Abstract
Many bacterial pathogens inject a cocktail of effector proteins into host cells through type III secretion systems. These effectors act in concert to modulate host physiology and immune signaling, thereby promoting pathogenicity. In search for additional Pseudomonas syringae effectors in suppressing plant innate immunity triggered by pathogen or microbe-associated molecular patterns (PAMPs/MAMPs), we identified P. syringae tomato DC3000 effector HopF2 as a potent suppressor of early immune response gene transcription and MAP kinase (MAPK) signaling activated by multiple MAMPs, including bacterial flagellin, elongation factor Tu, peptidoglycan, lipopolysaccharide and HrpZ1 harpin, and fungal chitin. The conserved surface-exposed residues of HopF2 are essential for its MAMP suppression activity. HopF2 is targeted to the plant plasma membrane through a putative myristoylation site, and the membrane association appears to be required for its MAMP suppression function. Expression of HopF2 in plants potently diminished the flagellin-induced phosphorylation of BIK1, a plasma membrane-associated cytoplasmic kinase, which is rapidly phosphorylated within one minute upon flagellin perception. Thus, HopF2 likely intercepts MAMP signaling at the plasma membrane immediately upon signal perception. Consistent with the potent suppression function of multiple MAMP signaling, expression of HopF2 in transgenic plants compromised plant nonhost immunity to bacteria P. syringae pv. phaseolicola, and plant immunity to a necrotrophic fungal pathogen Botrytis cinerea.
INTRODUCTION
Plants have evolved robust immune systems to defend against pathogen threats. The innate immune responses are launched by recognition of conserved pathogen or microbe-associated molecular patterns (PAMPs/MAMPs) through host pattern-recognition receptors (PRRs) (Jones and Dangl, 2006; Boller and Felix, 2009). In plants, the plasma membrane localized leucine-rich repeat-receptor-like kinases (LRR-RLKs) play a pivotal role in detecting MAMPs and mount PAMP-triggered immunity (PTI). Two well-characterized Arabidopsis MAMP receptors are FLS2 that perceives a conserved 22-amino acid peptide (flg22) from bacterial flagellin, and EFR that recognizes bacterial elongation factor EF-Tu (Gomez-Gomez and Boller, 2000; Zipfel et al., 2006). Upon flagellin perception, FLS2 associates with BAK1, another RLK with a short extracellular LRR domain (Chinchilla et al., 2007; Heese et al., 2007). BAK1 was originally identified as a BRI1-associated receptor kinase mediating plant hormone brassinosteroid (BR) signaling (Li et al., 2002; Nam and Li, 2002). FLS2-BAK1 heteromerization occurs almost instantaneously upon flg22 perception, which likely leads to the subsequent phosphorylation events of FLS2 and BAK1 (Schulze et al., 2010). Significantly, BAK1 is functionally involved in multiple MAMP signaling, and BAK1 heteromerizes with several LRR-RLKs including BRI1, FLS2 and EFR instantaneously after ligand binding (Schulze et al., 2010). Flg22 also rapidly induces the FLS2- and BAK1-dependent phosphorylation of a receptor-like cytoplasmic kinase BIK1 within a minute upon signal perception (Lu et al., 2010; Zhang et al., 2010). BIK1 forms a complex with FLS2/BAK1 in the plasma membrane, and transphosphorylates FLS2/BAK1 receptor complex (Lu et al., 2010). Downstream intracellular signaling events in PTI include changes in cytoplasmic Ca2+ levels, activation of MAP kinase (MAPK) cascades, induction of defense-related genes, production of reactive oxygen species, deposition of callose to reinforce the cell wall and stomatal closure to prevent pathogen entry (Ausubel, 2005; Boller and Felix, 2009).
To achieve a successful infection, host-adapted pathogens acquired the ability to interfere with plant immunity. Many bacterial effectors secreted through the type III secretion system (TTSS) have been shown to sabotage plant innate immunity. Great strides are being made in the understanding of the biological and enzymatic functions of individual effectors in the past few years with the development of new technologies and more sensitive and quantitative assays (Speth et al., 2007; Block et al., 2008; Gohre and Robatzek, 2008; Lewis et al., 2009; Hann et al., 2010). The P. syringae effector HopU1, a mono-ADP-ribosyltransferase (ADP-RT), modifies several Arabidopsis RNA-binding proteins, which represent a novel class of ADP-RT substrates (Fu et al., 2007). HopM1, another P. syringae effector, targets and degrades a member of the ARF family of guanine nucleotide exchange factors involved in vesicle trafficking (Nomura et al., 2006). We and several other groups discovered that two sequence distinct but functionally related effectors, AvrPto and AvrPtoB, are potent suppressors of multiple MAMP signaling by targeting MAMP receptor complexes (de Torres et al., 2006; He et al., 2006; Hann and Rathjen, 2007; Gohre et al., 2008; Shan et al., 2008; Xiang et al., 2008; Gimenez-Ibanez et al., 2009). In addition to effectors from P. syringae, DspA/E from Erwinia amylovora interacts with several RLKs (Meng et al., 2006), and XopN from Xanthomonas interacts with a tomato atypical RLK and suppresses PTI (Kim et al., 2009). A viral effector protein has also been reported to interact with RLKs (Fontes et al., 2004). Notably, the contribution of individual effectors to virulence is difficult to observe with bacterial deletion mutants likely due to the potential functional redundancy. It is not clear how these effectors cooperatively suppress host immune signaling in plant cells.
Although AvrPto and AvrPtoB are potent suppressors of plant PTI, deletion of AvrPto and AvrPtoB from P. syringae pv. tomato DC3000 only partially reduced its suppression activity on MAMP marker gene expression, suggesting additional effector(s) exhibiting redundant functions with AvrPto and AvrPtoB in suppressing PTI (He et al., 2006). We identified another DC3000 effector HopF2 that possesses similar function with AvrPto in suppressing plant PTI. HopF2 is a homolog of P. syringae pv. phaseolicola effector AvrPphF, which contributes to distinct avirulence and virulence functions in different bean and soybean cultivars (Jackson et al., 1999; Tsiamis et al., 2000; Shan et al., 2004). HopF2 from DC3000 possesses a putative myristoylation site that is important for its membrane localization and avirulence and virulence functions in tobacco and tomato (Robert-Seilaniantz et al., 2006). In Arabidopsis, HopF2 can block basal resistance as measured by increased vascular flow (Oh and Collmer, 2005), flg22-induced NHO1 induction (Li et al., 2005) and callose deposition (Guo et al., 2009), and HopA1-induced cell death (Jamir et al., 2004).
