4).
To assess pathogenicity of TTC21B variants by alternative objective means, we measured anatomical features labeled in situ with a cocktail of pax2, krox20, and myoD riboprobes on whole embryo flat-mounts. We have shown previously that the ratio of two dimensional embryo measurements spanning the distal ends of the fifth appreciable somites (w), versus the length of the notochord as indicated by adaxial cell labeling (l) is an effective approach to quantify gastrulation defects in ciliary morphants15. We randomly chose a subset of 15 different mutant rescue injected batches, age-matched at 9-10 somites, and calculated the w/l ratio for each embryo (n=7-10 embryos/injection). Upon comparison of mutant rescue versus wt w/l ratios, (once again, scored masked to injection cocktail) we observed that 14/15 were in agreement with the in vivo scores (Fig. 1c,d, Supplementary Table 2, Supplementary Table 5).
In vitro complementation assays
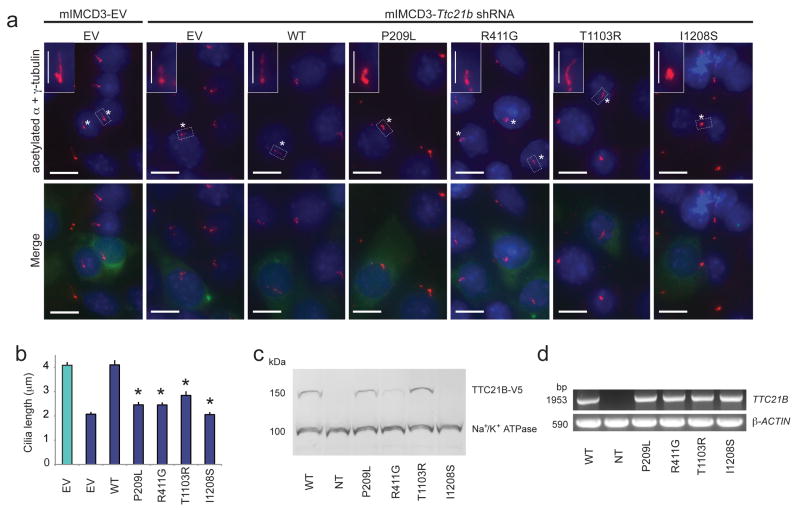
To validate further the zebrafish assays, we turned next to an in vitro mammalian cell model. Endogenous TTC21B localizes to the basal body and ciliary axoneme in ciliated murine inner medullary collecting duct (IMCD3) cells18, an observation which we reproduced with transient transfection of an expression vector harboring the full-length wt TTC21B open reading frame (ORF; Supplementary Fig. 2, Supplementary Fig. 3). Furthermore, mIMCD3 cells stably transfected with Ttc21b shRNA (resulting in 87.5% suppression of Ttc21b) display shortened cilia in comparison to wild-type cells18. We subsequently used these cell lines to test allele pathogenicity based on the ability of transiently transfected mutant constructs to rescue defects in cilia length (n=85-155 cilia measured/construct). Whereas wild-type TTC21B resulted in full rescue of cilia length in transiently transfected cells (p=0.94 vs. mIMCD3-empty vector (EV)), transient expression of 17 different mutant proteins (the same group examined in the zebrafish morphometric assay, in addition to L1002V and D1041N), indicated that four variants were indistinguishable from mIMCD3-Ttc21b shRNA cells alone (M280V, P753L, R939W, and I1208S) suggesting that they are functional nulls in this assay, and the other thirteen variants resulted in partial, but not complete rescue of cilia length indicating that they are hypomorphic alleles (Fig. 2a, b, Supplementary Table 2, Supplementary Table 6). Importantly, these results are concordant with our in vivo zebrafish scoring results. These observations are likely due to cellular mislocalization (abnormal for 15/16 variants tested; Supplementary Table 2, Supplementary Fig. 3) and/or aberrant protein levels (as determined by Western blot), (Fig. 2c,d; Supplementary Table 2; Supplementary Table 6). The L795P mutant protein was the only variant that was expressed at normal levels and localized correctly to the basal body, suggesting that its effect probably lies in the biochemical function of TTC21B.
Figure 2. In vitro rescue assay of cilia length defects in mIMCD3-Ttc21b shRNA cells.
a. Immunofluorescent staining of mIMCD3-EV or mIMCD3-Ttc21b shRNA cells transfected transiently with plasmids encoding wild-type (WT) or mutant versions of pCAG-V5-TTC21B-IRES-EGFP constructs demonstrate failure to rescue shortened cilia phenotypes. Cilia and centrosomes were detected with anti-acetylated α-tubulin and anti-γ-tubulin (red), green signal indicates transfected cells (GFP), and nuclei are stained with Hoechst dye (blue). Asterisks indicate cilia on transiently transfected cells, dashed boxes depict inset; EV, empty vector. Horizontal scale bars: 10μm; vertical scale bars: 4μm (insets).
b. Quantification of cilia length measurements. Whereas WT TTC21B rescues the cilia length defects induced by Ttc21b shRNA, mutant proteins result in either partial rescue (P209L, R411G, or T1103R) or are indistinguishable from shRNA cells alone (I1208S). t-test values for rescue vs. WT are denoted as (*), p<0.0001. Green bar, mIMCD3-EV cells; blue bars, mIMCD3- Ttc21b shRNA cells; error bars, standard error of the mean (SEM; see Supplementary Table 6 for measurement data).
c. Protein stability defects for some TTC21B missense variants. Compared to WT, P209L and R411G result in diminished levels of TTC21B; protein with the I1208S mutation is undetected (See Supplementary Table 7 for densitometry data). Na+/K+-ATPase was used as a loading control; NT, not transfected.
c. Transiently transfected pCAG-V5-TTC21B-IRES-EGFP constructs express at similar levels in mIMCD3 cells. RT-PCR data are shown for human TTC21B amplified from cDNA generated from total RNA extracted from mIMCD3 cells 72h post-transfection. Murine β-actin was used as a loading control.
