Abstract
Background
Previously, we described the heat shock response in dipteran species belonging to the family Stratiomyidae that develop in thermally and chemically contrasting habitats including highly aggressive ones. Although all species studied exhibit high constitutive levels of Hsp70 accompanied by exceptionally high thermotolerance, we also detected characteristic interspecies differences in heat shock protein (Hsp) expression and survival after severe heat shock. Here, we analyzed genomic libraries from two Stratiomyidae species from thermally and chemically contrasting habitats and determined the structure and organization of their hsp70 clusters.
Results
Although the genomes of both species contain similar numbers of hsp70 genes, the spatial distribution of hsp70 copies differs characteristically. In a population of the eurytopic species Stratiomys singularior, which exists in thermally variable and chemically aggressive (hypersaline) conditions, the hsp70 copies form a tight cluster with approximately equal intergenic distances. In contrast, in a population of the stenotopic Oxycera pardalina that dwells in a stable cold spring, we did not find hsp70 copies in tandem orientation. In this species, the distance between individual hsp70 copies in the genome is very large, if they are linked at all. In O. pardalina we detected the hsp68 gene located next to a hsp70 copy in tandem orientation. Although the hsp70 coding sequences of S. singularior are highly homogenized via conversion, the structure and general arrangement of the hsp70 clusters are highly polymorphic, including gross aberrations, various deletions in intergenic regions, and insertion of incomplete Mariner transposons in close vicinity to the 3'-UTRs.
Conclusions
The hsp70 gene families in S. singularior and O. pardalina evolved quite differently from one another. We demonstrated clear evidence of homogenizing gene conversion in the S. singularior hsp70 genes, which form tight clusters in this species. In the case of the other species, O. pardalina, we found no clear trace of concerted evolution for the dispersed hsp70 genes. Furthermore, in the latter species we detected hsp70 pseudogenes, representing a hallmark of the birth-and-death process.
Keywords: Diptera, hsp70 gene cluster, thermal adaptation, concerted evolution, Stratiomyidae
Background
The heat shock response is a universal phenomenon, and heat shock proteins (Hsps) form the most ancient and sophisticated cellular defense system present in all living organisms [1,2]. The Hsps are broadly classified on the basis of their molecular weights and functions into distinct families, with the Hsp70 family being the best studied and among the most important for cell protection [3,4]. Though the heat shock response is a universal phenomenon, various organisms even within the taxonomic groups of relatively low rank (e.g., order, family) often evolve different strategies to adapt to aggressive environments [4-8].
The mechanisms providing whole body adaptation of animals to various adverse conditions, including extreme thermal environments, have long attracted the attention of ecologists. After the description of Hsps in Drosophila and other organisms [1,9], we undertook systematic studies to establish a possible correlation between the general pattern of Hsp synthesis and whole-body adaptation to extreme conditions [7,10,11]. It is noteworthy that the predominant fraction of investigations on the stress response have largely (but not entirely) been previously performed on a small number of model organisms in the laboratory and, therefore, have limited value in an ecological context. Keeping this in mind we usually included in our investigation related species or geographical strains from nature, in particular those inhabiting biotopes or regions with strikingly contrasting average temperatures [8,11-14]. Using this approach, we, as well as other research groups, have accumulated abundant data showing that animals have evolved a variety of strategies to adjust their heat shock systems to changing environmental conditions [4,7]. Long-term and large-scale studies of the Hsps gene system in species and geographical strains that differ in the temperature of habitats, performed in various laboratories, have disclosed a few basic patterns in the functioning and evolution of this system in organisms living under adverse conditions [4,5,11,15-17].
In the course of these studies, it was repeatedly demonstrated that noticeable cellular levels of Hsps and/or corresponding RNA under normal physiological temperatures represent one of the most common adaptations to stressful conditions described in various organisms, including certain species of lizards, flies and ants [11,15,18].
In particular, in order to investigate how organisms have evolved to occupy contrasting habitats, we compared four species of Stratiomyidae (Diptera) whose larvae develop in naturally varying (semi)aquatic habitats, including highly aggressive conditions of hot volcanic springs [8]. Previously we have demonstrated that larvae of all these species, independently of their thermal history, show a high level of inducible Hsp70 under "normal" physiological conditions (constitutive or chronic expression), and that the level increases only 2 - 3 fold after a 30 minutes heat shock treatment and 24 hours of recovery. Larvae of these species are able to survive exceptionally heavy heat shock treatments (46 - 48°C), which are lethal to most of other dipteran species studied so far [19-21].
We also demonstrated that Oxycera pardalina Meigen, 1822, a Stratiomyidae species confined to cold and thermally constant environments, shows the highest relative heat-shock tolerance (i.e. can survive the largest shift in temperature as compared to that in its normal habitat). Whether and how the evolutionary alteration of Hsp70 expression and thermotolerance among the Stratiomyidae species involves changes to the hsp70 genes themselves remain unanswered questions.
Here we report the results of molecular analyses performed to compare the general arrangements and sequence of hsp70-containing loci in two Stratiomyidae species from contrasting thermal environments, O. pardalina and Stratiomys singularior Harris, 1776. For this purpose, we cloned and sequenced member genes of the hsp70 gene superfamily from the two species compared.
Our studies have demonstrated that the population of S. singularior that inhabits an area with very instable extreme larval habitats and strongly variable conditions, is characterized by a compact hsp70 cluster of 5 converting hsp70 genes. Contrastingly, the stable and constantly cold habitat-dwelling O. pardalina exhibits quite different distribution and spacing of hsp70 copies in the genome.
Results
General organization of hsp70 clusters in S. singularior and O. pardalina
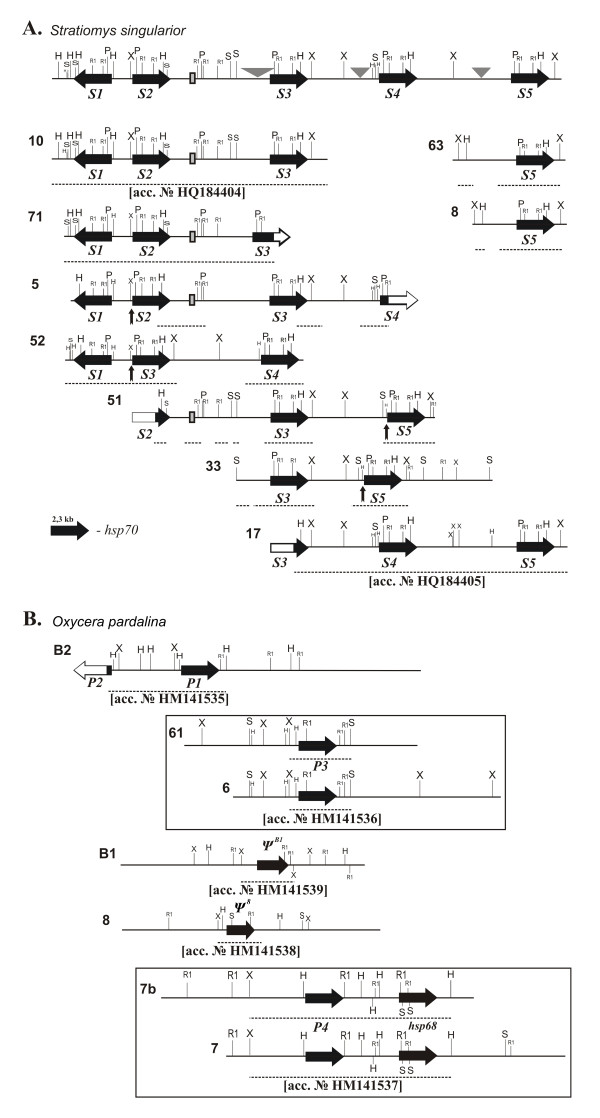
The detailed organization of hsp70 genes in S. singularior and O. pardalina emerges from the subcloning and sequencing of overlapping lambda clones isolated from genomic libraries (Figures 1A and 1B). We have isolated twenty lambda clones from a S. singularior genomic library, and after preliminary restriction analysis have chosen nine clones which apparently contain one or several complete hsp70 genes (Figure 1A). From these nine phages we have isolated and described four hsp70 copies in tandem orientation (hsp70S2-S5) and a fifth copy in inverse orientation (hsp70S1). Genes S2 - S5 are not identically spaced, and intergenic regions vary: approximately 4.7 kb between hsp70S3 and S4, 5.3 kb between hsp70S4 and S5, 5.9 kb between hsp70S2 and S3, and 1.2 kb between hsp70S1 and S2. The whole hsp70 cluster of S. singularior was sequenced, and surprisingly we failed to detect any significant homology among the intergenic regions between individual hsp70 genes.
Figure 1.
Nomenclature and organization of hsp70 genes in S. singularior (A) and in O. pardalina (B) genomes. Deduced arrangement of hsp70 cluster in S. singularior is given at the top. Names of the clones (phages) are given on the left. Individual hsp70 genes are represented by thick filled arrows with the names given underneath. The sequenced fragments are indicated by dotted lines underneath. Accession numbers for the deposited sequences are provided underneath the correspondent sequences. Vertical black arrows underneath phages 51, 52 and 33 indicate the sites of presumptive interlocus recombination. A typical copy of Hsp70 gene in a scale is given at the left side. Triangles indicate the sites where deletions have been detected in a few phages. Filled rectangle show the location of a fragment of Mariner transposable element. H - HindIII, P - PstI, R1 - EcoRI, S - SalI, X - XbaI.
