Abstract
Background
The World Wide Web plays a critical role in enabling molecular, cell, systems and computational biologists to exchange, search, visualize, integrate, and analyze experimental data. Such efforts can be further enhanced through the development of semantic web concepts. The semantic web idea is to enable machines to understand data through the development of protocol free data exchange formats such as Resource Description Framework (RDF) and the Web Ontology Language (OWL). These standards provide formal descriptors of objects, object properties and their relationships within a specific knowledge domain. However, the overhead of converting datasets typically stored in data tables such as Excel, text or PDF into RDF or OWL formats is not trivial for non-specialists and as such produces a barrier to seamless data exchange between researchers, databases and analysis tools. This problem is particularly of importance in the field of network systems biology where biochemical interactions between genes and their protein products are abstracted to networks.
Results
For the purpose of converting biochemical interactions into the BioPAX format, which is the leading standard developed by the computational systems biology community, we developed an open-source command line tool that takes as input tabular data describing different types of molecular biochemical interactions. The tool converts such interactions into the BioPAX level 3 OWL format. We used the tool to convert several existing and new mammalian networks of protein interactions, signalling pathways, and transcriptional regulatory networks into BioPAX. Some of these networks were deposited into PathwayCommons, a repository for consolidating and organizing biochemical networks.
Conclusions
The software tool Sig2BioPAX is a resource that enables experimental and computational systems biologists to contribute their identified networks and pathways of molecular interactions for integration and reuse with the rest of the research community.
Background
BioPAX is a protocol for the specification and representation of cell signaling pathways, gene-regulatory networks, protein-protein interactions and other types of biomolecular interaction data [1]. There are several software tools that use the BioPAX format for pathway visualization and analysis for hypotheses generation. For example, the popular tool Cytoscape allows customizable visualization and easy navigation of different types of networks [2]. Cytoscape plug-ins, including the popular BiNGO [3], and other plugins such as BiNoM [4], and cPath [5] further extend Cytoscape's capabilities for pathway analysis, data visualization, and data integration. BiNGO is a plugin that statistically analyzes a set of genes and their corresponding Gene Ontology functional annotations to determine which functional categories are overrepresented in that gene set. BiNGO uses Cytoscape's visualization capabilities to display the results. BiNoM is a plugin that performs structural analysis of networks, identifying strongly connected components, paths and cycles. cPath is an interaction database that can be included in Cytoscape as a plugin. The cPath database is a central repository for pathway and interaction datasets from multiple sources including MINT [6], IntAct [7], Reactome [8], and BioGRID[9]. The plugin allows for data retrieval from the central cPath database via an XML Web Services API, using the Cytoscape visualization engine for viewing biochemical networks. Interaction data stored in cPath are in BioPAX format.
BioPAX is one of several specification protocols that have been developed in an attempt to formally characterize biochemical regulatory molecular interactions. Some of these other specifications include the Proteomics Standard Initiative Molecular Interactions format (PSI-MI) [10] and the Systems Biology Markup Language (SBML) [11]. There are tools for conversion of some of these data formats into BioPAX. The previously mentioned Cytoscape plugin BiNoM also allows for conversion between BioPAX, SBML, and CellDesigner formats. However, most biochemical interaction data is not stored in one of these formats already, but rather stored in flat files, Excel spreadsheets, as network diagrams, or as tables in PDF format. While there are commercial products available to help researchers transform their flat text files into a general OWL format, to date, there are no tools available to transform flat files into BioPAX format. Such a tool would be useful because there are many pathway databases and networks that need to be converted for data sharing and reuse. Additionally, biologists that identify new interactions or describe new pathways in publications and do not have the technical expertise to convert their interaction data into BioPAX format will be able to do so with the tool.
Implementation
We created a Java-based tool, Sig2BioPAX, for the purpose of converting cell signaling pathways and interaction network datasets into BioPAX. The software package uses the Jena Library[12], which includes data structures for representation and modification of RDF and OWL files. In the implementation, BioPAX classes are represented as Java classes. The classes contain variables which store the settings and details necessary for proper instantiation of those classes. Program control is passed through a tree of functions which adds new instances of classes as necessary to represent the data from the input file. Reading the input flat text file line-by-line, Sig2BioPAX adds nodes to the OWL model sequentially. Before adding a new node, the program traverses the model to make sure that no identical node (that is, a node having the same children) already exists. Finally, the newly created model is written to an OWL file using the Jena library functions. See Figure 1 for a flow-diagram of the internals of the Sig2BioPAX program.
Figure 1.