TTC21B in the mammalian photoreceptor
Finally, we investigated the potential contribution of novel TTC21B changes in discrete endophenotypes of physiological relevance to our ciliopathy cohort. The alien mouse exhibits axial skeleton defects, polydactyly, and failure of neural tube closure which mirror hallmark features of the JATD and MKS individuals in our study24. However, neonatal lethality by E18.5 precludes us from studying the effects of Ttc21b suppression in postnatal onset phenotypes such as retinal degeneration. As part of our ongoing studies of proteins in the mouse photoreceptor sensory cilium20, we investigated the function of TTC21B in mammalian photoreceptors in a rodent model. First, we developed an antibody against murine TTC21B, and demonstrated that it localizes specifically to the transition zone of mouse photoreceptor sensory cilia (PSC) as evidenced by location just proximal to endogenous murine retinitis pigmentosa 1 (Rp1), which is located in the outer segment portion of the PSC axoneme25 (Supplementary Fig. 2c). Next, to test the effects of Ttc21b suppression in the retina, we generated a rodent-specific shRNA vector targeting Ttc21b transcript (shRNA-Ttc21b (2923)). Through in vivo electroporation of shRNA- Ttc21b (2923) in neonatal rat retinas, we observed significantly shortened, abnormally shaped PSC, a specific defect which could be rescued with co-injection of a human TTC21B expression vector, suggesting that Ttc21b is required for normal photoreceptor development (Supplementary Fig. 4).
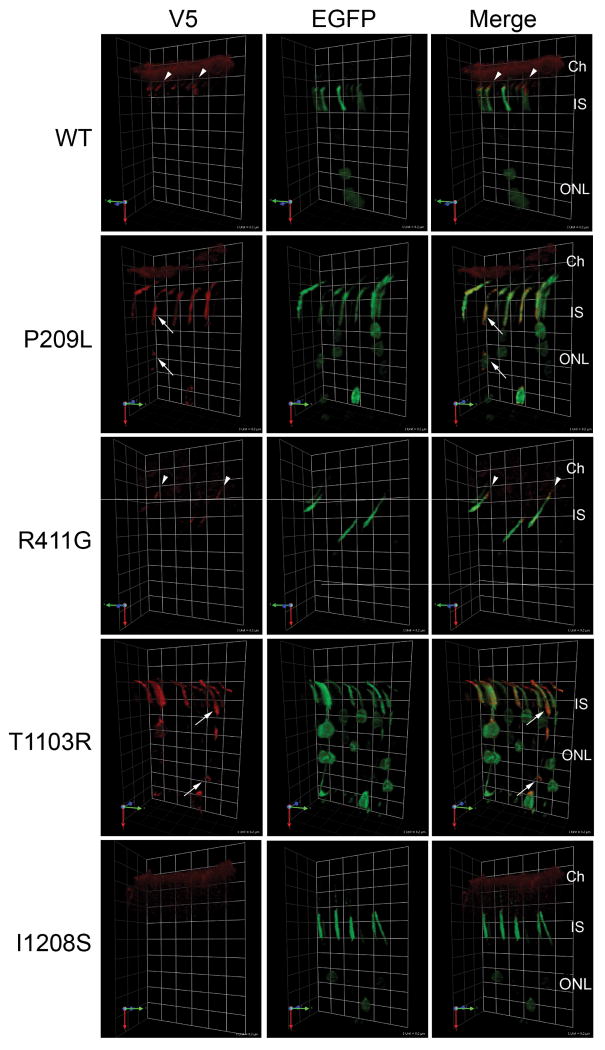
As yet another means to probe the pathogenicity of TTC21B variants found in our cohort, we used the in vivo retinal electroporation approach to assess mutant protein localization in rat PSC. We randomly selected six different nucleotide changes identified in patients and expressed them transiently in rat photoreceptors. Similar to the zebrafish studies, P209L, R411G, L1002V, T1103R, and D1041N mutant proteins mislocalize partially in the photoreceptor, consistent with the notion that they are hypomorphic alleles in vivo (Fig. 3, Supplementary Table 2) and partially rescue the ciliary length phenotypes in vitro. By contrast, I1208S protein is undetectable in transfected photoreceptor cells, which recapitulates both the in vitro Western analyses and completely fails to rescue ciliary length in vitro, suggesting it is functionally null probably from loss of protein stability or increased degradation (Fig. 2, Fig. 3, Supplementary Table 2, Supplementary Table 7).
Figure 3. TTC21B mutant proteins mislocalize in photoreceptor sensory cilia.
Neonatal rat retinas were transfected with expression plasmids encoding V5-tagged wild type (WT) and mutant TTC21B cDNAs and an IRES-EGFP cassette using in vivo electroporation. Four weeks post-transfection, the transfected portion of the retinas (EGFP-positive) were stained with anti-V5 antibody (red). The images shown are 3D reconstructions of confocal image stacks; grids are included to show perspective, grid size is 8.2mm in all images. Wild-type TTC21B localizes to the transition zones of photoreceptor cilia in transfected cells. The R411G mutant localizes predominantly to the transition zone of transfected cells, but more diffusely than wild-type protein. In contrast, P209L and T1103R mutant proteins are present both in transition zones and inner segments, as well as cell bodies of the transfected cells. I1208S protein is undetectable in transfected (GFP positive) photoreceptor cells. The choroid (Ch) is visible in some images due to detection of mouse immunoglobulin in choroidal blood vessels by the anti-mouse secondary antibody used. White arrows indicate examples; IS, inner segment; ONL, outer nuclear layer.