Recently, it has been shown that HopF2 targets RIN4, an important component in both PTI and effector-triggered immunity (ETI) (Wilton et al., 2010). HopF2 interfered with AvrRpt2-induced RIN4 degradation, thereby suppressing ETI mediated by AvrRpt2. HopF2 also targeted MAPK kinase 5 (MKK5) and likely other MKKs to inhibit PTI signaling in Arabidopsis (Wang et al., 2010). HopF2 possesses ADP-ribosyltransferase activity and ADP-ribosylated MKK5 in vitro. Interestingly, RIN4 was also able to be ADP-ribosylated by HopF2 (Wang et al., 2010). We report here that HopF2 suppresses diverse immune signaling triggered by multiple MAMPs at an immediately early step in PTI signaling. Importantly, HopF2 suppresses flg22-mediated BIK1 phosphorylation, an event upstream of MAPK cascade in PTI signaling. Expression of HopF2 in transgenic Arabidopsis compromised plant immunity to a non-adaptive bacterium and necrotrophic fungus. The putative myristoylation site of HopF2 is required for its MAMP suppression function.
RESULTS
HopF2 intercepts immune signaling activated by multiple MAMPs
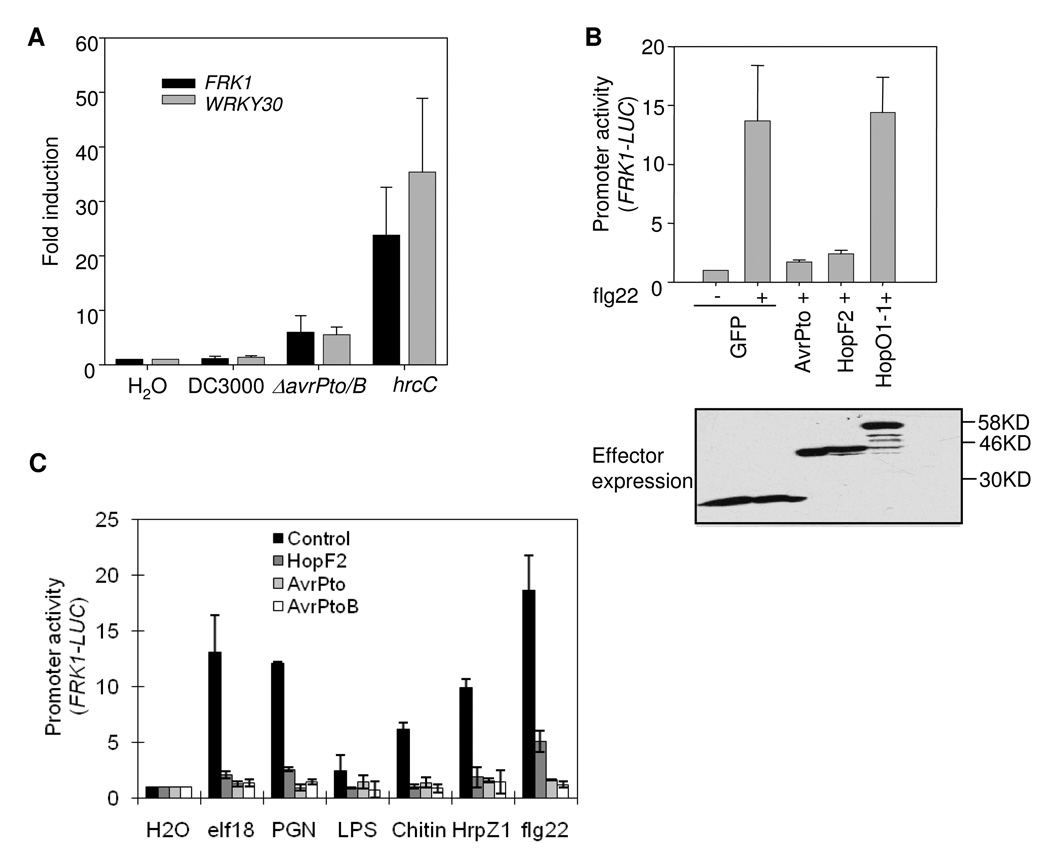
We have shown before that virulent bacteria DC3000, but not its TTSS mutant hrcC, suppresses MAMP-specific early defense gene activation (He et al., 2006). A cell-based genetic screen has identified AvrPto and AvrPtoB as potent suppressors of early MAMP signaling. However, the DC3000ΔavrPtoΔavrPtoB double mutant only partially reduced the suppression activity of DC3000 on MAMP marker gene expression (Fig. 1A). As shown by quantitative RT-PCR analysis, DC3000 hrcC strongly activated the expression of two early MAMP marker genes FRK1 and WRKY30 in Arabidopsis plants 6 hr after infiltration. The induction of these genes was not observed by DC3000 due to the collective suppression activity of type III effectors. Deletion of avrPto and avrPtoB from DC3000 partially restored FRK1 and WRKY30 expression, but to a much less extent than DC3000 hrcC. The data suggest that additional effectors likely exist in the suppression of early MAMP signaling.
Fig. 1. HopF2 suppresses immune signaling triggered by multiple MAMPs.
A. Real-time RT-PCR analysis of FRK1 and WRKY30 induction. Four-week-old Arabidopsis plants were inoculated with H2O, DC3000, DC3000 ΔavrPto/avrPtoB, or DC3000hrcC at 1 × 108 cfu/ml. The samples were collected 6 hr later for RNA isolation. The gene induction (fold change) by bacterial infiltration was compared with the expression level of H2O infiltration.
B. HopF2 suppresses flg22-induced FRK1-LUC activation. Protoplasts were co-transfected with GFP control or an effector and FRK1-LUC reporter. Three hours later, transfected protoplasts were treated with 10 nM flg22 for another 3 hr. The expression of effectors was detected by anti-GFP Western blot.
C. HopF2 suppresses multiple MAMP-mediated FRK1-LUC activation. Protoplasts were transfected with FRK1-LUC with or without HopF2, AvrPto or AvrPtoB for different MAMP treatments. The concentrations for different MAMPs are flg22, 10 nM; elf18, 10 nM; HrpZ1, 100 nM; PGN, 50 µg/ml; chitin, 50 µg/ml; and LPS, 50 µg/ml.
We cloned several additional bacterial effectors tagged with GFP in a plant expression vector and determined their effect on the flg22 activation of the FRK1-LUC reporter with Arabidopsis protoplast transient assay. Significantly, HopF2, but not HopO1-1, suppressed flg22 activation of FRK1-LUC. The effect of HopF2 on suppressing flg22-mediated gene transcription was almost as potent as AvrPto (Fig. 1B). The data also confirmed Wang et al.’s report about the suppression of flg22-induced FRK1 by HopF2 (Wang et al., 2010). Plants respond to multiple MAMPs and activate the convergent signaling. To determine whether HopF2 interrupts the immune response activated by other MAMPs in addition to flg22, we treated HopF2 transfected protoplasts with elf18 (the 18 amino acid peptide of EF-Tu), peptidoglycan (PGN), lipopolysaccharide (LPS), HrpZ1 harpin, and chitin. Similar to AvrPto and AvrPtoB, HopF2 dramatically inhibited the activation of FRK1 promoter by all these MAMPs (Fig. 1C). The results indicate that HopF2 suppresses the immune signaling triggered by multiple MAMPs and likely targets to a convergent component in plant innate immunity.