Characteristically, we detected various polymorphisms within the same intergenic regions isolated from individual clones. These polymorphisms include the insertion of a fragment (314 bp in length) of Mariner transposon in the hsp70S2-hsp70S3 intergenic region detected in all four phages containing these genes (phages 10, 71, 52 and 5). Furthermore, hsp70 copies in S. singularior cluster are subject to gross rearrangement, and although their order in most phages is S2, S3 and S4, in phages 33 and 51 the S5 gene is located after S3, while in phage 52 an inverted pair is formed by S1 and S3 genes (Figure 1A).
Interestingly, we detected deletions in almost all the hsp70 intergenic regions. In phage 8, there is a 1 kb deletion in the S4 - S5 region, phage 71 contains a 2 kb deletion in the hsp70S2 - hsp70S3 region, and phage 33 contains a 2kb deletion in the S3 - S5 intergenic region. In phage 71, a deletion in the 5'-flank of hsp70S3 includes the beginning of the transcribed region and essential heat shock regulatory elements; this deletion may make this hsp70S3 allele noninducible (Figure 1A).
The general organization of the hsp70 genes in O. pardalina characteristically differs from that of S. singularior (Figure 1). Preliminary Southern blot analysis of eleven lambda clones isolated from O. pardalina genomic DNA and carrying hsp70 sequences showed that most of the cloned fragments contained only a single hsp70 copy (data not shown). This implies that most hsp70 copies in this species, with two prominent exceptions described below, are unlinked or at least located at significantly larger distances from each other than the hsp70 genes in S. singularior. We performed a detailed restriction analysis of seven lambda clones isolated from the O. pardalina genomic library, which contained the complete sequences of one or two hsp70 family copies (Figure 1B); the other four phages were excluded from the detailed analysis, because they contained only fragments of hsp70 genes and did not carry overlapping sequences, judging by cross hybridization with their labeled terminal fragments (data not shown).
Phage B2 contains one complete copy of hsp70 (P1) and a 5'-fragment of another hsp70 copy (P2) in inverse orientation, apparently cut by Sau3A in the process of cloning (Figure 1B). Although we were able to detect two copies of hsp70 in inverse orientation in O. pardalina in phage B2, even in this case, the distance between these copies was 4.6 kb, i.e. four times as large as the correspondent intergenic region between the inverted S1 and S2 genes in S. singularior (Figure 1 A, B). The 5'-flanking regions of all hsp70 copies in O. pardalina exhibit extended homology upstream from transcription start (up to 650 bp including 5'-UTRs), while in S. singularior, 5'-flanking homology in the correspondent intergenic region is evident only within approximately 200 bp. This implies that in O. pardalina, the hsp70 genes undergo concerted evolution (i.e. interlocus recombination) in blocks containing more than 650 bp of 5'-flanking sequences, while in S. singularior concertedly evolving blocks include less than 200 bp of 5'-flanking sequences.
Figure 1B depicts four phages containing essentially the same hsp70 genes (i.e. clones 61 and 6 and 7b and 7). These phages are included into this figure to illustrate that in contrast to S. singularior, hsp70 genes in O. pardalina are not arranged in tight clusters, and indeed may be entirely unlinked. It is noteworthy that sequencing of hsp70 genes and intergenic regions from these phages did not reveal any differences. Hence, we conclude that phages 61 and 6 and 7b and 7 contain DNA from full sibs.
In O. pardalina, we observed two hsp70 pseudogenes (Figure 1B). The pseudogene located in phage B1 is 89% homologous to hsp70P1 and contains several short deletions resulting in ORF shifts. The other pseudogene, isolated from phage 8, is also homologous to hsp70P1 (86%) and represents a highly rearranged copy. It contains a long deletion (306 bp) and inversion (677 bp) in the coding region of the gene. This pseudogene does not contain the first ATG codon. Both pseudogenes lack HSE elements and fail to exhibit any homology with functional hsp70 genes of O. pardalina in the 5'-flanking regions. The O. pardalina hsp70 pseudogenes likely arose by retroposition, as suggested for similar duplicated hsp70 genes and pseudogenes in Drosophila [22,23]. Clones 7 and 7b contain homologous stretches of DNA carrying two tandem copies of genes belonging to the hsp70 family, located within a 3.5 kb interval. Interestingly, one copy is the inducible hsp70P4, and the second copy is probably hsp68 (see below). This is the first case to our knowledge when hsp70 and hsp68 genes were found to be so closely linked. Therefore, the detailed analysis of genomic libraries enabled us to describe five potentially functional hsp70 genes in S. singularior and five genes belonging to the hsp70 family, including a presumptive hsp68 gene, in O. pardalina. In order to make sure that we cloned and sequenced all or at least most of the hsp70 genes from the species compared, we performed a conventional Southern blot analysis using genomic DNA isolated from both species.
Southern analysis of S. singularior and O. pardalina genomic DNA
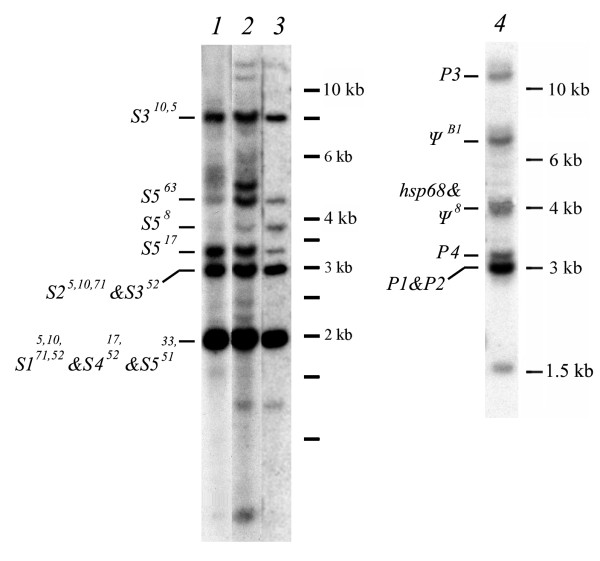
Figure 2 depicts the results of Southern blot analysis, performed to obtain independent data on the number of hsp70 copies in the genomes of the two species studied. Restriction digests of both species' DNA were hybridized to a probe specific for the 5'- and 3'-regions of the S. singularior hsp70 coding sequence. The data depicted in Figure 2 confirm the results of the genomic libraries analysis described above, and show that the S. singularior genome apparently contains five copies of hsp70, while O. pardalina contains four hsp70 copies. Moreover, based on sequencing data we were able to establish the correspondence between individual hsp70 copies and hybridizing bands (see below and Figure 2). Thus, we have cloned and sequenced most if not all copies of hsp70 genes present in the genomes of both species. The additional minor bands seen in genomic DNA of S. singularior isolated from different numbers of larvae (Figure 2) may be explained by restriction site polymorphism or polymorphism in the structure of hsp70 genes per se and/or their flanking regions.
Figure 2.
Southern blot analysis of HindIII restricted genomic DNA isolated from S. singularior (lanes 1, 2, 3) and O. pardalina (lane 4). Numbers on the right indicate the size of the markers in kb. Lane 1 - S. singularior DNA isolated from twenty larvae hybridized with 5' PCR fragment of S. singularior hsp70 gene (1 - 538 bp PCR fragment of complete cDNA copy); 2 - S. singularior, DNA isolated from twenty larvae with 3'-PCR fragment of S. singularior hsp70 gene (1250 - 2340 bp PCR fragment of complete cDNA copy); 3 - S. singularior DNA isolated from five larvae with 3'-PCR fragment of S. singularior hsp70 gene (1250 - 2340 bp PCR fragment of complete cDNA copy); 4 - O. pardalina, DNA isolated from twenty larvae hybridized with PCR fragment included 298 - 1851 bp of complete O. pardalina hsp70P1 gene. Presumptive correspondence of individual hsp70 family genes (according to sequencing data) to the hybridizing bands is indicated on the left of the lanes.
Preliminary 3'-RACE analysis of cDNA homologous to heat shock inducible RNA isolated from both species based on 3'-UTR heterogeneity in general has confirmed our conclusions about the hsp70 copy number estimated for both species, and indicates that all hsp70 copies isolated with the exception O. pardalina pseudogenes are apparently expressed, i.e. represented in the population of cDNA clones sequenced.
Sequence analysis of hsp70 promoters and regulatory elements in O. pardalina and S. singularior
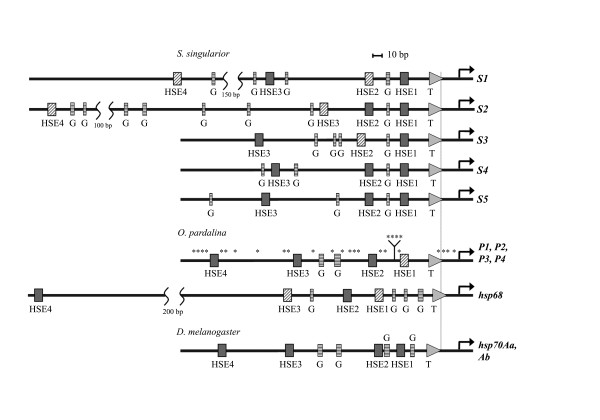
Sequence analysis revealed all components of hsp70 regulatory machinery in the 5'-UTR regions of most hsp70 genes of both species compared. The results of comparing promoter regions in both species are depicted in Figure 3. The general structure of the Drosophila melanogaster hsp70 promoter is given for comparison.
Figure 3.