Mutations in TTC21B are sufficient to cause JATD and isolated NPHP
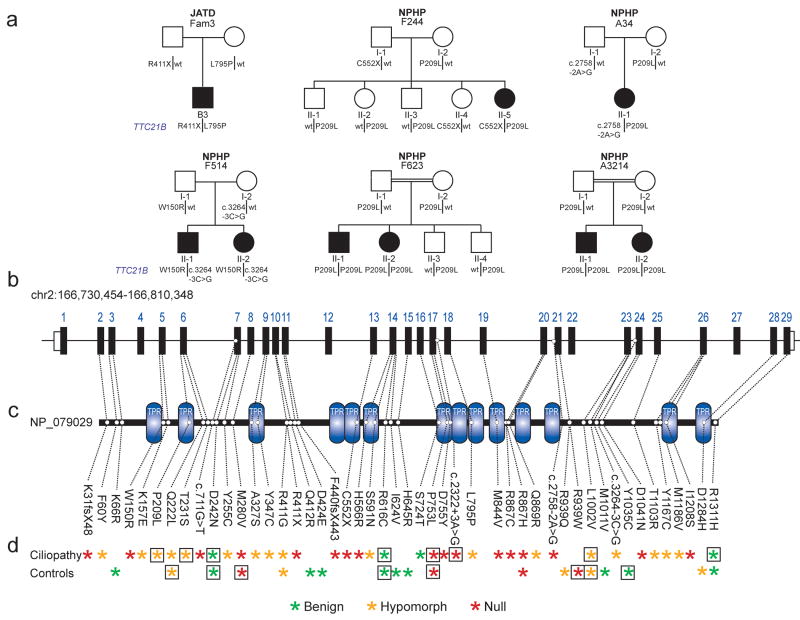
With functional data to inform the pathogenicity of each TTC21B allele detected in our cohorts, we first asked whether variants at that locus were sufficient to explain the clinical phenotypes in families with ciliopathies. In six pedigrees, we detected two pathogenic mutations that segregated under an autosomal recessive model (Fig. 4a). Representing the severe ciliopathies, JATD family 3 carried a heterozygous nonsense mutation and heterozygous nonsynonymous mutation encoding a highly conserved Leu795Pro change shown be hypomorphic by in vivo testing (Fig. 4a, Supplementary Table 2, Supplementary Table 8), which was neither seen in the 796 control chromosomes sequenced for the whole of TTC21B nor found in an additional 440 ethnically matched control chromosomes genotyped specifically for this allele (Table 1). Additionally, three families with early onset NPHP with extra-renal phenotypes were each compound heterozygotes with a pathogenic missense variant and either a splice junction variant or a truncating allele: Families F244 and A34 harbored a hypomorphic P209L encoding allele (as determined by all six lines of functional evidence; Supplementary Table 2) combined with either a nonsense mutation, or a splice junction variant predicted to result in deletion of a partial TPR domain respectively, with each variant absent from matched controls. Additionally, family F51426 carried a heterozygous W150R change shown to be functionally null in the in vivo zebrafish rescue assay, and a splice variant predicted to result in a 54-amino acid deletion (Fig. 4a, Table 1; Supplementary Table 2). Causative TTC21B lesions were also implicated in clinically milder ciliopathies; two consanguineous kindreds (F623 and A3214) with isolated NPHP each harbored the same P209L homozygous mutation (Fig. 4, Table 1). Of note, the 209L-encoding variant is a likely founder allele since we observed the same haplotype at coding regions spanning the locus in all P209L homozygotes (Supplementary Table 9).
Figure 4. Summary of all TTC21B variants detected.
a. Pedigrees of six ciliopathy families in which two TTC21B mutations are sufficient to manifest disease. JATD, Jeune Asphyxiating Thoracic Dystrophy; NPHP, Nephronophthisis. Filled circles or squares indicate individuals clinically diagnosed with a ciliopathy; unfilled indicates phenotypically normal.
b. Schematic of the human TTC21B genomic locus on chromosome 2; black boxes represent the 29 exons.
c. Human TTC21B protein is depicted as a black line with tetratricopeptide (TPR) domains shown in blue.
d. All novel variants detected by medical resequencing of TTC21B in our cohort of 753 ciliopathy patients and 398 controls are shown with respect to their genomic and protein locations (dashed lines from panels b. and c.). Overall pathogenicity of each variant as determined by in vivo functional assay (see Supplementary Table 2) is indicated with asterisks; green, benign; orange, hypomorphic; red, null. Boxes around asterisks indicate that a variant was detected in more than one individual. With the exception of P209L, all alleles were detected in heterozygosity.