HopF2 attenuates MAPK signaling
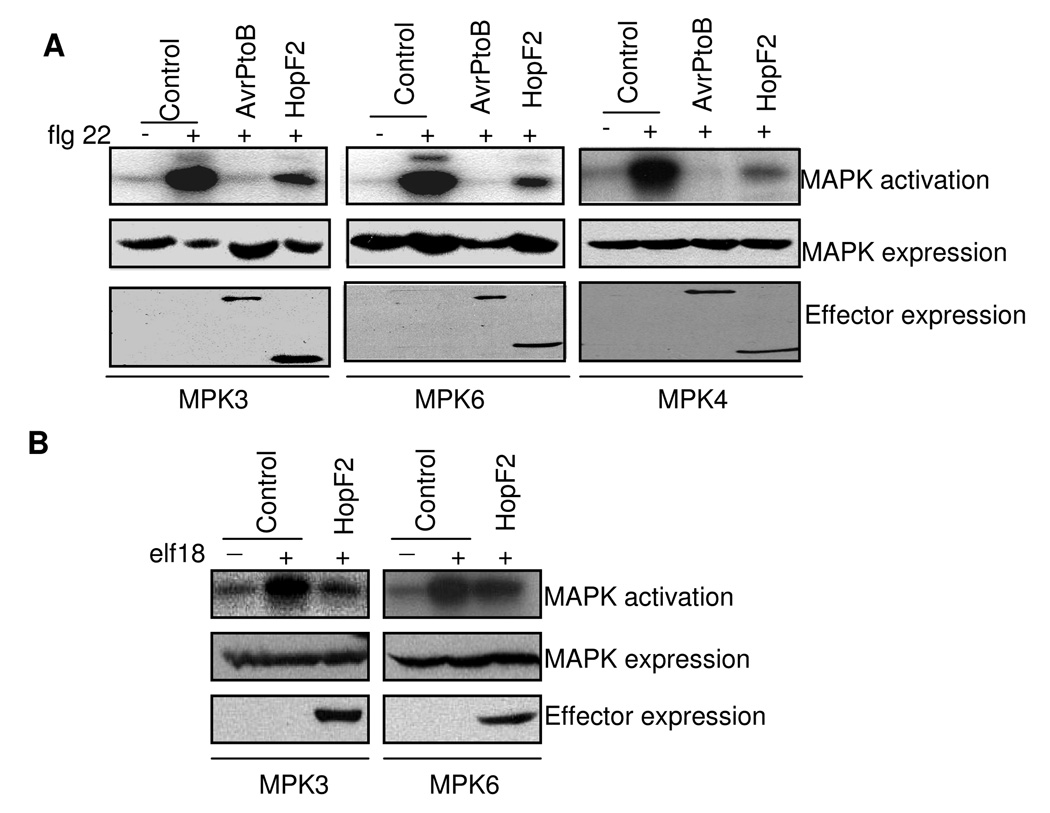
MAPK activation is a convergent early signaling event activated by different MAMPs in plants. It has been reported that flg22 activates MPK3, MPK4 and MPK6 in Arabidopsis (Asai et al., 2002; Suarez-Rodriguez et al., 2007; Rodriguez et al., 2010). To determine whether HopF2 suppresses the flg22 activation of MAPKs, we performed an immunocomplex kinase assay. Arabidopsis protoplasts were co-transfected with HA-tagged MAPK and GFP-tagged effectors. The MAPK activation was stimulated with flg22 treatment for 10 min. Consistent with previous report, expression of AvrPtoB completely blocked the activation of MPK3 and MPK6 by flg22 (Fig. 2A). Similarly, expression of HopF2 in protoplasts significantly diminished the flg22 activation of MPK3 and MPK6. In addition, both AvrPtoB and HopF2 suppressed the activation of MPK4 by flg22 (Fig. 2A). These results confirmed the recent report by Wang et al. (2010). The activation of MPK3 and MPK6 by elf18 was also largely abolished by the expression of HopF2 (Fig. 2B). The data indicate that HopF2 suppresses MAMP-induced early gene transcription and MAPK activation.
Fig. 2. HopF2 attenuates MAP kinase signaling.
A. HopF2 suppresses flg22-mediated MPK3, MPK4 and MPK6 activation. HA-tagged MPK3, MPK4 or MPK6 was co-expressed with GFP-tagged effectors. Transfected protoplasts were incubated for 6 hr before 1 µM flg22 treatment for 10 min. An anti-HA antibody was used for immunoprecipitation of MAPKs. Kinase activity was detected by an in-vitro kinase assay (top). Protein expression is shown for MAPKs (middle) and effectors (bottom).
B. HopF2 suppresses elf18-mediated MPK3 and MPK6 activation. Transfected protoplasts were treated with 1 µM elf18 for 10 min.
HopF2 intercepts flg22-induced BIK1 phosphorylation
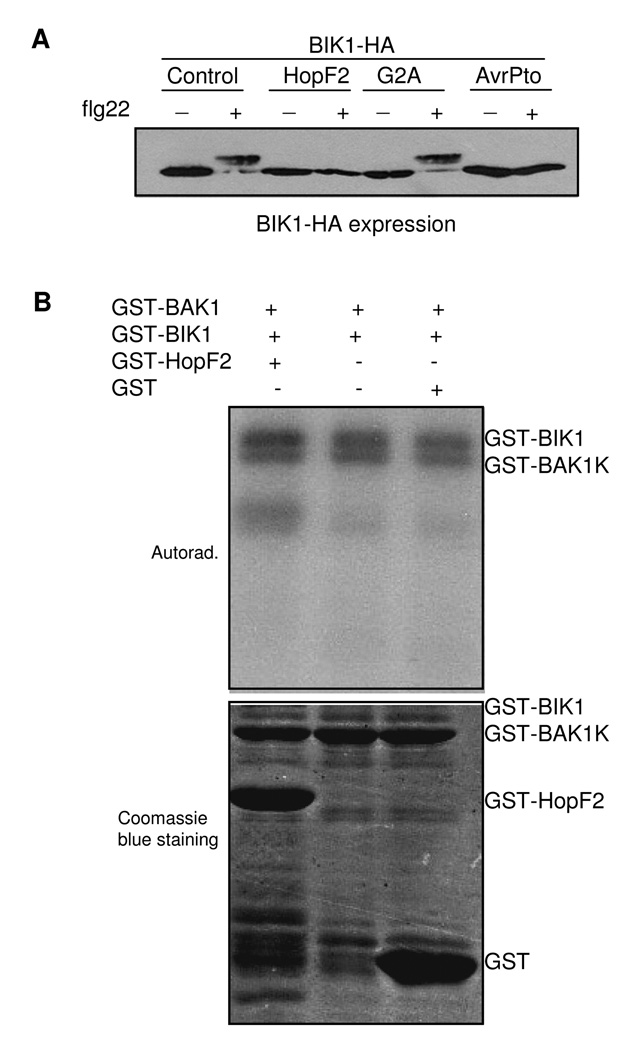
We recently showed that flg22 rapidly induces a cytoplasmic kinase BIK1 phosphorylation within a minute upon signal perception (Lu et al., 2010). BIK1 is a plasma membrane localized protein forming a complex with FLS2/BAK1(Veronese et al., 2006; Lu et al., 2010). Significantly, flg22-induced BIK1 phosphorylation was suppressed by expression of HopF2 in protoplasts (Fig. 3A). BIK1 phosphorylation was indicated as mobility shift upon flg22 stimulation. This mobility shift was not observed when BIK1 was co-expressed with HopF2 in protoplasts (Fig. 3A). Expression of AvrPto also completely blocked BIK1 phosphorylation. The data are consistent with the plasma membrane localization of HopF2, and suggest that HopF2 suppresses MAMP-mediated signaling at an immediately early step. We further tested whether HopF2 could potentially target and/or de-phosphorylate BIK1 phosphorylation in vitro. HopF2 did not interact with BIK1 in a co-immunoprecipitation assay (Fig. S1) and yeast two-hybrid assay (data not shown). Furthermore, HopF2 did not affect BIK1 autophosphorylation and its phosphorylation on BAK1with an in vitro kinase assay (Fig. 3B), indicating that HopF2 does not directly target and inhibit BIK1 kinase activity.