General arrangement of promoter regions of S. singularior and O. pardalina. The locations of HSEs are indicated by filled horizontal rectangles. TATA-boxes are indicated by triangles (T). The shaded rectangles indicate the positions of non-canonical HSEs. Vertical rectangles indicate the positions of GAGA sites (G). The black bended arrows mark the transcription start and direction. D. melanogaster hsp70 promoter (from 70A region) is given at the bottom for comparison. Asterisks above the O. pardalina regulatory region mark single nucleotide polymorphisms.
We aligned the promoters of the hsp70S genes separately, since divergence among the genes becomes significant at approximately 40 bp upstream of the TATA signal (See Additional files 1, 2, 3, 4 and 5: Figures S1, S2, S3, S4 and S5 for individual alignments of the S. singularior hsp70 gene promoters). In S. singularior, promoters of individual hsp70 genes vary (Figure 3). Four candidate heat shock elements ("HSEs") are evident in the promoters of hsp70S1 and hsp70S2. In hsp70S3, hsp70S4, and hsp70S5, only three candidate HSEs are observed. Additionally, the promoters of hsp70S3, S4, and S5 contain indels and rearrangements. hsp70S352 contains a repetitive 39 bp insertion upstream of the TATA signal that ablates all but the most proximal of the HSEs (See Additional file 3: Figure S3). hsp70S533 has a similar type of 60 bp insertion upstream of the TATA signal, flanked by an hsp70S4-derived conversion tract (See Additional file 5: Figure S5). Whether these indels and rearrangements modulate hsp70 expression, as has been observed for Drosophila hsp70 genes [24,25], remains to be tested. Interestingly, in the same phages (52, 51 and 33) we observed gross rearrangements of the whole cluster (i.e. hsp70S3 in the place of hsp70S2 in phage 52, and hsp70S5 in the place of hsp70S4 in phages 51 and 33). The insertions of repetitive DNA thus likely represent the footprints of transposable elements responsible for the reshuffling of the hsp70 cluster observed in these particular phages. Intralocus unequal recombination likely occurred, because the promoter region of gene S3 in phage 52 is highly homologous to gene S2, including conversion tracts. Similarly, the promoter region of gene S5 from phages 33 and 51 is practically identical to that of S4. The sites of presumptive recombination breaks are shown by vertical arrows in Figure 1A.
The transcription start point was found to be identical in both species studied (see Figure 3). We identified the hsp70 transcription start by 5'-RACE using RNA isolated after HS from the larvae of both species, as described in Methods.
In all S. singularior hsp70 genes, the distance between TATA and transcription start point (AGT) is 24 bp, and the start codon is located 230 bp further downstream. In this species, all hsp70 TATA-boxes are represented by 5'-TATATATA-3' nucleotides, and GAG or GAGA corresponds to the position of the presumptive GAGA-factor binding site.
We aligned approximately 660 bp of promoter sequence from the four Oxycera hsp70 genes. These promoters are 93% identical to one another, and each contains four candidate HSEs in homologous positions ranging from 27 - 28 to 222 - 224 bp upstream of the TATA signal, which is represented by sequence TATAAATA (Figure 3).
The promoter of presumptive hsp68, cloned from O. pardalina, also contains a canonical TATA-box (TATAAATG), several GAG sequences, and four HSEs (Figure 3).
It is evident that essential regulatory elements located in the promoter regions of the two species hsp70 genes (i.e. HSEs and GAG or GAGA) vary in number and spacing when comparing between genes and species. In general, the spacing of major regulatory elements (HSEs and GAGA) in O. pardalina is more similar to that of D. melanogaster (Figure 3). All the above supports that promoters of both Stratiomyidae species studied are functional heat-inducible promoters.
Sequence analysis and evolutionary relationships of hsp70 transcriptional units in O. pardalina and S. singularior
Oxycera pardalina: We recovered the full sequence of one allele for each of the hsp70P1, hsp70P2, and hsp70P4 genes. These genes are intronless and contain a 1920 bp CDS.
The three whole CDSs are on average 98.4% identical, diverging by 20 - 27 silent and 5 - 10 replacement substitutions (See Additional file 6: Table S1 for numbers of silent and replacement fixed differences). The 5'-UTRs of the three intact hsp70 genes are 252 - 260 bp long and 91% identical (data not shown).
We recovered 325 - 340 bp of alignable sequence downstream of the three cloned intact hsp70 CDSs; sequence identity in this region is 77%. In contrast to the near identity of hsp70P1, P3, and P4, two hsp70 pseudogenes, denoted ψhsp70B1 and ψhsp708, bear numerous indels and substitutions and do not encode full-length proteins. The UTRs and promoter regions of both pseudogenes were fully sequenced and aligned. However, we found neither potential HSE and TATA boxes nor any significant homology between these regions (data not shown).
Stratiomys singularior: We sequenced multiple alleles of all five hsp70 genes: hsp70S1, 3 alleles; hsp70S2, 2; hsp70S3, 4; hsp70S4, 2; hsp70S5, 5. Alleles are named according to the clones from which they were obtained (Figure 1A). All alleles contain an intronless, 1917 bp CDS; no pseudogenes were recovered. The CDSs are more similar to one another than was seen in O. pardalina; an average of seven fixed differences distinguish any pair of S. singularior hsp70 CDSs (See Additional file 7: Table S2 for MK tests results), for an average pairwise identity of 99.6%. Comparing the hsp70S genes to one another via MacDonald-Kreitman style tests reveals that in all cases, the number of polymorphisms is much greater than the number of fixed differences, and that both polymorphism and divergence are similarly skewed towards silent substitutions (Additional file 7: Table S2). These results are consistent with concerted evolution and purifying selection.
The mechanism of concerted evolution among the hsp70S CDSs is likely gene conversion, observed for other insect hsp70 genes [22,23]. There are 50 single-nucleotide polymorphisms ("SNPs") shared among the hsp70S genes (black bars in Additional file 8, Figure S6). Considering only the silent polymorphisms, to avoid confounding any effects of positive or balancing selection, we examined the probabilities of shared SNPs arising among the hsp70S genes via independent mutation or gene conversion using the method of Rozas and Aguade [26]. Table 1 indicates that in all pairwise comparisons of hsp70S genes, we can reject the null hypothesis of independent mutation at P < 10-5.
Table 1.
Conversion-mediated shared polymorphism at the Stratiomys hsp70 genes
| Gene | S2 | S3 | S4 | S5 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | ησ | ssSNPs | p | N | ησ | ssSNPs | p | N | ησ | ssSNPs | p | N | η | ssSNPs | p | |
| S1 | 889 | 37 | 6 (+1) | <10-6 | 889 | 35 | 13 (+5) | <10-6 | 890 | 49 | 11 (+3) | <10-6 | 890 | 68 | 13 (+1) | <10-6 |
| S2 | 888 | 30 | 6 (+2) | <10-6 | 889 | 45 | 5 (+0) | <10-5 | 890 | 70 | 8 (+0) | <10-6 | ||||
| S3 | 889 | 46 | 7 (+2) | <10-6 | 890 | 68 | 10 (+3) | <10-6 | ||||||||
| S4 | 891 | 74 | 17 (+0) | <10-6 | ||||||||||||
N, total number of silent sites. ηs, total number of silent mutations. ssSNPs, number of shared silent single-nucleotide polymorphisms (numbers in parentheses indicate additional number of shared replacement single-nucleotide polymorphisms; not considered in probability estimation). p, probability of ssSNPs arising via parallel mutation, estimated from hypergeometric distribution.
We recovered and aligned 262 bp of 5'-UTR sequence from all the hsp70S genes/alleles whose CDSs were completely sequenced, plus one additional allele for each of hsp70S3, hsp70S4, and hsp70S5 (Additional file 9: Figure S7). Only the first 15 bp of the hsp70S371 5'-UTR could be aligned; the UTR is disrupted by large blocks of repetitive sequences upstream of this position, consistent with insertion and/or rearrangement via transposition. Whether this rearrangement alters the expression of hsp70S371 is unknown. The homogeneity observed in the hsp70S CDSs diminishes in the 5'-UTRs. The 5'-UTRs are 82% identical overall; linkage disequilibrium is higher than in the CDSs, and four blocks of linked, polymorphic sites that are shared among genes are strongly suggestive of conversion tracts (See grey bars in Additional file 9: Figure S7). Seventeen individual SNPs are shared among multiple genes, again consistent with gene conversion (black bars in Additional file 9: Figure S7).
The 3'-UTRs of the S. singularior hsp70 genes are more divergent than those of O. pardalina. In S. singularior the distance of a typical PolyA signal (AATAAA) from the stop codon, determined by 3'-RACE, varies from 193 to 247 bp in individual hsp70 copies.
In O. pardalina the position of a candidate polyadenylation signal slightly varies in individual copies of hsp70 genes and falls 183 - 186 bp downstream of the stop codon.