Table 1. Pathogenic TTC21B variants detected in ciliopathy patients.
| Clinical Diagnosis | Patient ID | Ethnicity | TTC21B Allele 1 | TTC21B Allele 2 | Control Allele Frequencies | Other Ciliopathy Alleles | |||
|---|---|---|---|---|---|---|---|---|---|
| N. European | Other | ||||||||
| JATD | B3 | N. European | R411X | L795P | 0/440 | 0/880 | 0/356 | 0/356 | |
| JATD | A2343M | N. European | T231S | 0/440 | 0/356 | ||||
| JATD | PB36 | Pakistani | D755Y | 0/440 | 0/356 | ||||
| MKS-like | MKS-542 | N. European | F60Y | 0/440 | 0/356 | BBS7: Y671C het | |||
| MKS | MKS-015 | Tunisian | T231S | 0/440 | 0/356 | TMEM216: L114R hom | |||
| MKS-like | MKS-060143 | N. European | M280V/Q222L | 0/440 | 4/356 | RPGRIP1L: A1183G het | |||
| MKS | MKS-410 | Turkish | A327S | 0/440 | 0/356 | CC2D2A: F1195SfsX11 hom | |||
| MKS | B36 | Indian (Gujarati) | Y347C/D1041N | 0/440 | 0/356 | ||||
| MKS-like | MKS-95 | Pakistani | P753L | 0/440 | 5/356 | BBS1:G559D het; CEP290: R2120S het | |||
| MKS | 39 | Pakistani (Mirpuri) | M844V | 0/440 | 0/356 | ||||
| MKS | MKS-10bis | Guadeloupe Martinique | R867H | 0/440 | 1/356 | CC2D2A: R1049X hom | |||
| MKS-like | MKS-742 | N. European | Q869R | 0/440 | 0/356 | ||||
| MKS-like | MKS-640 | N. European | L1002V | 2/440 | 1/356 | ||||
| JBTS | MTI-140 | N. European | S591N | 0/440 | 0/356 | ||||
| JBTS | MTI-556 | Turkish | R867C | 0/440 | 0/356 | ||||
| JBTS | MTI-163 | Irish/Japanese | M1186V | 0/440 | 0/356 | ||||
| BBS | AR775-03 | N. European | F440fsX443/L1002V | 0/440 | 0/356 | MKS1: D286G het; RPGRIP1L: A229T het | |||
| BBS | 12620/V.15 | N. European | c.2322+3A>G | 0/440 | 0/356 | BBS1: M390R hom | |||
| BBS | 13881/VI.24 | N. European | c.2322+3A>G | 0/440 | 0/356 | BBS2:L209fsX229 hom | |||
| BBS | 13155/III.17/BBA | N. European | K157E | 0/440 | 0/356 | BBS12: F90fsX112 het; BBS12:T501M het | |||
| BBS | 17142 IX.5 | France | P209L | 0/440 | 0/356 | BBS4: Q218fsX228 het; BBS4: c.151+1G>T het | |||
| BBS | AR873-03 | Iraqi | T231S | 0/440 | 0/356 | ||||
| BBS | 9524/I.14 | N. European | T231S | 0/440 | 0/356 | BBS12:P159L het; BBS12:I346T het | |||
| BBS | 18136/X.5-BBJ21 | Portuguese | T231S | 0/440 | 0/356 | ||||
| BBS | 18136/X.5-BBJ22 | Portuguese | T231S | 0/440 | 0/356 | ||||
| BBS | 10172/I.25 | N. European | c.711G>T/Y255C | 0/440 | 0/356 | BBS10: C91fsX95 het; BBS10: R34P het | |||
| BBS | 16584/VIII.16 | Algerian | R411G | 0/440 | 1/356 | BBS7:S270fsX298 hom | |||
| BBS | 17733/IX.29 | N. European | L1002V | 2/440 | 1/356 | ||||
| BBS | AR8-03 | N. European | L1002V | 2/440 | 1/356 | ||||
| BBS | 10216/I.27 | N. European | T1103R | 0/440 | 0/356 | BBS6: D143fsX158 het; BBS6:S479X het | |||
| BBS | 18136/X.5-BBJ27 | Portuguese | I1208S | 0/440 | 0/356 | BBS1: R146X het; BBS1: M390R het | |||
| NPHP | F623-II1 | Portuguese | P209L | P209L | 0/440 | 0/356 | |||
| NPHP | A3214-II1 | Egyptian | P209L | P209L | 0/440 | 0/356 | |||
| NPHP+ | A34-II1 | Portuguese | P209L | c.2758-2 A>G | 0/440 | 0/440 | 0/356 | 0/356 | |
| NPHP+ | F244-II1 | Turkish | C552X | P209L | 0/440 | 0/440 | 0/356 | 0/356 | NPHP4: P1160L het; NPHP4: R735W het |
| NPHP+ | F514 | N. European | W150R | c.3264-3C>G | 0/440 | 0/440 | 0/356 | 0/356 | |
| NPHP | F1369-II1 | N. European | K31fsX48 | 0/440 | 0/356 | ||||
| NPHP | F925-II1 | N. European | c.2322+3A>G | 0/440 | 0/356 | ||||
| NPHP | A3123-II1 | N. European | T231S | 0/440 | 0/356 | ||||
| NPHP | A749-II1 | Turkish | T231S | 0/440 | 0/356 | ||||
| NPHP | F1385-II1 | N. European | H566R | 0/440 | 0/356 | ||||
| NPHP+ | A1152-II1 | India | P753L | 0/440 | 5/356 | ||||
| NPHP+ | A1502-II1 | N. European | L1002V | 2/440 | 1/356 | ||||
| NPHP | A1285-II1 | Turkish | Y1167C | 0/440 | 0/356 | ||||
Functional studies unmask a significant enrichment of TTC21B changes in ciliopathies
Because an additional 38 unrelated patients (5%) harbored heterozygous TTC21B changes that were detrimental to protein function (Table 1), we wondered if there was a specific enrichment for such TTC21B lesions in ciliary disease. First, we returned to our medical resequencing data and, to exclude bias due to differences in ethnicity or consanguinity, we analyzed only the cases and controls of N. European descent. Among cases and controls (n=1110 and 610 (440 plus an additional 85 individuals from HapMap) chromosomes respectively; Supplementary Table 1), we noted no significant difference in the frequency of novel nucleotide changes per chromosome analyzed in the coding regions of TTC21B (cases = 3.4% (38/1110), controls = 2.0% (12/610), p= 0.06; Supplementary Table 2). However, upon consideration of pathogenic changes only, we observed a significant enrichment of pathogenic variants in N. European cases versus controls (cases = 2.5% (28/1110), controls = 0.06% (4/610), p=0.003; Supplementary Table 2); this result was reproduced upon permutation analysis, in which genotypes were assigned randomly without replacement in the two cohorts, as reported previously15.