Fig. 3. HopF2 suppresses flg22-induced BIK1 phosphorylation in vivo.
A. HopF2 blocks flg22-induced BIK1 phosphorylation. Protoplasts were co-transfected with BIK1-HA and GFP-tagged HopF2, HopF2G2A or AvrPto for 6 hr and treated with 1 µM flg22 for 10 min.
B. HopF2 did not affect BIK1 autophosphorylation and phosphorylation on BAK1. An in vitro kinase assay was performed by incubating GST-BIK1, GST-BAK1K with or without GST-HopF2. Proteins were separated with SDS-PAGE and analyzed by autoradiography (top panel). The top band is autophosphorylated GST-BIK1, and then phosphorylated GST-BAK1K. The protein loading control was shown by Coomassie blue staining (bottom panel).
AvrPphC does not affect HopF2 MAMP suppression function in Arabidopsis
In some bean cultivars, the effector AvrPphC from P. syringae pv. phaseolicola suppresses AvrPphF avirulence function mediated by the R1 resistance protein (Tsiamis et al., 2000). We tested whether AvrPphC could interfere with HopF2 MAMP suppression function. We co-expressed HopF2 and AvrPphC, and tested the activation of FRK1-LUC upon flg22 treatment in protoplasts. As expected, HopF2 significantly suppressed flg22-induced FRK1-LUC activation. AvrPphC itself had no effect on FRK1-LUC induction. The expression of AvrPphC did not ameliorate the strong suppression by HopF2 on FRK1-LUC induction (Fig. S2), suggesting that AvrPphC did not affect HopF2 virulence in Arabidopsis.
Myristoylation and conserved surface residues of HopF2 are essential for its MAMP suppression activity
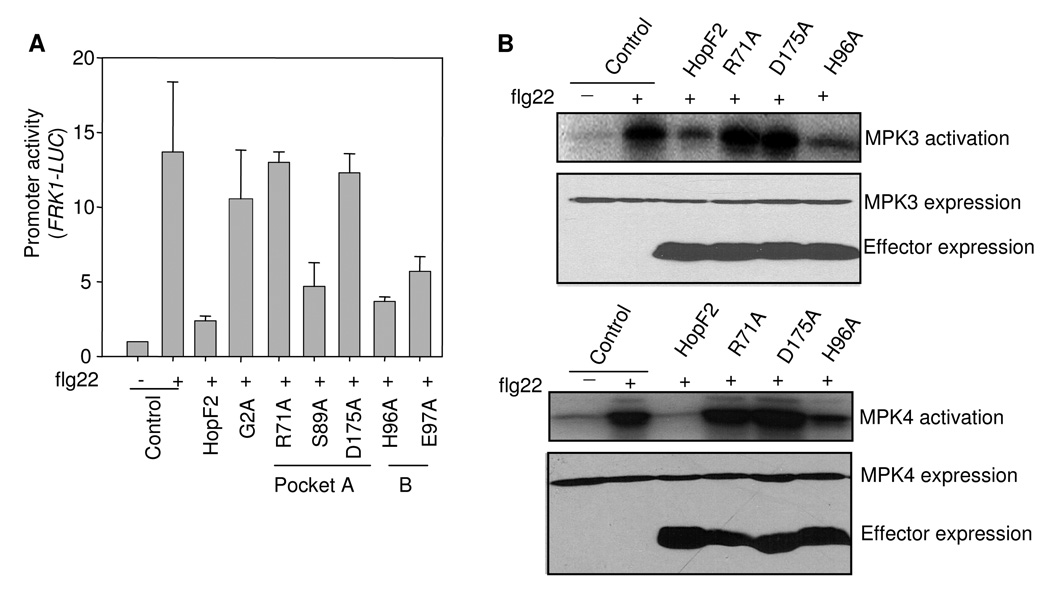
HopF2 has a putative myristoylation site at its N-terminus, and mutation of this site disrupts its membrane localization (Robert-Seilaniantz et al., 2006). More importantly, the putative myristoylation site is required for HopF2 avirulence and virulence functions in tobacco and tomato. We examined whether the putative myristoylation site is essential for HopF2 suppression function in MAMP signaling. We substituted the putative myristoylation site Gly with Ala (G2A) and tested its effect on flg22-induced FRK1-LUC activation. As shown in Fig. 4A, HopF2 G2A mutation was unable to suppress flg22 induction of FRK1-LUC. In addition, HopF2 G2A also lost the ability to block flg22-mediated BIK1 phosphorylation (Fig. 3A). Taken together, the data suggest that plasma membrane localization of HopF2 is required for its MAMP suppression activity and HopF2 likely targets to a membrane-associated component for its MAMP suppression function.
Fig. 4. Myristoylation and conserved surface residues of HopF2 are required for its MAMP-suppression function.
A. G2, R71 and D175 are required for HopF2 suppression of FRK1 induction. Protoplasts were co-transfected with HopF2 or its mutants and FRK1-LUC reporter, and incubated for 3 hr before treated with 10 nM flg22 for another 3 hr.