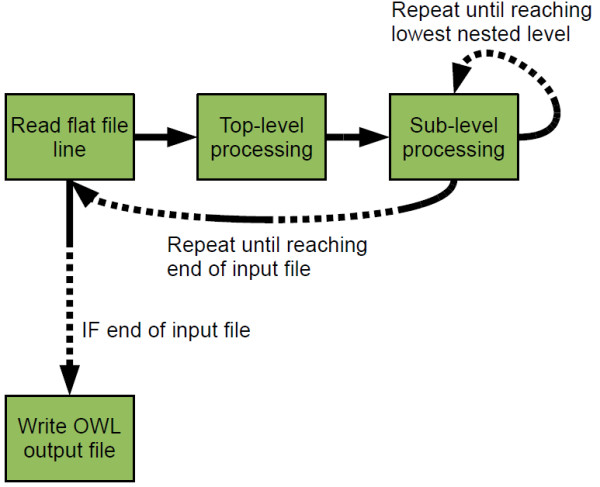
A flow diagram of Sig2BioPAX. The input file is processed by the Top-level processing unit and then by Sub-level units until reaching the lowest level of processing iteratively. Once all lines from the input file were processed the program outputs an OWL file.
There are a finite number of reaction types that can be processed and converted into the BioPAX format. These reactions are described in Table 1. Sig2BioPAX mainly supports cell signaling reaction types including protein-protein binding interactions, protein kinase phosphorylations and phosphatase dephosphoryations, guanine nucleotide exchange reactions, and ubiquitinations. Since BioPAX level 3 has added support for transcriptional regulatory interactions, Sig2BioPAX can be used to convert protein/DNA interactions in the form of transcription-factors binding to regulatory regions of genes. Sig2BioPAX categorizes the input reaction lines based on user-supplied rules. These rules determine the logic used to convert the flat files to the BioPAX level 3 format. The rules must be specified in a rules file using a custom language which is fully described in the program's instruction manual and also available within the GUI version of the program by clicking the question mark button. A default rules.txt is included with the program's release. Users may modify the included rules.txt or create their own rules file from scratch.
Table 1.
A description of the reaction types processed by Sig2BioPAX
| Reaction Type | Description |
|---|---|
| Binding | Molecules bind to form a complex |
| Kinase Phosphorylation | Kinase catalyzes addition of phosphate group to target molecule |
| Dephosphorylation | Catalyst initiates removal of phosphate group from target molecule |
| Guanine Nucleotide Exchange | GDP removed from complex and replaced with GTP |
| GTPase activating protein | GTP bound to compound becomes GDP |
| Ubiquitination | Ubiquitin molecule is added to target compound |
| Deubiquitination | Ubiquitin molecule is removed from target compound |
| Sumoylation | Small ubiquitin-related modifier is attached to target molecule |
| Cleavage with Phospholipase C | PLC cleaves PIP2 into IP3 and DAG |
| Cleavage on Cysteine | Deactivating cleavage on a cysteine residue of target protein |
| Inactivating Cleavage | Deactivating cleavage of target molecule |
| Activating Cleavage | Cleavage of pro-protein into active form |
| Protein-protein Interaction | Otherwise unspecified reaction between two proteins |
| Transcription | Protein activates or inhibits transcription of gene products |
| Transcription Factor Promoter Binding | Transcription factor and protein bind to create complex |
One of Sig2BioPAX's accepted input parameter is the input file template type, which is a descriptor of the format of the input file. Four different input formats are accepted by the current version. The default type, sig, is described in Table 2. The second type, called source_target, forgoes describing the source and target types (i.e. kinase) and only describes the reaction type between the source and target (i.e. phosphorylation). The third template type is tf_target, used to describe transcription factor binding to the promoter of target genes as determined by ChIP-seq or ChIP-chip types of experiments [13]. The fourth template type is sif, or simple interaction format, which is a format specifying the target and source names and the interaction type. In the implementation, input templates are stored within the Line.java source file, which contains the function ReadLine(). This function is composed of IF statements, one for each input template type. In this code file, the testing condition for the IF statement are defined. The input line is parsed through a series of statements. By modifying the order and content of these statements, the user can customize the input templates.
Table 2.
A description of an input line using the default input template, sig, and the meaning of the individual elements on the input line.
| sig template: SN SH SM ST SL TN TH TM TT TL E TI ID | |
|---|---|
| SN | Source compound name |
| SH | Source Swiss-Prot human accession number |
| SM | Source Swiss-Prot mouse accession number |
| ST | Source Type of compound |
| SL | Source cellular location |
| TN | Target compound name |
| TH | Target Swiss-Prot human accession number |
| TM | Target Swiss-Prot mouse accession number |
| TT | Target Type of compound |
| TL | Target cellular location |
| E | Effect of source on target compound - Activating, inhibiting, or neutral |
| TI | Type of interaction |
| ID | Pubmed ID of reference article |
Each element must be separated by whitespace, and the line is terminated by the newline character.
Results
Sig2BioPAX is packaged as an executable JAR file. The tool is available in both command line and graphical user interface (GUI) versions. The command line version of the program, accessible by using the command line argument -cmd, accepts input parameters including the input file name, output file name, rules file name, option to overwrite output file, and an input-template type. In the command line version of the program, these options are passed as command line arguments. For example, to use input file foo.txt, output file bar.owl, the rules file rulesfile.txt, and the overwrite option, the command is: 'java -jar sig2biopaxv4.jar - cmd -in:foo.txt -out:bar.owl -r:rulesfile.txt -o'. The sig input template type is the default input template type. In the GUI version of the program, the input, output, and rules files can be selected from a file browser and the other options can be set interactively (Figure 2). Sig2BioPAX can be downloaded from the Google Code project hosting site at http://code.google.com/p/sig2biopax/
Figure 2.