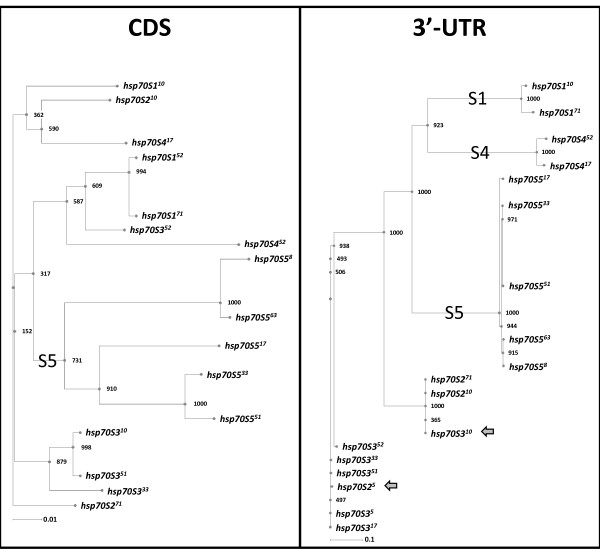
As for the 5'-UTRs, we sequenced 3'-UTRs of more alleles than just those for which we obtained full-length CDSs; more than 2 alleles of each hsp70S gene 3'-UTR were completely sequenced (Figure 1A). In contrast to the 5'-UTRs, which could all be aligned as one group, we aligned the 3'-UTRs of hsp70S1, hsp70S4, and hsp70S5 separately for visualization since their shared sequence is less than 100 bp long. Multiple candidate polyadenylation signals are found in all the 3'-UTRs. Only hsp70S2 and hsp70S3 share significant sequence identity. In general, the 3'-UTRs vary to a greater extent than both the CDSs and 5'-UTRs, with sequence identity <80% (See Additional files 10, 11, 12 and 13: Figures S8, S9, S10 and S11 for alignments of the 3'-UTRs). Hsp70SS352 has a deletion eliminating most of the 3'-UTR, approximately 47 bp downstream of the stop codon; the effects of this deletion were not tested but are consistent with rearrangements outside the hsp70 CDSs observed in the 5'-UTR and promoter (see above, below). Only hsp70S2 and hsp70S3 show evidence of gene conversion in the 3'-UTR: in clone 5, hsp70S2's 3'-UTR has been converted over at least 300 bp to exactly resemble the 3'-UTR of hsp70S3 (see grey bars in Additional file 11: Figure S9). Similarly, in clone 10, hsp70S3's 3'-UTR has been converted with hsp70S2's. These less frequent, larger, and unbroken conversion tracts, along with the overall higher degrees of divergence among and polymorphism within 3'-UTRs, indicate less homogenization via gene conversion. This apparent reduction in conversion is more pronounced than is observed in the 5'-UTRs, which show both large tracts and the multiple shared SNPs indicative of homogenizing gene conversion. The net effect of conversion among CDSs with divergence among 3'-UTRs is evident in comparative phylogenetic trees (Figure 4). In the CDS tree, only hsp70S5 resolves as a clade and is supported by a majority of bootstrap trials. In contrast, the 3'-UTR tree contains hsp70S1, hsp70S4, and hsp70S5 clades, all with strong bootstrap support, and only the wholly-converted hsp70S2/hsp70S3 alleles from clones 5 and 10 prevent the assemblage of hsp70S2 and hsp70S3 clades. Thus, the hsp70S CDSs evolve in concert; the 3'-UTRs evolve largely independently.
Figure 4.
Neighbor-joining phylograms of Stratiomys hsp70 coding sequences (left) and 3'-UTRs (right). Alleles named by phage number (superscript). Node labels indicate supportive number of 1000 bootstrap trials. Gene-specific clades labeled by gene abbreviation (e.g., "S1" = hsp70S1). Rulers indicate genetic distance.
The overall pattern of conversion-mediated homogeneity among hsp70 that decays with distance from CDS, and polymorphic flanks that contain large indels, is consistent with duplicated hsp70s in Drosophila.
Inducible proteins encoded by hsp70 family genes in S. singularior and O. pardalina
Most of the varying sites in hsp70 genes of both species are silent and, hence, the deduced Hsp70s at amino acid level are highly homologous. Thus S. singularior individual Hsp70s exhibit almost 97% homology, while their homology with O. pardalina and D. melanogaster Hsp70s reaches 93% and 82%, respectively. Furthermore, the homology between deduced individual Hsp70s of O. pardalina reaches 99%. Characteristically, most of the varying sites in both species are localized in the C-domain of Hsp70, which is expected based on the above-presented data and the results of other authors showing a higher level of divergence at the 3'-ends of hsp70 genes in Drosophila [27,23]. The molecular weights of individual S. singularior Hsp70s (deduced by Vector NTI) are 69.89 - 70.18 kD and pI 5.48 - 5.75.
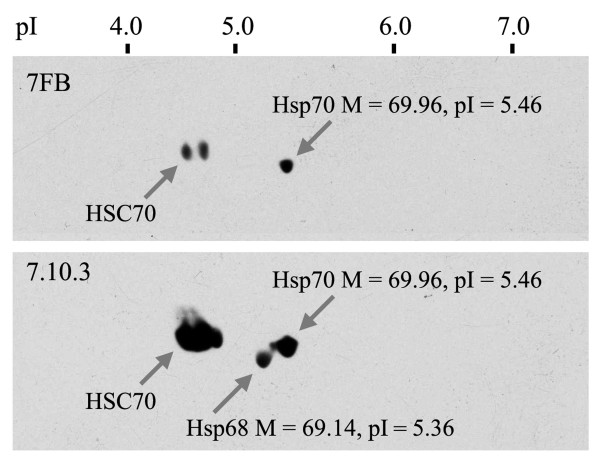
In order to further characterize Hsp70 family members in the species compared, we have performed two-dimensional electrophoresis of total proteins from O. pardalina and S. singularior with subsequent immunoblotting with the 7FB antibody, which in D. melanogaster reacts only with inducible Hsp70s, and with the 7.10.3 antibody, which reacts with all members of the Hsp70 family in most Diptera species examined including Hsp68 [8].
The molecular weights of individual O. pardalina Hsp70 members deduced by Vector NTI (69.85 - 69.96 kD) and pI (5.45 - 5.46) vary and practically coincide with the results of 2D electrophoresis of O. pardalina proteins (Figure 5).
Figure 5.
The pattern of O. pardalina proteins as revealed by two-dimensional electrophoresis. Western blotting of sections of 2D blots containing Hsp70 family members using 7FB (A) and 7.10.3 (B) antibodies. Constitutive (HSC70), inducible (Hsp70), and Hsp68 proteins are indicated by arrows.
The 7.10.3, but not 7FB, antibody revealed one lower molecular weight protein, which characteristically has a slightly more acidic isoelectric point than the presumptive Hsp70 (Figure 5). We hypothesized that this particular protein corresponded to D. melanogaster Hsp68. Importantly, the molecular weight of this protein and pI revealed by 2D (69.1 and 5.4 correspondently) practically coincide with those of Hsp68 deduced basing on the presumptive hsp68 gene sequence located in phage 7 and 7b of O. pardalina (69.14 kD and 5.36).
Using Coomassie stained gels we have isolated spots of expected pI for the Hsp70 family members and performed fingerprinting and microsequencing analysis as described previously [8]. In both species, inducible proteins of interest are represented by a few isoforms, which are well separated on the gel and easy to isolate (Figure 5). After in-gel trypsin digestion and fingerprinting analysis using MALDI-ToF, the National Center for Biotechnology Information database was searched with the Profound search engine. This search has established that as expected the excised 70 kDa spots in both species correspond to members of the Hsp70 family previously described in various Diptera species (Additional file 1, Figure S12).
The predicted Hsp70 polypeptide of S. singularior and O. pardalina consists of 638 a.a. and 639 a.a., respectively, and presents all classic Hsp70 family signatures, including the presence of the serine residue in the ATPase domain (Additional file 14: Figure S12 for comparison with paralogs from several organisms) that characterizes the inducible HSP70 proteins of invertebrates [28]. We also detected a few amino acids residues specific for the both Stratiomyidae species and other highly thermotolerant organisms e.g. Locusta migratoria (Additional file 15: Figure S12). Conceptual translation showed that Stratiomyidae Hsp70 sequences had 72% identity with human Hsp70 and 84 - 85% identity with D. melanogaster Hsp70.
Our microsequencing data together with phylogenetic analysis and description of typical hsps promoters enable to conclude that all hsp70 genes isolated, with the prominent exception of the two O. pardalina pseudogenes, definitely represent inducible members of hsp70 family.
Previously we have shown that all four Stratiomyidae species exhibit high thermotolerance, correlated with high constitutive cellular levels of Hsp70. Surprisingly, Northern hybridization experiments failed to demonstrate hsp70 expression under normal physiological conditions, and only using a highly sensitive RT-PCR technique we were able to detect a very weak leakage of hsp70 genes in the species studied [8]. Surprisingly, the accumulation of Hsps in the cells of these species after heat shock challenge continues for many hours after HS treatment and, therefore, inducible Hsp70s in these species are probably more stable in comparison with Drosophila and many other Diptera species [29]. Based on these data, it was of significant interest to compare the amino acid sequence of Stratiomyidae Hsp70s with the sequence of Hsp70s isolated from other animals. The results of this analysis are shown in Figure 6.
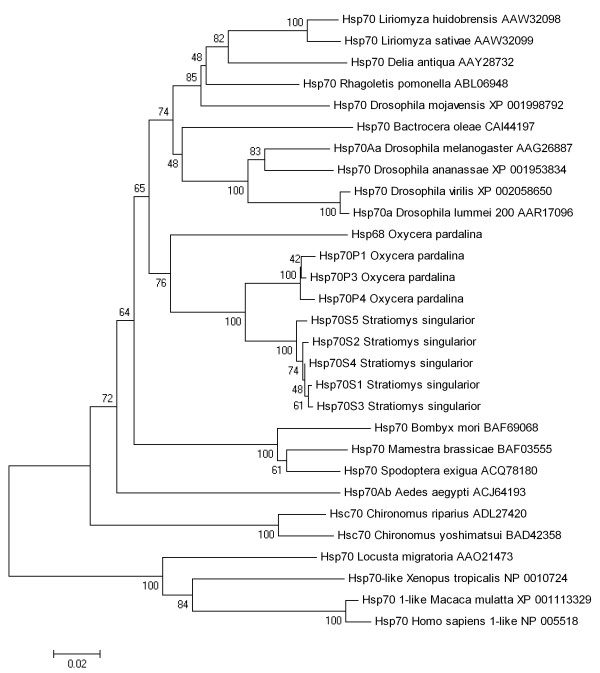
Figure 6.