B. R71 and D175 are required for HopF2 suppression of MAPK activation. HA-tagged MAPK was co-expressed with HA-tagged HopF2 or its mutants for 6 hr before 1 µM flg22 treatment for 10 min. Kinase activity was detected by an in-vitro kinase assay (top). Protein expression is shown for MAPKs and effectors (bottom).
Structural analysis of AvrPphF identified two clusters of conserved surface-exposed residues based on certain structural similarity to the catalytic domain of bacterial ADP-RTs (Singer et al., 2004). Mutational analysis of AvrPphF indicated that R72 and D174 in pocket A are required for its virulence function in susceptible bean cultivar Tendergreen and its avirulence function in resistance cultivar Red Mexican (Singer et al., 2004). S90 in pocket A, and H97 and E98 in pocket B are less essential for AvrPphF virulence and avirulence functions. We individually substituted the corresponding residues in HopF2 with Ala and tested its ability to suppress flg22 activation of FRK1-LUC. Significantly, the R71A and D175A mutations of HopF2 in pocket A, which are the equivalent of R72 and D174 in AvrPphF, completely lost the ability of suppressing flg22 activation of FRK1-LUC (Fig. 4A). The results confirmed the recent report by Wang et al. (2010). The mutations in S89, H96 and E97 of HopF2 had little or no effect on its MAMP suppression activity. We further tested whether R71 and D175 are required for HopF2 to suppress flg22-induced MAPK activation with an immunocomplex kinase assay. As shown in Fig 4B, although wild-type HopF2 dramatically suppressed flg22-activated MPK3 and MPK4, HopF2 R71A and D175A were no longer able to suppress MPK3 and MPK4 activation by flg22. H96A still kept the ability to block MAPK activation. The protein expression levels of different HopF2 mutants were comparable with that of wild-type HopF2 (Fig. 4B).
HopF2 is a suppressor of plant nonhost immunity
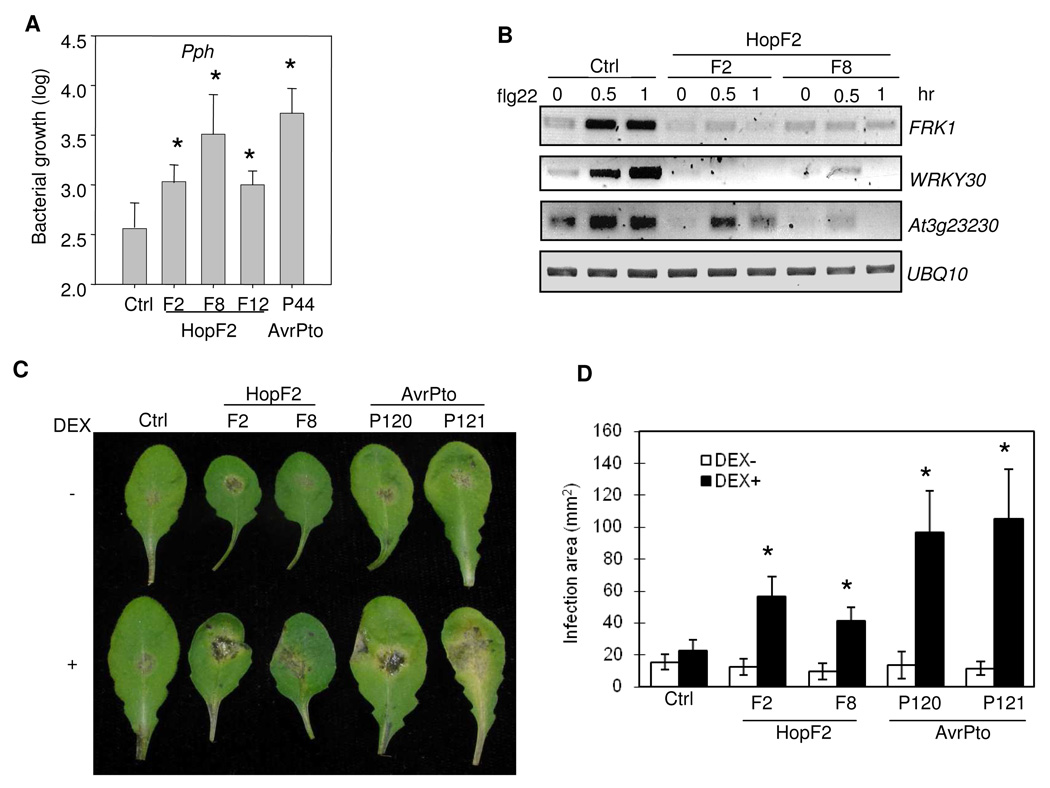
Plant nonhost immunity is in part attributed by plant PTI (Li et al., 2005; He et al., 2006; Ham et al., 2007). To test whether HopF2 could compromise plant nonhost immunity, we generated HopF2 transgenic plants under the control of the dexamethasone-inducible promoter. We examined the growth of a non-adaptive bacterial strain P. syringae pv. phaseolicola (Pph) in wild-type plants and transgenic plants expressing HopF2. Three days after infection, bacterial populations in three independent lines of HopF2 transgenic plants were about 5- to 10-fold higher than that in wild-type control plants (Fig. 5A). The susceptibility of HopF2 transgenic plants to Pph infection was comparable to that of AvrPto transgenic plants (Fig. 5A).
Fig. 5. HopF2 suppresses plant nonhost resistance.
A. HopF2 transgenic plants support non-adaptive bacteria growth. Four-week-old plants (Col-0 control, HopF2 or AvrPto transgenic plants) were sprayed with 10 µM DEX for 24 hr before bacterial inoculation. Arabidopsis leaves were inoculated with Pph NPS3121 (Pph) at 5 × 105 cfu/ml. The bacterial counting was performed 3 days after inoculation. * indicates a significant difference with p<0.05 when compared with data from control plants based on the results of an unpaired Student’s t-test.
B. RT-PCR analysis of MAMP marker gene induction. Ten-day-old seedlings were treated with 10 µM DEX for 24 hr, then treated with 10 nM flg22 for 0.5 and 1 hr.
C. Susceptibility to botrytis. Four-week-old plants were sprayed with 10 µM DEX for 24 hr before Botrytis cinerea strain BO5 were sprayed at concentration of 105 spores/ml. Disease symptom was recorded 2 days after infection.
D. Lesion development of B. cinerea infection. The area of infection was measured 4 days postinoculation. * indicates a significant difference with p<0.05.
Consistent with transient expression of HopF2 in protoplasts, the expression of HopF2 in stable transgenic plants suppressed flg22-activation of endogenous MAMP marker genes by RT-PCR analysis (Fig. 5B). FRK1, WRKY30 and At3g23230 were strongly induced 0.5 and 1 hr after flg22 treatment in control plants, while their induction was largely diminished in HopF2 transgenic plants (Fig. 5B). Expression of HopF2 in plant cells suppressed the immune responses triggered by MAMPs from both bacteria and fungi. We found that HopF2 transgenic plants were more susceptible to a necrotrophic fungal pathogen Botrytis cinerea than wild-type control plants (Fig. 5C and Fig. 5D). Compared with wild-type plants, HopF2 transgenic plants showed increased disease symptoms with chlorosis and necrosis (Fig. 5C) and enlarged infection areas (Fig. 5D) after Botrytis cinerea infection. The similar results were observed with AvrPto transgenic plants (Fig. 5C and Fig. 5D).