Genetic interaction of TTC21B with other ciliopathy loci
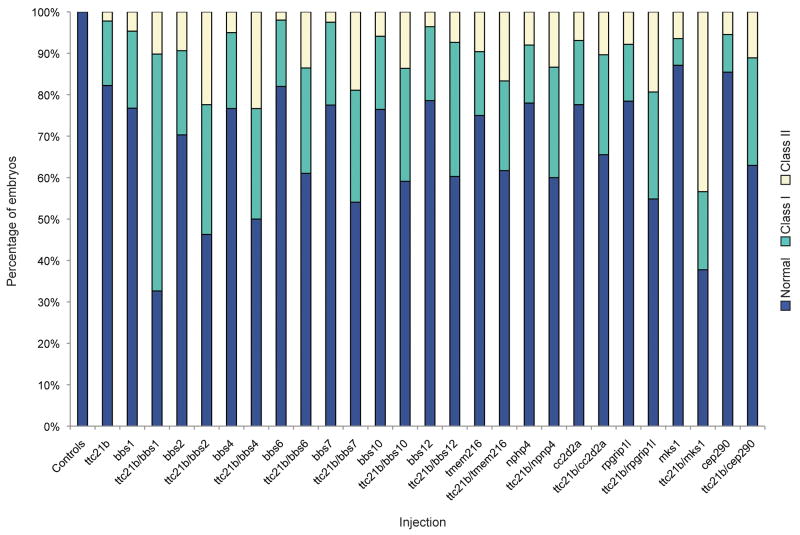
Taken together, we found likely pathogenic alleles in ∼5% of ciliopathy patients, suggesting that TTC21B might be a common contributor to the total mutational load in ciliopathies, not least because over one third of patients with heterozygous TTC21B alleles harbor causal trans alleles in 13 known causal ciliopathy genes (Table 1). As a first test of this concept, we asked whether injection of a subeffective dose of ttc21b tb-MO would alter the severity/penetrance of mid-somitic phenotypes caused by the suppression of each of the 13 endogenous ciliopathy transcripts. Consistent with our genetic predictions, gastrulation phenotypes were markedly exacerbated in the presence of a subeffective dose of ttc21b tb-MO compared to single injection of either MO alone for 13/13 double injections (Fig. 5). Importantly, the distinct effects on phenotypic severity observed across co-injections likely reflect the specificity of each ttc21b -trans interaction: some pairwise combinations gave a reproducible (n=3 injections; 50-75 embryos/injection, scored blind) significant increase in the overall percentage of affected embryos (e.g. bbs1/ttc21b), whereas other combinations increased the percentage of severely affected embryos (e.g. bbs4/ttc21b; Fig. 5).
Figure 5. In vivo modeling of TTC21B genetic interaction with other ciliopathy loci.
ttc21b interacts genetically with all loci known to contribute to disease in our ciliopathy cohort including bbs1, bbs2, bbs4, bbs6, bbs7, bbs10, bbs12, tmem216, nphp4, cc2d2a, rpgrip1l, mks1, and cep290 (see Table 1). Co-injection of a subeffective dose of ttc21b morpholino with any other single ciliopathy morpholino markedly exacerbates either the overall penetrance of gastrulation phenotypes (in comparison to the same doses injected alone) or a specific severity class.
Discussion
Genetic lesions affecting the function of ciliary and basal body proteins are beginning to offer the opportunity to model an important genetic paradigm. Cumulative phenotypic and genotypic data from the ∼30 known causal ciliopathy loci in clinically diverse cohorts have indicated the presence of numerous pathogenic mutations that can both drive recessive phenotypes and contribute likely second-site modifiers. This model offers the opportunity to improve the predictive power of genotypic information since a functionally-derived annotation of the total mutational load of the ciliopathy module might improve genotype-phenotype correlations.
The identification of causal and contributory/epistatic mutations in TTC21B supports this model. Importantly, this is the first locus for which systematic ciliopathy resequencing was undertaken in the absence of prior genetic data linking this locus to a human disorder. The rarity of the discovered alleles, almost all of which are private mutations, coupled to the phenotypic variability of our patient cohorts precludes, at present, the association of specific alleles with discrete ciliopathy endophenotypes. Comparison of mouse and human data provide some limited insights, in that homozygous null TTC21B is embryonic lethal and unlikely to be found in patients, yet a hypomorphic allele coupled to a null mutation appears sufficient to cause JATD. However, a similar genotype (c.2758-2A>G/P209L or C552X/P209L) leads to early-onset NPHP, while P209L homozygosity leads to apparently isolated NPHP. Interestingly, the same P209L allele was also found in the heterozygous state in a BBS patient with cystic renal disease (prevalence in BBS <24%)27, leading us to speculate that the presence of this mutation might contribute to the development of renal phenotypes. It is also interesting to note that, for five of the alleles tested in the zebrafish gastrulation system, injection of mutant mRNA exacerbated the MO phenotype. Although this might suggest that these alleles are dominant negatives, injection of increased amounts of mutant mRNA alone did not induce any phenotypes in wild-type embryos (data not shown). This raises the possibility that these alleles might exert a toxic effect only when the levels of wt protein fall below a critical threshold; a better appreciation of the biochemical properties of wt and mutant TTC21B will be required to understand this mechanism.