Phylogenetic relationships of the Hsp70 proteins from S. singularior, O. pardalina and several other species. Accession numbers are given after species name. The evolutionary history was inferred using the neighbor-joining method. The percentage of replicate trees, in which the associated taxa clustered together in the bootstrap test (500 replicates), are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset.
Roughly, the dendrogram reflects the accepted phylogenetic relationships between the taxa included [30], with some deviations. In Figure 6, species of Stratiomyidae (family belonging to the suborder Orthorrhapha of the order Diptera) form a separate cluster, as well as all species of another suborder, Cyclorrhapha (the families Drosophilidae, Agromyzidae, Tephritidae), with one exception, Bactrocera oleae (Tephritidae). These two branches were clustered together. Contrastingly, members of the "more primitive" suborder Nematocera (Chironomidae, Culicidae) have the much lower similarity to other Diptera; formally, they were grouped with the cluster (Lepidoptera + all other Diptera) and did not form a separate cluster (Figure 6).
Interestingly, heat shock proteins (Hsp70) from both Stratiomyidae species grouped with paralogous sequences from several insect species including the ones known to be highly thermotolerant such as Bombyx mori [10], Locusta migratoria [31] and Drosophila mojavensis [29]. However, although even a few mammalian Hsp70s were included into the tree, it is premature to speculate on any functional relatedness of these proteins because such speculation will require the inclusion and analysis of many more sequences.
Discussion
In this paper we investigated the structure and general arrangement of hsp70 genes from two Stratiomyidae species from strikingly contrasting habitats.
According to field observations and measurements, S. singularior has highly variable and instable larval habitats in the arid steppe zone of the Crimea. Strong changes in the shore larval habitats of S. singularior were observed since 2005 to 2009, both within the season and between the years. These changes were related mostly to hydrological regime, i.e. varying precipitation and the water level of the lakes. Generally, in the periods of the higher water table, many larval habitats were flooded or strongly influenced by hypersaline lake water (mineralization reached 150 - 280‰ and higher). The decreasing water level of the lake coincided with a decrease of mineralization (up to 80 - 35‰ in some larval habitats); in these periods, the respective habitats were moistened mostly with groundwater of lower salinity. It should be also mentioned that the abundance of S. singularior larvae varied greatly along the shoreline and between the (micro)habitats. In favorable places, e.g. in some groundwater-fed pools above the lake water margin, the larvae reached high abundance (hundreds and thousands individuals per m2), but typically, they were completely absent or found only as single individuals along the most part of the shore.
It seems that in the study area, S. singularior is represented by a number of micropopulations mostly confined to some shore localities, which may be at a distance of several km from each other. Many of these favorable breeding places are temporary and completely dry out in drier and warmer years. However, in this case adult flies are able to cover the distance of several kilometers easily in search of new places for oviposition.
Therefore, our preliminary observations on the Crimean larval habitats of S. singularior testify to a variability in the conditions between the habitats, strong variation of conditions within the habitats occurring with time, together with the temporary nature of many habitats and their possible isolation from each other. All these features may result in a high polymorphism of hsp70 demonstrated in this particular species.
We demonstrated a high level of polymorphism by Southern blot hybridization with genomic DNA digests, depending on the number of the larvae taken for analysis (Figure 2). Furthermore, sequencing of several phages containing the same hsp70 genes revealed polymorphism in the intergenic regions including long deletions. In a few cases we detected deletions of entire individual hsp70 genes (in phages 52, 33 and 51), likely a result of unequal recombination.
These deletions may result from an evolutionary trend to make the hsp70 cluster in this species more compact. Previously we have described more compact organization of the hsp70 genes cluster in a more thermotolerant low-latitude species Drosophila virilis in comparison with the closely related higher-latitude D. lummei from the temperate climatic zone [13] and, therefore, the observed deletions may represent certain pattern. Alternatively, the deletions described in S. singularior may be due to a high overall rate of DNA loss in this species, a phenomenon previously demonstrated for two Drosophila species [32].
In contrast to the S. singularior biotope, the studied larval habitat of O. pardalina - a cold spring near St. Petersburg observed from 2004 to 2010 - was very stable in mineralization, temperature, water regime, and in other major conditions both within season and interannually. This spring and headwater stream seems to be an island habitat for the O. pardalina population. Another similar habitat is situated at about 3 km, and it is unlikely that O. pardalina adults migrate regularly between these localities. The abundance of larvae is not high, not exceeding 10 - 50 individuals per m2 of semiaquatic shore substrate (moss etc.). Thus, habitats of the two species differ dramatically from each other not only in the average parameters such as temperature and mineralization, but also in the spatial organization and in the degree of variability in time.
Therefore, in the case of the latter species we have a comparatively small and probably highly homozygous population. Southern blot hybridization analysis (Figure 2) did not reveal any polymorphisms in O. pardalina hsp70 genes and, hence, confirms this conclusion. Furthermore, individual phages containing the same fragments of DNA did not exhibit any polymorphisms either in coding or in intergenic regions of the same hsp70 genes. Since these phages were isolated from two independently obtained genomic libraries they cannot contain the same DNA but rather likely represent highly homologous sibs.
Previously, when we investigated inbred laboratory strains of D. virilis and D. lummei, we also demonstrated apparent identity of the overlapping hsp70-containing clones, which is typical for highly homozygous strains [13].
In spite of strikingly different habitats, the larvae of all Stratiomyidae species studied have very high tolerance to elevated temperature in comparison with most of other Diptera species investigated [8]. Furthermore, although tolerances were in general correlated with the typical habitat temperatures of the species, even O. pardalina larvae inhabiting cold springs with constant temperature regime (5 - 10°C), significantly exceeded all Drosophila species in tolerance and exhibited a huge gap between the natural temperatures (5 - 10°C) and the critical temperature (43°C).
Importantly, larvae of all four Stratiomyidae species investigated have exhibited high constitutive levels of Hsp70 proteins which are induced only 2 - 3-fold after temperature elevation [8]. In this respect, Stratiomyidae species resemble Locusta migratoria, an insect species adapted to arid tropical and subtropical climate, where temperature frequently exceeded 40°C [31].
Additionally, in contrast to Drosophila, the accumulation of Hsp70 continues in the cells of the treated larvae of all Stratiomyidae species investigated for many hours after heat shock and plateaus only approximately 24 - 36 h after the treatment [8].
The results obtained may be explained by constitutive weak transcription (leakage) of hsp70 genes under normal physiological conditions, and possibly by comparatively higher stability of Hsps in Stratiomyidae species in comparison with other insect species studied.
In order to further characterize the molecular basis underlying this pattern of hsp70 expression and differences in thermotolerance in the Stratiomyidae and other Diptera, as well as between the Stratiomyidae species, we compared the general arrangement of hsp70 genes clusters of the two Stratiomyidae species from thermally different habitats. Surprisingly, the analysis revealed quite different organization of hsp70 genes in the two species compared.
Although the hsp70 copy number does not differ significantly between less tolerant O. pardalina and more thermotolerant S. singularior, the general organization of hsp70 genes is divergent in these two species. While in S. singularior all hsp70 genes are confined to one genomic section of about 29 kb in length, in O. pardalina most of hsp70 genes are scattered in the genome and apparently located rather far from each other if linked at all. However, in both species we have found two hsp70 copies in an inverted orientation (a fundamental and probably ancient hsp70 gene unit) [22]. Furthermore, in O. pardalina we cloned a fragment carrying one copy of inducible hsp70 and one copy of presumptive hsp68 gene in tandem orientation separated by only 3.5 kb.
Previously we described two hsp68 copies in inverted orientation in species of the virilis group of Drosophila [13]. We speculated that hsp68 in these species arose by duplication of a basic unit of hsp70 genes typically arranged in Diptera species in inverted orientation. However, in the virilis group species hsp70 and hsp68 are located in the same chromosome but are separated by a large distance [13]. Presumptive hsp68 gene of O. pardalina is closely linked to hsp70 gene (Figure 1B) and exhibits higher homology with hsp70 genes of this species than with hsp68 genes from other Diptera species (data not shown). These data enable us to suggest that O. pardalina hsp68 also represents a duplication of hsp70 gene. At the present time we can not discriminate between two hypotheses explaining the appearance of hsp68. The appearance of the hsp68 by duplication of hsp70 may occur very early in Diptera evolution and predate the split of Drosophilidae and Stratiomyidae. Alternatively hsp68 may appear independently in the evolution of Drosophila and Stratiomyidae and acquire similar functions and structure by convergence.
It is noteworthy that even for the genes located in inverted orientation, which apparently represents the primitive state of hsp70 in Diptera, the distance between the inverted copies in O. pardalina is four times as large as between the inverted copies in S. singularior (Figure 1A and 1B).
The different spacing of hsp70 genes in the species compared may resemble different abilities of their genome to efficiently respond to stress. The homogenized coding sequences and highly compact organization of hsp70s in S. singularior may be necessary for fast and efficient transcriptional response and thereby thermotolerance of the larvae to extreme and rapidly changing conditions of the environment.