DISCUSSION
To combat the pathogenic invaders, plants have evolved strategies and tactics to instantaneously recognize an array of MAMPs and launch a complex network of immune signaling. We report here that P. syringae effector HopF2 potently suppresses diverse immune signaling events triggered by conserved microbial components of bacteria and fungi. The suppression apparently occurs at a very early and essential step immediately upon MAMP perception on the plasma membrane. HopF2 is a membrane-localized protein with a putative myristoylation site (Robert-Seilaniantz et al., 2006). The membrane association is required for its MAMP suppression function. Consistently, HopF2 intercepts flg22-mediated phosphorylation of BIK1, a plasma membrane localized kinase interacting with FLS2/BAK1 complex (Lu et al., 2010; Zhang et al., 2010). Apparently, HopF2 does not directly interact with BIK1 or inhibit BIK1 kinase activity (Fig. 3B and S1). BAK1, likely functioning upstream of BIK1 (Lu et al., 2010), is required for multiple MAMP responses and appears to function in distinct receptor signaling complexes to integrate MAMP perception into downstream signaling events (Chinchilla et al., 2007; Heese et al., 2007; Schulze et al., 2010). It is likely that HopF2 also targets BAK1 or other convergent component residing in the plasma membrane in transducing MAMP signaling.
Host-adapted pathogens employ various means to dampen host immunity. P. syringae injects about 30 virulence effectors into plant cells (Lindeberg et al., 2006; Lindeberg et al., 2009). These effectors deploy diverse biochemical activities to jeopardize various plant cellular processes. Many elegant studies have reconciled a general pattern that a single effector may target multiple host factors at critical steps in key host processes and distinct effectors can converge on a specific host target or cellular compartment to achieve the virulence robustness and redundancy (Lewis et al., 2009; Hann and Rathjen, 2010). It remains challenge to address how the discrete activities of these effectors are precisely coordinated and temporally controlled. Several Salmonella effectors (SipA, SipC, SopB, SopE, SopE2 and SptP) and translocase SipB are all delivered to the cell plasma membrane, suggesting that plasma membrane is a critical interface for effector-effector interplay and effector-target interaction during bacterial entry (Cain et al., 2004; Raffatellu et al., 2005; McGhie et al., 2009). Sophisticated functional interactions between these Salmonella effectors following their delivery into the host cells were revealed by a systemic experimental screen (Cain et al., 2008). Localization studies suggest that several P. syringae effectors localize to plasma membrane in plant cells through the putative myristoylation site (Nimchuk et al., 2000; Shan et al., 2000; Robert-Seilaniantz et al., 2006), implying the plant plasma membrane as a battlefield for coordinated effector actions to promote bacterial invasion. Indeed, several plasma membrane localized effectors, including HopF2, AvrPto and AvrRpm1, are potent suppressors of PTI signaling (Kim et al., 2005; He et al., 2006; Wang et al., 2010). It is of great interest to study how these effectors work in harmony in suppressing plant immune signaling under natural infection condition.
Recently, HopF2 has been shown to directly interact with Arabidopsis RIN4, a virulence target of three additional P. syringae effectors, AvrRpm1, AvrRpt2 and AvrB (Wilton et al., 2010). RIN4 is a plasma membrane localized protein interacting with two resistance proteins RPM1 and RPS2 (Mackey et al., 2002; Axtell and Staskawicz, 2003; Mackey et al., 2003; Liu et al., 2009). RIN4 is phosphorylated upon infection by bacteria carrying AvrRpm1 or AvrB, whereas RIN4 is cleaved by AvrRpt2 to induce effector-triggered immunity (ETI). Expression of HopF2 in transgenic plants suppressed AvrRpt2-induced RIN4 cleavage and AvrRpt2-mediated ETI (Wilton et al., 2010). Importantly, HopF2 promoted P. syringae growth in a RIN4-dependent manner, suggesting that RIN4 is a virulence target of HopF2 in Arabidopsis (Wilton et al., 2010). Apparently, targeting RIN4 does not count for the full virulence function of HopF2 since it has not been reported that RIN4 is deficient in flg22-induced MAPK activation and early gene transcription. Instead, RIN4 has been proposed to be a negative regulator of PTI (Mackey et al., 2002; Kim et al., 2005).
During the preparation of this manuscript, Wang et al. reported that HopF2 inhibited flg22 signaling by targeting MAPK kinase 5 (MKK5) and other MKKs. HopF2 ADP-ribosylates MKK5 in vitro (Wang et al., 2010). HopF2 is a membrane localized protein, and membrane association is required for its MAMP suppression and virulence and avirulence functions in different plant species. It remains unknown where MKKs localize and how HopF2 interacts with diverse MKKs in plant cells. Apparently, flg22 signaling constitutes MAPK-dependent and MAPK-independent pathways (Asai et al., 2002; Boudsocq et al., 2010). Given the potent virulence function of HopF2 in suppressing multiple MAMP signaling, it is likely that HopF2 targets both MAPK-dependent and MAPK-independent pathways or a convergent upstream component immediately after MAMP perception. Consistent with this, expression of HopF2 inhibited flg22-mediated phosphorylation of BIK1, which is phosphorylated within a minute upon flg22 perception. Accumulating evidence suggests that BIK1 likely functions upstream or independent of MAPK cascade in plant innate immunity (Veronese et al., 2006; Lu et al., 2010; Zhang et al., 2010). In particular, it has been shown that the MKK inhibitor did not suppress flg22-induced BIK1 phosphorylation and active MKK5 did not phosphorylate BIK1, suggesting that MKK5 is not required for BIK1 phosphorylation (Lu et al., 2010). Thus, our data implicate that HopF2 targets a plasma membrane localized component upstream of BIK1 phosphorylation and MAPK cascade in PTI signaling. Together with the data by Wang et al. (2010) and Wilton et al. (2010), it appears that HopF2 targets multiple host factors in suppressing plant PTI and ETI signaling.