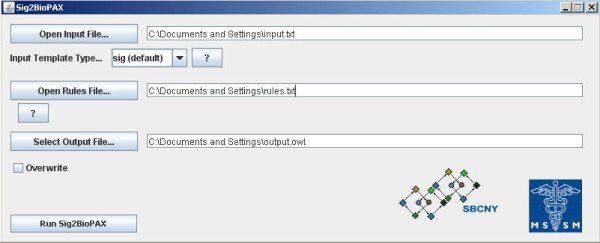
A screenshot of the GUI version of Sig2BioPAX. Users are provided with the ability to interactively specify the input file, the input file format, the rules file, and an output OWL file.
We used Sig2BioPAX to convert several network datasets into the BioPAX format, and some of these networks are available on Pathway Commons [14], an international collaborative database of biomolecular pathways. The datasets we converted to BioPAX are original networks we extracted from the literature for the projects: the presynaptome [15], representing protein-protein interactions present in presynaptic nerve terminals of mammalian neurons, and the neuronal signalome [16], representing cell signaling interactions extracted from neuroscience literature describing combined cell signaling pathways in mammalian neurons, the adhesome [17], a network of interaction in focal adhesions; and a kinase-substrate network we constructed for the program KEA [18], kinase enrichment analysis, and ChEA [19], which stand for chip-seq/chip enrichment analysis.
Conclusions
As the bulk and complexity of genome-wide molecular data increases, methods for sharing and exchanging data need to be further developed. Effective standard representation of data enables seamless data exchange across platforms, tools and databases. However, converting existing and new data into such exchange formats is not trivial. The Sig2BioPAX tool will further enable researchers to easily convert their flat file interaction data into the computable BioPAX format so that their data can be reused and interpreted by other researchers worldwide.
Availability and Requirements
Project name: Sig2BioPAX
Project home page: http://code.google.com/p/sig2biopax/
Operating system(s): Platform independent
Programming language: Java
Other requirements: Jena 2.6.2 or higher
License: GNU GPL
Any restrictions to use by non-academics: none
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AM initiated and managed the project. RLW implemented and tested the Sig2BioPAX Java tool. AM and RLW wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Ryan L Webb, Email: Ryan.Webb@mssm.edu.
Avi Ma'ayan, Email: avi.maayan@mssm.edu.
Acknowledgements
This research was supported by NIH Grants P50GM071558-01A27398, R01DK088541, R01GM054508, R01DA15446, and KL2RR029885-0109. We would like to thank Emek Demir from MSKCC for assistance with the BioPAX specification.
References
- Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D'Eustachio P, Schaefer C, Luciano J. et al. The BioPAX community standard for pathway data sharing. Nat Biotechnol. 2010;28(9):935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B. et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics. pp. 3448–3449. [DOI] [PubMed]
- Zinovyev A, Viara E, Calzone L, Barillot E. BiNoM: a Cytoscape plugin for manipulating and analyzing biological networks. Bioinformatics. 2008;24(6):876–877. doi: 10.1093/bioinformatics/btm553. [DOI] [PubMed] [Google Scholar]
- Cerami E, Bader G, Gross B, Sander C. cPath: open source software for collecting, storing, and querying biological pathways. BMC Bioinformatics. 2006;7(1):497. doi: 10.1186/1471-2105-7-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanzoni A, Montecchi-Palazzi L, Quondam M, Ausiello G, Helmer-Citterich M, Cesareni G. MINT: a Molecular INTeraction database. FEBS Letters. 2002;513(1):135–140. doi: 10.1016/S0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J. et al. The IntAct molecular interaction database in 2010. Nucleic Acids Research. 2010;38(suppl 1):D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastrik I, D'Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, de Bono B, Gillespie M, Jassal B, Lewis S. et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biology. 2007;8(3):R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2010. [DOI] [PMC free article] [PubMed]
- Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von Mering C. et al. The HUPO PSI's molecular interaction format--a community standard for the representation of protein interaction data. Nat Biotechnol. 2004;22(2):177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A. et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Jena – A Semantic Web Framework for Java. http://jena.sourceforge.net/
- Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2010. [DOI] [PMC free article] [PubMed]
- Abul-Husn NS, Bushlin I, Morón JA, Jenkins SL, Dolios G, Wang R, Iyengar R, Ma'ayan A, Devi LA. Systems approach to explore components and interactions in the presynapse. PROTEOMICS. 2009;9(12):3303–3315. doi: 10.1002/pmic.200800767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma'ayan A, Jenkins SL, Neves S, Hasseldine A, Grace E, Dubin-Thaler B, Eungdamrong NJ, Weng G, Ram PT, Rice JJ. et al. Formation of regulatory patterns during signal propagation in a Mammalian cellular network. Science. 2005;309(5737):1078–1083. doi: 10.1126/science.1108876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9(8):858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A, Ma'ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25(5):684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. pp. 2438–2444. [DOI] [PMC free article] [PubMed]