On the other hand, O. pardalina inhabiting cold running waters with comparatively stable thermal conditions may not require such tight hsp70 cluster organization and, hence, underwent dispersal and pseudogenization of a few hsp70 copies in the process of evolution. Two hsp70 pseudogenes detected in the genome of this species in our studies may represent the remains of previously active genes. Previously we have shown that larvae of O. pardalina produce significantly less Hsp70 in response to heat shock in comparison with all other Stratiomyidae species investigated [8]. The observed drastic differences in the organization of hsp70 genes in S. singularior and O. pardalina may account for the differences in the thermotolerance and Hsp70 levels after HS treatment observed between these species.
Furthermore, HSE1 of O. pardalina is not canonical and contains only two 5 bp units. Therefore, the first canonical element (HSE2) in O. pardalina is located at significantly larger distance from the transcription start than in S. singularior (Figure 3). The differences in the number of canonical HSEs and their spacing may also contribute to different activity of hsp70 genes in the two species [33] and explain why O. pardalina exhibits lower thermotolerance in comparison with all other Stratiomyidae species studied so far [8].
It is also evident from Figure 3 that in the case of O. pardalina, the arrangement of promoter regions of all hsp70 genes is practically identical, while in the case of S. singularior the promoters of all members of hsp70 cluster differ significantly. These differences result from fine tuning of an hsp70 "battery" necessary for adaptation of the latter species to highly variable environmental conditions. Therefore, our results indicate that O. pardalina hsp70 genes may be locally adapted, whereas higher hsp70 polymorphism may have important evolutionary consequences for S. singularior.
The evolution of hsp70 genes in the species compared is quite different in many respects. The MK tests demonstrate that the divergence between individual S. singularior hsp70 genes is very low, especially considering the age of the duplications. This is consistent with concerted evolution in this species, and also strikingly different from the divergence between paralogs in O. pardalina (See Additional file 6: Table S1). In all cases, the number of shared polymorphisms between S. singularior hsp70 genes is highly unlikely to have occurred via parallel mutation (Table 1). The presence of multiple conversion-mediated shared SNPs and large conversion tracts in the alleles of the same hsp70 members support gene conversion as a mechanism of concerted evolution (and thus sharing polymorphism) among duplicated hsp70 genes of S. singularior species [34].
The overall pattern of conversion-mediated homogeneity among hsp70 genes in S. singularior that decays with distance from CDS, and polymorphic flanks that contain large indels, is consistent with previously described duplicated hsp70s in Drosophila [22]. We cannot exclude the occurrence of gene conversion in O. pardalina, because in this species we did not isolate multiple alleles of individual hsp70 genes. However, since the level of fixed differences between hsp70 genes in O. pardalina is several fold higher than in S. singularior (Additional file 6: Table S1), one may conclude that in the former species the conversion process is less efficient if present at all. The large distances between individual hsp70 genes in O. pardalina genome likely decrease the frequency of their conversion [35,36]. Whether the large distances also decrease their coordinate expression is an interesting question.
Several authors justly outlined many logical and statistical problems associated with using two-species comparison for studying adaptations [37], and we are well aware that unless similar correlation is obtained for other Stratiomyidae species or geographical populations, any generalizations are premature. However, a similar pattern of hsp70 organization has been previously described when studying two closely-related species of the virilis group of Drosophila, which replace one another along a latitudinal cline [14]. The less thermotolerant high-latitude species D. lummei, similar to O. pardalina, exhibits several-fold larger distance between tandemly arranged hsp70 copies in the cluster, in comparison with the closely-related more tolerant low-latitude D. virilis [14]. Interestingly, along these lines one of the hsp70 copies described in D. lummei was shown to be a nonfunctional pseudogene lacking the first 300 nucleotides of coding sequence. Similarly, two likely nonfunctional pseudogenes have been described in the present study in the less thermotolerant O. pardalina. Dispersed genomic arrangement is not absolutely required for hsp70 degeneration. The tight hsp70 clusters of the tropical island endemic D. mauritiana segregate multiple pseudogenes, although in this case a founder effect rather than relaxation of selection may modulate hsp70 pseudogene frequencies [22]. In spite of the approximately identical hsp70 copy number in Stratiomyidae and Drosophila virilis group species, there are several clear-cut differences in the organization and evolution of hsp70 cluster between these phyla. In species of the virilis group, not only the hsp70 transcriptional units but also the 3'-flanking sequences are highly conserved within D. virilis and between D. lummei and D. virilis. Thus, the 3'-flanking sequence of several D. virilis hsp70 genes is near identical for more than 1600 bp. Such an arrangement suggests tandem duplication of hsp70 copies by unequal crossing over in the course of Drosophila species evolution [13].
In contrast, our analysis detected only short, varying length regions of significant homology in the intergenic regions in S. singularior hsp70 cluster. Similarly, we detected only 650 bp of significant homology in the intergenic regions located at the vicinity of 5'-ends of hsp70 genes in O. pardalina.
At the present time, we can only speculate concerning the molecular mechanisms which may underlie the different arrangement of hsp70 genes in the species studied, although the pattern of compactness and conversion in S. singularior vs. dispersion and divergence in O. pardalina is clear. Several large deletions in S. singularior, including those spanning entire individual hsp70 genes, may be indicative of an evolutionary trend towards compactness to provide a highly coordinate response to the changing environmental conditions. The deletions may also be a consequence of conversion; the more homogenized the individual hsp70 units, the more likely they are also to participate in unequal crossover. On the other hand, the dispersed arrangement and interlocus divergence of hsp70 copies in O. pardalina may result from long-range recombination and relaxed selection on hsp70 sequences in a species living in very stable thermal conditions.
The analysis of hsp70 arrangement in other species and natural populations of Stratiomyidae and related taxa (work in progress) should help to further evaluate the evolution of hsp70 genes in the ecological context.
Conclusions
The evolution of hsp70 genes in the species compared is quite different in many respects.
Although the hsp70 copy number does not differ significantly between less tolerant O. pardalina and more thermotolerant S. singularior, the general organization of hsp70 genes is divergent in these two species. While in S. singularior all hsp70 genes are confined to one genomic section of about 29 kb in length, in O. pardalina most of hsp70 genes are scattered in the genome and apparently located rather far from each if linked at all. However, in both species we have found two hsp70 copies in an inverted orientation, which represents a fundamental and probably ancient hsp70 gene unit. The divergence between individual S. singularior hsp70 genes is very low, especially considering the age of the duplications. This is consistent with concerted evolution in this species, and also strikingly different from the divergence between paralogs in O. pardalina. The observed drastic differences in the organization of hsp70 genes in S. singularior and O. pardalina may contribute to the differences in the thermotolerance between these species, and thus recapitulate the history of their thermal adaptation.
Methods
Insects and habitats
In this study, we have investigated two stratiomyids, Stratiomys singularior Harris, 1776 and Oxycera pardalina Meigen, 1822. Stratiomys singularior is a very common and widely distributed Transpalaearctic species [38] whose larvae develop in various shallow aquatic and semiaquatic habitats of standing waters. For this study, S. singularoir larvae were collected from the water margin zone of the highly mineralized chloride coastal Lake Koyashskoe in the Crimea. The measured temperature in the Crimean habitat of S. singularior in May-August varies from 15 to 30°C, but apparently, it is much lower in winter and can be significantly higher in summer. Water mineralization in the S. singularior habitat varied from 35 to 280 g/L, and could fluctuate around the highest values for weeks and months. Oxycera pardalina is also widely distributed in Western and Central Europe. The larvae of this species are confined to clear-water cold carbonate springs and streams [38]. The larvae were collected in the water margin zone of a spring near St. Petersburg characterized by a year-round stable low water temperature (5 - 10°C). The habitats of these populations and the techniques of sampling, identification and experiments have been described in detail in a previous paper [8].
Preparation of RNA and cDNA synthesis
Total RNA from S. singularior larvae was prepared by the standard method with TRIZOL (Invitrogen). A cDNA library has been obtained from S. singularior total RNA using MINT cDNA kit (Evrogen) according to manufacturer's manual.
Constructing and screening of cDNA library
Total double-stranded cDNA was cloned into the pAL-TA vector (Evrogen) using T4 DNA ligase (Promega). E. coli DH5α strain competent cells were transformed with ligation mix and inoculated onto LB agar Petri dishes. After overnight incubation at 32°C, colonies were transferred onto Hybond-N membranes (Amersham) and in situ hybridization was performed as described [39]. A full-size hsp70 gene of D. melanogaster was used as a labeled probe to screen cDNA library.
5'- and 3'-RACE analysis
The MINT cDNA kit was used for the first strand cDNA synthesis and amplification of 5'- and 3'-ends of cDNAs, as described by the manufacturer. For 5'- and 3'-end amplification outwards primers to 5'- and 3'-fragments of S. singularior and O. pardalina hsp70 coding region were used (See Additional file 15: Table S3 for primers used in the study). PCR-fragments obtained were cloned into pAL-TA vector (Evrogen) for sequencing.
2D electrophoresis and immunoblotting
Protein extraction and two-dimensional gel electrophoresis were performed as described by O'Farrell [40], with slight modifications [41]. In brief, lysates were prepared from larvae after exposure to various temperatures. After 2D electrophoresis of 50 μg of total protein, the proteins were transferred from gel into a nitrocellulose membrane (Hybond ECL; Amersham) according to the manufacturer's protocol. We used a monoclonal antibody specific to D. melanogaster inducible Hsp70 (antibody 7FB) and a monoclonal antibody which recognizes all members of D. melanogaster Hsp70 family (antibody 7.10.3) including inducible Hsp70, Hsp68 and HSC70 (the antibodies were kind gift of Dr. Susan Lindquist, Massachusetts Institute of Technology). Immune complexes were detected via chemoluminescence (ECL kit, Amersham) with appropriate peroxidase-conjugated anti-rat secondary antibodies (Millipore). Protein concentrations were normalized using BCA kit (Pierce) according to manufacturer's manual.