MATERIALS AND METHODS
Plant growth and pathogen assays
Arabidopsis plants were grown in a growth chamber at 23°C with a 13 hr photoperiod for 30 days before RNA isolation, bacterial inoculation or protoplast isolation. Different P. syringae strains were grown overnight at 28°C in the KB medium with appropriate antibiotics. The final concentration of antibiotics as follows: Kanamycin (Km, 50mg/l), Tetracycline (Tc, 20mg/l), Streptomycin (Sm, 100mg/l), Spectinomycin (Sp, 50mg/l) and Gentamycin (Gm, 10mg/l). Bacteria were pelleted by centrifugation, washed and diluted to the desired density with 10 mM MgCl2. Arabidopsis leaves were infiltrated with bacteria using a needleless syringe for RNA isolation and bacterial counting. To measure bacterial growth, two leaf discs were ground in 100 µl H2O and serial dilutions were plated on KB medium with appropriated antibiotic. Bacterial colony forming units (cfu) were counted 2 days after incubation at 28°C. Each data point is shown as triplicates.
Botrytis cinerea strain BO5 was cultured on Potato Dextrose Agar (Difco) and incubated at room temperature. Conidia were resuspended in distilled water and spore concentration adjusted to 105 spores/ml. Gelatin (0.5%) was added to conidial suspension before inoculation. Spores were sprayed onto whole Arabidopsis plants and disease development monitored over a period of 5 days.
Generation of HopF2 transgenic plants
HopF2 transgenic plants were generated by Agrobacterium-mediated transformation with the HopF2 construct under the control of the dexamethasone-inducible promoter with an HA epitope tag (McNellis et al., 1998). To induce HopF2 expression, transgenic plants were sprayed with 10 µM dexamethasone containing 0.025% silwet L-77 one day before infiltration with flg22 or bacteria for RT-PCR and disease assays.
Plasmid constructs and point mutations
The effector constructs were made by cloning PCR fragments from Pst DC3000 genomic DNA into a plant expression vector with a GFP tag at the C terminus. HopF2 was amplified with primers 5’-CGGGATCCATGGGTAATATTTGCGGCACC-3’ and 5’-GAAGGCCTGACCCTTTCGACCGGCACTTTC-3’, in which the ATA start codon was replaced with ATG; HopO1-1: 5’-CGGGATCCATGGGTAATATTTGTGGTAC-3’ and 5’-GAAGGCCTCTCGTCAGAGCTCTCTGC-3’; AvrPphC: 5’-CATGCCATGGGAAATGTTTGTTTCCG-3’ and 5’-GAAGGCCTCTGAGGGGGCCGCTCAAAAAG-3’. The other effector, reporter and MAPK constructs were reported previously (He et al., 2006).
Point mutations were generated by site-specific mutagenesis kit (Stratagene) using following primers, 5’-CGGGATCCATGGCTAATATTTGCGGCACC-3’ and 5’-GGTGCCGCAAATATTAGCCATGGATCCCG-3’; R71A: 5’-GATACCGAGCTTTTCGCAACGACGGATAGTCGC-3’ and 5’-GCGACTATCCGTCGTTGCGAAAAGCTCGGTATC-3’; S89A: 5’-GCGGGCAATCCACAAGCCATGGCGAGTATCC-3’ and 5’-GGATACTCGCCATGGCTTGTGGATTGCCCGC-3’; H96A: 5’-CAGGTGCCCAACCAGCCGAAGCAAGGGCGTAC-3’ and 5’-GTACGCCCTTGCTTCGGCTGGTTGGGCACCTG-3’; E97A: 5’-GTGCCCAACCACACGCAGCAAGGGCGTACG-3’ and 5’-CGTACGCCCTTGCTGCGTGTGGTTGGGCAC-3’; D175A: 5’-GCAAGGTCTATGCCGCCGCTTCGTCTGTAGC-3’ and 5’-GCTACAGACGAAGCGGCGGCATAGACCTTGC-3’.
Arabidopsis protoplast transient expression and MAPK assays
Protoplast transient expression and in-vitro MAPK assays were carried out as described previously (He et al., 2006; He et al., 2007). UBQ10-GUS was cotransfected with FRK1-LUC as an internal control, and the promoter activity was presented as LUC/GUS ratio. Protoplasts were collected 6 hr after transfection for protein expression, kinase activity and promoter activity assays. Protoplasts transfected with plasmid DNA without effectors were used as controls.
RT-PCR analysis
Total RNA was isolated from leaves after bacterial or flg22 treatment by using TRIzol Reagent (Invitrogen). Complementary DNA was synthesized from 1.5 µg of total RNA using 0.1 µg oligo(dT) primer and reverse transcriptase (Invitrogen). RT-PCR was run for 35 cycles. Real-time RT-PCR analysis was carried out with an ABI 7900HT sequence detection system. The expression of FRK1, WRKY30, or At3g23230 was normalized to the expression of UBQ10. WRKY30 and At3g23230 were amplified with primers, WRKY30 5’-GCAGCTTGAGAGCAAGAATG-3’ and 5’-AGCCAAATTTCCAAGAGGAT-3’; At3g23230 5’-CTCAGCGGTCTCAAATGTCC-3’ and 5’-AGGAGCAGCAACAACCAATC-3’. The primers for FRK1 and UBQ10 was reported (He et al., 2006).
Supplementary Material
ACKNOWLEDGMENTS
This work was initiated in Jen Sheen’s laboratory at Massachusetts General Hospital and Harvard Medical School with the support by grant from the NIH (R01GM70567) to J.S. The work in L.S and P.H’s laboratories was supported by the Texas A&M University start-up funds to L.S and P.H, and NIH (R01GM092893) to P.H and NSF (IOS-1030250) to L.S.