Given recent advances in sequencing technology, these data not only encourage efforts to conduct saturated resequencing of the ciliary proteome28, but to also implement a similar paradigm to other definable functional modules of the morbid human genome. However, we note that, bereft of functional annotation, sequencing alone might not always be sufficient to provide appropriate resolution. As such, efforts to develop functional assessment of the biochemical and/or cellular output of discrete modules should be integral parts of patient exome and genome resequencing.
Methods
Ciliopathy cases, controls, and mutational screening
Ciliopathy DNAs were ascertained from ten different collection points in North America and Europe, and control DNAs were obtained from healthy individuals of N. European or S.E. Asian descent with no clinical ciliopathy criteria (retinal degeneration, polydactyly or other skeletal malformations, renal dysfunction, mental retardation or other central nervous system defects) from two collection points (Supplementary Table 1) following informed consent. We amplified the exon sequence and intron/exon junctions of TTC21B from DNA extracted from lymphoblast cell lines or lymphocytes according to standard protocols. PCR products were sequenced with BigDye Terminator v3.1 chemistry on an ABI 3100 or ABI 3730 (Applied Biosystems), sequences were aligned with PolyPhred, finished with Consed, and variants were confirmed by resequencing and visual assessment of chromatograms. Primer sequences and PCR conditions are available upon request.
DNA constructs
We amplified the human TTC21B ORF from the pENTR-Myc-hTHM1 vector (a gift from David Beier, Harvard Medical School), cloned it into a Gateway entry vector to eliminate the Myc tag, and confirmed the ORF by direct sequencing. The insert was then moved into the pCS2+ (zebrafish studies) or pCAG-V5-cDNA-IRES-EGFP-pA (in vitro mIMCD3 rescue and localization, and in vivo photoreceptor localization studies) Gateway destination expression vectors using LR clonase II (Invitrogen) mediated recombination. Missense mutations were introduced into the TTC21B ORF using the QuikChange site directed mutagenesis kit (Stratagene) according to manufacturer's instructions, and confirmed by direct sequencing. shRNAs targeting Ttc21b were cloned into the pCAG-mir30-puro vector; shRNA-Ttc21b (2923) is specific for rodent Ttc21b and provides 70-80% knockdown of Ttc21b mRNA levels following transfection into mIMCD3 cells (data not shown).
Zebrafish embryo manipulation and morpholino injection
Translation-blocking and splice-blocking MOs (Gene Tools) were diluted to appropriate concentrations in deionized, sterile water (2, 4, 6, 8, and 10ng/nl for the dose response, 10ng/nl for rescue experiments, and 4ng/nl for genetic interaction studies) and injected into wt zebrafish embryos at the two-cell stage as described29. To rescue morphant phenotypes, we transcribed mRNA from linearized pCS2+-TTC21B vector with the SP6 mMessage mMachine kit (Ambion). Morpholinos targeting other ciliopathy genes have been characterized previously (bbs123, bbs223, bbs423, bbs623, bbs723, bbs1023, bbs1223, tmem21613, rpgrip1l,15mks114, and cep29014. We carried out in situ hybridization on whole embryos fixed with 4% paraformaldehyde with riboprobes against pax2, krox20, and myoD using standard protocols. Images were captured at 8× magnification and measurements were taken of the width spanning the 5th somites from the anterior end, and the length of the notochord as defined by myoD staining of adaxial cells. Endogenous ttc21b expression was determined by extracting total RNA from 0.5dpf embryos with Trizol (Invitrogen) according to manufacturer's instructions. Oligo-dT-primed total RNA was reverse transcribed using SuperScriptIII reverse transcriptase (Invitrogen) and the resulting cDNA was PCR amplified.
TTC21B Antibodies
Anti-TTC21B antibodies were generated against two synthetic peptides in rabbits. The peptide sequences used were: Ms #77∼90: CLMYVHKMSPNPDRE-amide and #1290∼1304: CQVLEAHPTYPKIRKD-amide. The resulting antisera were affinity purified using the peptide antigen (YenZym Antibodies, South San Francisco, CA).
Cell Culture and Transfection
The mIMCD3-EV and mIMCD3-Ttc21b shRNA stable cell lines (a gift from Dr. David Beier, Harvard Medical School; previously referred to as EV4 and R1-4 respectively18), mIMCD-SSTR3-EGFP stable cell line (a gift from Dr. Gregory J. Pazour, University of Massachusetts Medical School), and mIMCD3 cells were maintained in DMEM:F12 media supplemented with 10% FBS, and 0.5 mM sodium pyruvate (Invitrogen). Cells were plated in 6-well plates (on glass cover slips for immunofluorescent procedures) and transfected at 70-80% confluency using Lipofectamine 2000 (Invitrogen) with 4 μg DNA/well. Cells were serum starved in 0.5% FBS at 24h post-transfection until harvest and were fixed 48 h after transfection for immunocytochemistry or harvested 72 h after transfection for Western blot. For cilia measurement assays, cells were cotransfected with phrGFP-IIN (Stratagene) as a transfection control.