Genomic libraries construction, screening and clone analyses
Genomic libraries from S. singularior and O. pardalina were prepared by partial Sau3A digestion of genomic DNA with subsequent ligation into the BamHI site of lambda Dash phage arms (Stratagene). Before ligation, restriction mixture was separated by ultracentrifugation in sucrose gradient for removal of short restriction fragments, and resulting fraction contained fragments with 14 - 23 kb length used for cloning. Gradient parameters were: 10 - 40% sucrose on 1 M NaCl, 20 mM Tris-HCl pH 8.0 and 5 mM EDTA with ultracentrifugation at 26 000 g during 24 h. Ligated DNA was packaged into phage particles using lambda packaging extracts (Stratagene). Recombinant phages were selected, amplified and screened using E. coli XL-Blue MRA (P2) host strains. For genomic libraries screening, S. singularior hsp70 full-length cDNA was used as a probe.
Isolated phages containing hsp70 genes were investigated by restriction analysis and hybridization, and subcloned into KS-Bluescript (pBluescript SK+) for subsequent sequencing.
Southern blotting
Southern blot analysis of S. singularior and O. pardalina genomic DNA was performed as described [39]. Five micrograms of each DNA sample was digested with restriction endonucleases. After agarose gel electrophoresis, each gel was treated for 15 min in 0.25 M HCl and then incubated twice in denaturing buffer (1.5 M NaCl, 0.5 M NaOH) for 30 min. After 30 min incubation in neutralization buffer (1.5 M NaCl, 0.5 M Tris-HCl pH 7.5), gels were capillary-blotted onto nylon membranes and fixed by UV cross-linking using the UV Stratalinker 2400 (Stratagene) protocol. Standard high-stringency hybridization and wash conditions were used for Southern blot analysis. To detect hsp70 sequences in Southern blots and genomic libraries, different EcoRI fragments of cloned S. singularior hsp70 full-length cDNA (2340 nt in length) were labeled by random priming and used as probes.
The original screen of all libraries, individual recombinant clones analyses and Southern blots were performed using S. singularior hsp70 full-length cDNA, while Southern blots of O. pardalina genomic DNA were probed with 1250 - 2340 bp PCR-fragment of coding region of O. pardalina hsp70 gene.
PCR conditions and primers
To obtain S. singularior hsp70 intergenic regions, we used primers outward from 5'- and 3'- hsp70 fragments. To obtain the hsp70-hsp68 intergenic region of O. pardalina, we used primers outward from the 3'-end of hsp70 and 5'-flanking fragment of hsp68 gene. To obtain the O. pardalina region located between hsp70 genes found in inverse orientation, we used primer outward from the 5'-end of hsp70. For amplification of the O. pardalina hsp70 coding region fragment used to probe Southern blots, inward primers for 5'- and 3'-ends of the gene were used (see Additional file 1, Table S3). PCR conditions depended on primer annealing time and temperature. All reactions contained 1.25 units of Encyclo DNA polymerase (Evrogen) per probe, 1.5 - 2.5 mM MgCl2, 0.2 mM of each dNTP, suitable quantity of DNA and 10 pM of each primer in 50 μl (total volume).
Sequence analysis
Clones were sequenced with Sequenase (Amersham) and ABI 377 sequencers. Sequences were assembled manually and aligned using NCBI Blast and Vector NTI. Relevant sequence information has been deposited in GenBank. GenBank accessions numbers for hsp70 sequences are as following: S. singularior cDNA used as a probe for genomic libraries screen, EU523048; S. singularior genomic contigs assembled basing on several sequenced clones, HQ184404, HQ184405; O. pardalina genomic contigs, HM141535, HM141536, HM141537, HM141538, HM141539.
Whenever possible, hsp70 sequences were divided by eye into four segments: coding sequence ("CDS"), 5'- and 3'-untranslated regions ("UTRs"), and upstream promoter regions. The 5'-UTR was delineated by the TATA signal and last bases upstream of the start codon. The 3'-UTR was extended from the first bases downstream of the stop codon through the polyadenylation signal when possible. The promoter region was approximated as 400 - 600 bp upstream of the TATA signal.
Sequences were aligned using CLUSTALX [42], trimmed to eliminate end gaps, and unalignable sequences were removed (i.e., sequences containing large indels or rearrangements, see below). BLAST (http://www.ncbi.nlm.nih.gov/BLAST) was utilized to assess identity of repetitive sequences around indels and rearrangements. Sequences were grouped across genes when they shared high homology (e.g. CDS, 5'-UTRs) or within genes when they diverged (e.g. most 3'-UTRs). After realignment of the trimmed, grouped sequences, custom perl scripts (available upon request) parsed alignments to generate figures and tally substitutions. For phylogenetic analysis, neighbor-joining tree building and 1000 bootstrap trials were conducted using CLUSTALX, MEGA [43] and Vector-NTI.
Analysis of polymorphism and divergence was conducted using DNASP [44]. Testing the probability of shared polymorphisms arising via gene conversion vs. independent mutation utilized the approximation of the hypergeometric distribution (implemented in DNASP; method described in [26]).
List of abbreviations
Hsp70: heat shock protein 70.
Authors' contributions
DG participated in the design of the study and genomic library analysis. IY participated in genomic libraries analysis and sequence alignments. BB has been involved in drafting of the manuscript and statistical analysis of the data. OZ participated in the design of the study and clones analysis. AP collected all biological material, assessed habitat conditions and determined the species, participated in drafting of the manuscript. ME participated in the design of the study, drafting of the manuscript and coordination of the whole project. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Alignment of hsp70S1 promoter sequences.
Figure S2. Alignment of hsp70S2 promoter sequences.
Figure S3. Alignment of hsp70S3 promoter sequences.
Figure S4. Alignment of hsp70S4 promoter sequences.
Figure S5. Alignment of hsp70S5 promoter sequences.
Table S1. Numbers of silent and replacement fixed differences between O. pardalina hsp70 genes.
Table S2. Results of MK (McDonald-Kreitman) tests.
Figure S6. Alignment of Stratiomys hsp70 coding sequences.
Figure S7. Alignment of Stratiomys hsp70 5'-UTR sequences.
Figure S8. Alignment of hsp70S1 3'-UTR sequences.
Figure S9. Alignment of hsp70S2 and hsp70S3 3'-UTR sequences.
Figure S10. Alignment of hsp70S4 3'-UTR sequences.
Figure S11. Alignment of hsp70S5 3'-UTR sequences.
Figure S12. Deduced amino acids sequence of Hsp70S1 (S. singularior), Hsp70P1 and Hsp68 (both from O. pardalina). Comparison with paralogs from several organisms exhibiting homology to Stratiomyidae Hsp70.
Table S3: Primers for PCR and RACEs, used in the study.
Contributor Information
David G Garbuz, Email: Dgarbuz@yandex.ru.
Irina A Yushenova, Email: irinayushenova@mail.ru.
Olga G Zatsepina, Email: ozatsepina@yandex.ru.
Andrey A Przhiboro, Email: dipteran@mail.ru.
Brian R Bettencourt, Email: bbettencourt@alnylam.com.
Michael B Evgen'ev, Email: misha572001@yahoo.com.
Acknowledgements
Work was supported by the Russian Foundation for Basic Research, project No 09-04-00643 and 09-04-00660, project from "Genofond dynamics" program, Grant of the Program of Molecular and Cellular Biology RAN to M.E. and Russia President Grant to young scientists to D.G. No MK-1418.2010.4. We are grateful to Dr. E. Nudler and Dr. K. Shatalin from NYU for their advice and help. We thank Evrogen Company for PCR kits and kits for cDNA-synthesis.
References
- Schlesinger M, et al, editor. Heat Shock: from Bacteria to Man. New York, Cold Spring Harbor Lab; 1982. [Google Scholar]
- Lindquist S. The heat-shock response. Ann Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Ann Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Ann Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Dong Y, Miller LP, Sanders JG, Somero GN. Heat-shock protein 70 (Hsp70) expression in four limpets of the genus Lottia: interspecific variation in constitutive and inducible synthesis correlates with in situ exposure to heat stress. Biol Bull. 2008;2:173–181. doi: 10.2307/25470698. [DOI] [PubMed] [Google Scholar]
- Podrabsky JE, Lopez JP, Fan TW, Higashi R, Somero GN. Extreme anoxia tolerance in embryos of the annual killifish Austrofundulus limnaeus: insights from a metabolomics analysis. J Exp Biol. 2007;210:2253–2266. doi: 10.1242/jeb.005116. [DOI] [PubMed] [Google Scholar]
- Evgen'ev MB, Garbuz DG, Shilova V, Zatsepina OG. Molecular mechanisms underlying thermal adaptation of xeric animals. J Biosci. 2007;32:489–499. doi: 10.1007/s12038-007-0048-6. [DOI] [PubMed] [Google Scholar]
- Garbuz DG, Zatsepina OG, Przhiboro AA, Yushenova I, Guzhova IV, Evgen'ev MB. Larvae of related Diptera species from thermally contrasting habitats exhibit continuous up-regulation of heat shock proteins and high thermotolerance. Mol Ecol. 2008;17:4763–4777. doi: 10.1111/j.1365-294X.2008.03947.x. [DOI] [PubMed] [Google Scholar]
- Tissières A, Mitchell HK, Tracy UM. Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. J Mol Biol. 1974;84:389–398. doi: 10.1016/0022-2836(74)90447-1. [DOI] [PubMed] [Google Scholar]
- Evgen'ev MB, Sheinker VS, Levin AV. Molecular mechanisms of adaptation to hyperthermia in higher organisms. Synthesis of heat-shock proteins in cell cultures and larvae of certain silkworm species. Mol Biol. 1987;21:484–494. [Google Scholar]
- Ulmasov KA, Shammakov S, Karavaev K, Evgen'ev MB. Heat shock proteins and thermoresistance in lizards. Proc Natl Acad Sci USA. 1992;89:1666–1670. doi: 10.1073/pnas.89.5.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatsepina OG, Ulmasov KA, Beresten SF, Molodtsov VB, Rybtsov SA, Evgen'ev MB. Thermotolerant desert lizards characteristically differ in terms of heat-shock system regulation. J Exp Biol. 2000;203:1017–1025. doi: 10.1242/jeb.203.6.1017. [DOI] [PubMed] [Google Scholar]
- Evgen'ev MB, Zatsepina OG, Garbuz DG, Lerman DN, Velikodvorskaia VV, Zelentsova ES, Feder ME. Evolution and arrangement of the hsp70 gene cluster in two closely related species of the virilis group of Drosophila. Chromosoma. 2004;113:223–232. doi: 10.1007/s00412-004-0312-6. [DOI] [PubMed] [Google Scholar]
- Garbuz DG, Zatsepina OG, Feder ME, Evgen'ev MB. Evolution of termotolerance and the heat-shock response: evidence from inter/intra specific comparison and interspecific hybridization in the Drosophila virilis species group. I/Thermal phenotype. J Exp Biol. 2003;206:2399–408. doi: 10.1242/jeb.00429. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Wehner R. Heat shock protein synthesis and thermotolerance in Cataglyphis, an ant from the Sahara desert. Proc Natl Acad Sci USA. 1995;92:2994–2998. doi: 10.1073/pnas.92.7.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Hofmann GE. Thermal acclimation changes DNA-binding activity of heat shock factor 1 (HSF1) in the goby Gillichthys mirabilis: implications for plasticity in the heat-shock response in natural populations. J Exp Biol. 2002;20:3231–3240. doi: 10.1242/jeb.205.20.3231. [DOI] [PubMed] [Google Scholar]
- Tomanek L. The importance of physiological limits in determining biogeographical range shifts due to global climate change: the heat-shock response. Physiol Biochem Zool. 2008;81:709–717. doi: 10.1086/590163. [DOI] [PubMed] [Google Scholar]
- Feder ME, Krebs RA. In: Stress, Adaptation, and Evolution. Bijlsma R, Loeschcke V, editor. Basel: Birkhuser Verlag; 1997. Ecological and evolutionary physiology of heat-shock proteins and the stress response in Drosophila: complementary insights from genetic engineering and natural variation; pp. 155–173. [DOI] [PubMed] [Google Scholar]
- Pritchard G. Insects in thermal springs. Mem Entomol Soc Can. 1991;155:89–106. [Google Scholar]
- Yin X, Wang S, Tang J, Hansen JD. Thermal resistance of fifth-instar Cydia pomonella (L.) (Lepidoptera: Tortricidae) as affected by pretreatment conditioning. J Stor Prod Res. 2006;42:75–85. doi: 10.1016/j.jspr.2004.11.004. [DOI] [Google Scholar]
- Duggan IC, Boothroyd IKG, Speirs DA. Factors affecting the distribution of stream macroinvertebrates in geothermal areas: Taupo Volcanic Zone, New Zealand. Hydrobiologia. 2007;592:235–247. doi: 10.1007/s10750-007-0748-9. [DOI] [Google Scholar]
- Bettencourt BR, Feder ME. Hsp70 duplication in the Drosophila melanogaster species group: how and when did two become five? Mol Biol Evol. 2001;18:1272–1282. doi: 10.1093/oxfordjournals.molbev.a003912. [DOI] [PubMed] [Google Scholar]
- Bettencourt BR, Feder ME. Rapid concerted evolution via gene conversion at the Drosophila hsp70 genes. J Mol Evol. 2002;54:569–586. doi: 10.1007/s00239-001-0044-7. [DOI] [PubMed] [Google Scholar]
- Lerman DN, Feder ME. Naturally occurring transposable elements disrupt hsp70 promoter function in Drosophila melanogaster. Mol Biol Evol. 2005;22:776–783. doi: 10.1093/molbev/msi063. [DOI] [PubMed] [Google Scholar]
- Haney RA, Feder ME. Contrasting patterns of transposable element insertions in Drosophila heat-shock promoters. PLoS One. 2009;29:e8486. doi: 10.1371/journal.pone.0008486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Aguade M. Gene conversion is involved in the transfer of genetic information between naturally occurring inversions of Drosophila. Proc Natl Acad Sci USA. 1994;91:11517–11521. doi: 10.1073/pnas.91.24.11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhotia SC, Prasanth KV. Tissue- and development-specific induction and turnover of hsp70 transcripts from loci 87A and 87C after heat shock and during recovery in Drosophila melanogaster. J Exp Biol. 2002;205:345–358. doi: 10.1242/jeb.205.3.345. [DOI] [PubMed] [Google Scholar]
- Kourtidis A, Drosopoulou E, Nikolaides NE, Natzi VI, Chintiroglou CC, Scouras ZG. Identification of several cytoplasmic Hsp70 genes from the Mediterranean mussel (Mytilus galloprovincialis) and their long-term evolution in Mollusca and Metazoa. J Mol Evol. 2006;62:446–459. doi: 10.1007/s00239-005-0121-4. [DOI] [PubMed] [Google Scholar]
- Krebs RA. A comparison of Hsp70 expression and thermotolerance in adults and larvae of three Drosophila species. Cell Stress Chaperones. 1999;4:243–249. doi: 10.1379/1466-1268(1999)004<0243:ACOHEA>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi D, Engel MS. Evolution of the insects. Cambridge, New York, Melbourne: Cambridge University Press; 2005. [Google Scholar]
- Qin W, Tyshenko MG, Wu B, Walker V, Robertson R. Cloning and characterization of a member of the hsp70 gene family from Locusta migratoria, a highly thermotolerant species. Cell Stress Chaperones. 2003;8:144–152. doi: 10.1379/1466-1268(2003)008<0144:CACOAM>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D, Hartl DH. High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species group. Mol Biol Evol. 1998;15:293–302. doi: 10.1093/oxfordjournals.molbev.a025926. [DOI] [PubMed] [Google Scholar]
- Tian S, Haney RA, Feder ME. Phylogeny disambiguates the evolution of heat-shock cis-regulatory elements in Drosophila. PLoS One. 2010;5:e10669. doi: 10.1371/journal.pone.0010669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H. A method for estimating the mutation, gene conversion and recombination parameters in small multigene families. Genetics. 2002;161:865–872. doi: 10.1093/genetics/161.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorák J, Jue D, Lassner M. Homogenization of tandemly repeated nucleotide sequences by distance-dependent nucleotide sequence conversion. Genetics. 1987;116:487–498. doi: 10.1093/genetics/116.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin G. Characterization of the gene conversions between the multigene family members of the yeast genome. J Mol Evol. 2002;55:14–23. doi: 10.1007/s00239-001-0085-y. [DOI] [PubMed] [Google Scholar]
- Garland T, Adolph S. Why not to do two-species comparative studies: Limitations on inferring adaptation. Physiol Zool. 1994;67:797–828. [Google Scholar]
- Rozkošný R. A Biosystematic Study of the European Stratiomyidae (Diptera) Vol. 2. Junk Publishers, The Hague, The Netherlands; 1983. [Google Scholar]
- Sambrook J, Frisch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York; 1989. [Google Scholar]
- O'Farrell PH. High resolution two-dimentional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Zatsepina OG, Garbuz DG, Shilova V, Karavanov AA, Tornatore P, Evgen'ev MB. Use of surface-enhanced laser desorption ionization - time-of-flight to identify heat shock protein 70 isoforms in closely related species of the virilis group of Drosophila. Cell Stress Chaperones. 2005;10:12–16. doi: 10.1379/CSC-71.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Alignment of hsp70S1 promoter sequences.
Figure S2. Alignment of hsp70S2 promoter sequences.
Figure S3. Alignment of hsp70S3 promoter sequences.
Figure S4. Alignment of hsp70S4 promoter sequences.
Figure S5. Alignment of hsp70S5 promoter sequences.
Table S1. Numbers of silent and replacement fixed differences between O. pardalina hsp70 genes.
Table S2. Results of MK (McDonald-Kreitman) tests.
Figure S6. Alignment of Stratiomys hsp70 coding sequences.
Figure S7. Alignment of Stratiomys hsp70 5'-UTR sequences.
Figure S8. Alignment of hsp70S1 3'-UTR sequences.
Figure S9. Alignment of hsp70S2 and hsp70S3 3'-UTR sequences.
Figure S10. Alignment of hsp70S4 3'-UTR sequences.
Figure S11. Alignment of hsp70S5 3'-UTR sequences.
Figure S12. Deduced amino acids sequence of Hsp70S1 (S. singularior), Hsp70P1 and Hsp68 (both from O. pardalina). Comparison with paralogs from several organisms exhibiting homology to Stratiomyidae Hsp70.
Table S3: Primers for PCR and RACEs, used in the study.