Immunofluorescence Procedures
Cells were fixed in 100% methanol (cilia length rescue assays) or 4% paraformaldehyde for 10 min (localization studies), permeabilized with 0.5% Triton X-100 in PBS for 10 min and blocked with 1% bovine serum albumin (BSA) and 0.2% Triton X-100 in PBS for 10 min. The cells were then incubated with a cocktail of anti-acetylated α-tubulin and anti-γ-tubulin antibodies (Ab, both 1:1000, Sigma; cilia length rescue assays) or a monoclonal anti-V5 Ab (1:2,500, Invitrogen; localization studies) in blocking buffer for 1 h. After washing three times, 10 min each in PBS, a secondary Ab incubation, Alexa Fluor 555–conjugated goat anti-mouse IgG (1:1000, Invitrogen) was carried out for 1 h. Cells were subsequently washed once in PBS for 10 min, incubated with Hoechst dye (1:1000, Invitrogen) for 5 min and washed twice (10 min each). For colocalization, mIMCD3 cells were incubated with a polyclonal rabbit Ab against γ-tubulin (1:4000, Sigma) for 1 h and with a secondary Ab, Alexa Fluor 488–conjugated goat anti-rabbit IgG (1:1000, Invitrogen) for 1 h. Cover slips were mounted using Fluoromount (Electro Microscopy Sciences, Hatfield, PA). Fluorescent signals were visualized using either a Nikon 90i or Nikon TE300 fluorescent microscope.
Cilia length measurements
For each transfection condition, 18 different fields were randomly chosen from the cover slip for imaging at 60× magnification. All cilia on transfected cells, as evidenced by cytoplasmic GFP, were measured using Nikon NIS Elements software. Pairwise comparisons were conducted using a student's t-test.
Western blot
Cells were lysed with 2× LDS sample buffer (Invitrogen) and total proteins (100 μg/well) were run on a precast NuPAGE 4-12% Bis-Tris Gel (Invitrogen) and transferred to a PVDF membrane. The membrane was incubated with a blocking buffer (LI-COR) for 1 h at room temperature, and subsequently with a monoclonal V5 antibody (Invitrogen; 1:5,000) with gentle rocking overnight at 4°C. The membrane was washed 4 times with PBS/0.1% Tween-20 and then incubated with a secondary antibody (IRDye goat anti-mouse IgG, 1:10,000) for 1 h at room temperature. After washing with PBS/0.1% Tween-20 and PBS only, the signals were detected with an infrared imager (Odyssey, LI-COR). The loading control antibody to Na+/K+-ATPase was used at 1:5,000. The protein levels (fluorescent units) were quantified with software provided by the LI-COR.
In vivo electroporation of rat retinas
We used the in vivo electroporation technique to either knockdown endogenous Ttc21b or express human TTC21B proteins in rat photoreceptor cells. Injected plasmids were electroporated into retinal cells using tweezer-type electrodes in the right eyes of neonatal rats (n=4-6 rats/injection) as described30,31. For suppression experiments, 0.5μl of endotoxin-free shRNA-Ttc21b (with or without human TTC21B cDNA) were injected sub-retinally and transfections with non-targeted shRNA were performed in parallel. A plasmid encoding arrestin-EGFP was included in each transfection to allow for evaluation of PSC structure. Four weeks following transfection, the pupils of the rats were dilated with 1% tropicamide, and the rats exposed to 15K lux light for 30 minutes to ensure that the arrestin-EGFP was located in the outer segments. 50μm thick sections of the transfected portions of retinas were then prepared and evaluated using confocal microscopy. Three-dimensional reconstructions of the confocal image stacks were generated and analyzed using Volocity 3D imaging software (Improvision, Waltham, MA). In addition to evaluating the qualitative appearance of PSCs in these studies, we also measured photoreceptor outer segment length using the Quantitation module of the Volocity 3D imaging software. The lengths of at least 10 outer segments were measured in sections from at least 3 separate animals. For expression studies, pCAG-V5-cDNA-IRES-EGFP plasmids containing the wt or mutant ORF were electroporated into neonatal rat retinas. Animals were sacrificed 4 weeks following transfection, and frozen sections prepared from the portions of the eyecups with EGFP signal. Sections were stained with anti-V5 antibodies (Invitrogen), followed by Alexa-555 conjugated secondary antibodies (Invitrogen). The location of V5-tagged proteins in photoreceptor cells was evaluated by fluorescence microscopy using Volocity 3D imaging software.
Supplementary Material
Acknowledgments
We are grateful to the ciliopathy patients and their families for their continued participation and enthusiasm. We thank Nadia Elkhartoufi for technical assistance, David Parker for critical reading of the manuscript, and Yorkshire Regional Genetics Service for help in obtaining control DNA samples. This research was supported by grants from National Institutes of Health grant R01HD04260 from the National Institute of Child Health and Development (N.K.), R01DK072301, R01DK075972 (N.K.), R01DK068306, R01DK064614, R01DK069274 (F.H.) and NRSA fellowship F32 DK079541 (E.E.D.) from the National Institute of Diabetes, Digestive and Kidney disorders, RO1EY12910 from the National Eye Institute (E.A.P.), the Macular Vision Research Foundation (N.K.) the Foundation-Fighting Blindness (N.K., E.A.P., Q.L.), the F.M. Kirby Foundation (E.A.P.), the Rosanne Silbermann Foundation (E.A.P.), the PKD Foundation (C.B.), German Kidney Foundation (C.B.), German research foundation (DFG BE 3910/5-1 and SFB/TRR57; C.B.) and an MRC research training fellowship (J.H.). This work was also supported in part by the Intramural Research Program of the National Human Genome Research Institute. F.H. is an Investigator of the Howard Hughes Medical Institute, a Doris Duke Distinguished Clinical Scientist, and the Frederick G. L. Huetwell Professor. NK is a Distinguished George W. Brumley Professor.
Footnotes
Accession codes. GenBank: Homo sapiens TTC21B mRNA, NM_024753; Homo sapiens TTC21B amino acid, NP_079029; Danio rerio ttc21b mRNA, NM_001128258.
Author Contributions: Experiments were designed by E.E.D., E.A.P., N.K. Mutational screening, analysis, and confirmation was conducted by E.E.D., J.H., C.S., K.S., G.R., C.V.L., D.M.M., A.C.Y., D.A.W., P.C., M.M., L.R.L., P.C., B.M., N.F.H., J.C.M., R.W.B., G.G.B., NISC Comparative Sequencing Program, G.G., E.A.O., J.G.G., T.A.B., C.A.J., E.D.G., R.A.G. Ciliopathy patient samples were provided by J.H., S.R., B.T., I.K., N.A.S., T.J.N., K.J.S., H.K., T.E.G., R.A.L., C.B., S.S., P.J.S., P.L.B., J.G.G., E.R.M., T.A.B., H.D., C.A.J., F.H., N.K. In vivo and in vitro functional studies were carried out by E.E.D., Q.Z., Q.L., B.H.D., L.M.D. The manuscript was written by E.E.D., Q.Z., E.A.P., N.K. with helpful comments from C.B., J.G.G., E.R.M., T.A.B., C.A.J., F.H.
References
- 1.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–48. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 2.Zaghloul NA, Katsanis N. Functional modules, mutational load and human genetic disease. Trends Genet. 2010;26:168–76. doi: 10.1016/j.tig.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoefele J, et al. Evidence of oligogenic inheritance in nephronophthisis. J Am Soc Nephrol. 2007;18:2789–95. doi: 10.1681/ASN.2007020243. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann C, et al. Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal-hepatic-pancreatic dysplasia. Am J Hum Genet. 2008;82:959–70. doi: 10.1016/j.ajhg.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otto EA, et al. Hypomorphic Mutations in Meckelin (MKS3/TMEM67) cause Nephronophthisis with Liver Fibrosis (NPHP11) J Med Genet. 2009 doi: 10.1136/jmg.2009.066613. [DOI] [PubMed] [Google Scholar]
- 6.Baala L, et al. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am J Hum Genet. 2007;81:170–9. doi: 10.1086/519494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baala L, et al. The Meckel-Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. Am J Hum Genet. 2007;80:186–94. doi: 10.1086/510499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delous M, et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat Genet. 2007;39:875–81. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- 9.Gorden NT, et al. CC2D2A is mutated in Joubert syndrome and interacts with the ciliopathy-associated basal body protein CEP290. Am J Hum Genet. 2008;83:559–71. doi: 10.1016/j.ajhg.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sayer JA, et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38:674–81. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 11.Valente EM, et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat Genet. 2006;38:623–5. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 12.Wolf MT, et al. Mutational analysis of the RPGRIP1L gene in patients with Joubert syndrome and nephronophthisis. Kidney Int. 2007;72:1520–6. doi: 10.1038/sj.ki.5002630. [DOI] [PubMed] [Google Scholar]
- 13.Valente EM, et al. Mutations in TMEM216 perturb ciliogenesis and cause Joubert, Meckel and related syndromes. Nat Genet. 2010;42:619–25. doi: 10.1038/ng.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leitch CC, et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet. 2008;40:443–8. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- 15.Khanna H, et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet. 2009;41:739–745. doi: 10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louie CM, et al. AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat Genet. 2010;42:175–80. doi: 10.1038/ng.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piperno G, et al. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J Cell Biol. 1998;143:1591–601. doi: 10.1083/jcb.143.6.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran PV, et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet. 2008;40:403–10. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li JB, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–52. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 20.Liu Q, et al. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mykytyn K, et al. Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet. 2001;28:188–91. doi: 10.1038/88925. [DOI] [PubMed] [Google Scholar]
- 22.Ansley SJ, et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–33. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 23.Zaghloul NA, et al. Functional analyses of variants reveal a significant role for dominant negative and common alleles in oligogenic Bardet-Biedl syndrome. Proc Natl Acad Sci U S A. 2010;107:10602–7. doi: 10.1073/pnas.1000219107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herron BJ, et al. Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat Genet. 2002;30:185–9. doi: 10.1038/ng812. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q, Zuo J, Pierce EA. The retinitis pigmentosa 1 protein is a photoreceptor microtubule-associated protein. J Neurosci. 2004;24:6427–36. doi: 10.1523/JNEUROSCI.1335-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuhaus TJ, Stallmach T, Leumann E, Altorfer J, Braegger CP. Familial progressive tubulo-interstitial nephropathy and cholestatic liver disease -- a newly recognized entity? Eur J Pediatr. 1997;156:723–6. doi: 10.1007/s004310050699. [DOI] [PubMed] [Google Scholar]
- 27.Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA. New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet. 1999;36:437–46. [PMC free article] [PubMed] [Google Scholar]
- 28.Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–2. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- 29.Badano JL, et al. Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature. 2006;439:326–30. doi: 10.1038/nature04370. [DOI] [PubMed] [Google Scholar]
- 30.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A. 2007;104:1027–32. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.