LITERATURE CITED
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- Block A, Li G, Fu ZQ, Alfano JR. Phytopathogen type III effector weaponry and their plant targets. Curr Opin Plant Biol. 2008;11:396–403. doi: 10.1016/j.jbi.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain RJ, Hayward RD, Koronakis V. The target cell plasma membrane is a critical interface for Salmonella cell entry effector-host interplay. Mol Microbiol. 2004;54:887–904. doi: 10.1111/j.1365-2958.2004.04336.x. [DOI] [PubMed] [Google Scholar]
- Cain RJ, Hayward RD, Koronakis V. Deciphering interplay between Salmonella invasion effectors. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000037. e1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- de Torres M, Mansfield JW, Grabov N, Brown IR, Ammouneh H, Tsiamis G, Forsyth A, Robatzek S, Grant M, Boch J. Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J. 2006;47:368–382. doi: 10.1111/j.1365-313X.2006.02798.x. [DOI] [PubMed] [Google Scholar]
- Fontes EP, Santos AA, Luz DF, Waclawovsky AJ, Chory J. The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev. 2004;18:2545–2556. doi: 10.1101/gad.1245904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–288. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- Gohre V, Robatzek S. Breaking the barriers: microbial effector molecules subvert plant immunity. Annu Rev Phytopathol. 2008;46:189–215. doi: 10.1146/annurev.phyto.46.120407.110050. [DOI] [PubMed] [Google Scholar]
- Gohre V, Spallek T, Haweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol. 2008;18:1824–1832. doi: 10.1016/j.cub.2008.10.063. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Guo M, Tian F, Wamboldt Y, Alfano JR. The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol Plant Microbe Interact. 2009;22:1069–1080. doi: 10.1094/MPMI-22-9-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham JH, Kim MG, Lee SY, Mackey D. Layered basal defenses underlie non-host resistance of Arabidopsis to Pseudomonas syringae pv. phaseolicola. Plant J. 2007;51:604–616. doi: 10.1111/j.1365-313X.2007.03165.x. [DOI] [PubMed] [Google Scholar]
- Hann DR, Rathjen JP. Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J. 2007;49:607–618. doi: 10.1111/j.1365-313X.2006.02981.x. [DOI] [PubMed] [Google Scholar]
- Hann DR, Rathjen JP. The long and winding road: virulence effector proteins of plant pathogenic bacteria. Cell Mol Life Sci. 2010 doi: 10.1007/s00018-010-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann DR, Gimenez-Ibanez S, Rathjen JP. Bacterial virulence effectors and their activities. Curr Opin Plant Biol. 2010 doi: 10.1016/j.pbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- He P, Shan L, Sheen J. The use of protoplasts to study innate immune responses. Methods Mol Biol. 2007;354:1–9. doi: 10.1385/1-59259-966-4:1. [DOI] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nurnberger T, Sheen J. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 2006;125:563–575. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci U S A. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RW, Athanassopoulos E, Tsiamis G, Mansfield JW, Sesma A, Arnold DL, Gibbon MJ, Murillo J, Taylor JD, Vivian A. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc Natl Acad Sci U S A. 1999;96:10875–10880. doi: 10.1073/pnas.96.19.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamir Y, Guo M, Oh HS, Petnicki-Ocwieja T, Chen S, Tang X, Dickman MB, Collmer A, Alfano JR. Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 2004;37:554–565. doi: 10.1046/j.1365-313x.2003.01982.x. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kim JG, Li X, Roden JA, Taylor KW, Aakre CD, Su B, Lalonde S, Kirik A, Chen Y, Baranage G, McLane H, Martin GB, Mudgett MB. Xanthomonas T3S Effector XopN Suppresses PAMP-Triggered Immunity and Interacts with a Tomato Atypical Receptor-Like Kinase and TFT1. Plant Cell. 2009 doi: 10.1105/tpc.108.063123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MG, da Cunha L, McFall AJ, Belkhadir Y, DebRoy S, Dangl JL, Mackey D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Guttman DS, Desveaux D. The targeting of plant cellular systems by injected type III effector proteins. Semin Cell Dev Biol. 2009;20:1055–1063. doi: 10.1016/j.semcdb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Li X, Lin H, Zhang W, Zou Y, Zhang J, Tang X, Zhou JM. Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci U S A. 2005;102:12990–12995. doi: 10.1073/pnas.0502425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M, Cunnac S, Collmer A. The evolution of Pseudomonas syringae host specificity and type III effector repertoires. Mol Plant Pathol. 2009;10:767–775. doi: 10.1111/j.1364-3703.2009.00587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M, Cartinhour S, Myers CR, Schechter LM, Schneider DJ, Collmer A. Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol Plant Microbe Interact. 2006;19:1151–1158. doi: 10.1094/MPMI-19-1151. [DOI] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G. RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000139. e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci U S A. 2010;107:496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol. 2009;12:117–124. doi: 10.1016/j.mib.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Bonasera JM, Kim JF, Nissinen RM, Beer SV. Apple proteins that interact with DspA/E, a pathogenicity effector of Erwinia amylovora, the fire blight pathogen. Mol Plant Microbe Interact. 2006;19:53–61. doi: 10.1094/MPMI-19-0053. [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell. 2000;101:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- Oh HS, Collmer A. Basal resistance against bacteria in Nicotiana benthamiana leaves is accompanied by reduced vascular staining and suppressed by multiple Pseudomonas syringae type III secretion system effector proteins. Plant J. 2005;44:348–359. doi: 10.1111/j.1365-313X.2005.02529.x. [DOI] [PubMed] [Google Scholar]
- Raffatellu M, Wilson RP, Chessa D, Andrews-Polymenis H, Tran QT, Lawhon S, Khare S, Adams LG, Baumler AJ. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells. Infect Immun. 2005;73:146–154. doi: 10.1128/IAI.73.1.146-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Shan L, Zhou JM, Tang X. The Pseudomonas syringae pv. tomato DC3000 type III effector HopF2 has a putative myristoylation site required for its avirulence and virulence functions. Mol Plant Microbe Interact. 2006;19:130–138. doi: 10.1094/MPMI-19-0130. [DOI] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- Schulze B, Mentzel T, Jehle AK, Mueller K, Beeler S, Boller T, Felix G, Chinchilla D. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem. 2010;285:9444–9451. doi: 10.1074/jbc.M109.096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Thara VK, Martin GB, Zhou JM, Tang X. The pseudomonas AvrPto protein is differentially recognized by tomato and tobacco and is localized to the plant plasma membrane. Plant Cell. 2000;12:2323–2338. doi: 10.1105/tpc.12.12.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nurnberger T, Martin GB, Sheen J. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Oh HS, Chen J, Guo M, Zhou J, Alfano JR, Collmer A, Jia X, Tang X. The HopPtoF locus of Pseudomonas syringae pv. tomato DC3000 encodes a type III chaperone and a cognate effector. Mol Plant Microbe Interact. 2004;17:447–455. doi: 10.1094/MPMI.2004.17.5.447. [DOI] [PubMed] [Google Scholar]
- Singer AU, Desveaux D, Betts L, Chang JH, Nimchuk Z, Grant SR, Dangl JL, Sondek J. Crystal structures of the type III effector protein AvrPphF and its chaperone reveal residues required for plant pathogenesis. Structure. 2004;12:1669–1681. doi: 10.1016/j.str.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Speth EB, Lee YN, He SY. Pathogen virulence factors as molecular probes of basic plant cellular functions. Curr Opin Plant Biol. 2007;10:580–586. doi: 10.1016/j.pbi.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF, Krysan PJ. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 2007;143:661–669. doi: 10.1104/pp.106.091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiamis G, Mansfield JW, Hockenhull R, Jackson RW, Sesma A, Athanassopoulos E, Bennett MA, Stevens C, Vivian A, Taylor JD, Murillo J. Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J. 2000;19:3204–3214. doi: 10.1093/emboj/19.13.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 2006;18:257–273. doi: 10.1105/tpc.105.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li J, Hou S, Wang X, Li Y, Ren D, Chen S, Tang X, Zhou JM. A Pseudomonas syringae ADP-Ribosyltransferase Inhibits Arabidopsis Mitogen-Activated Protein Kinase Kinases. Plant Cell. 2010 doi: 10.1105/tpc.110.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton M, Subramaniam R, Elmore J, Felsensteiner C, Coaker G, Desveaux D. The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc Natl Acad Sci U S A. 2010;107:2349–2354. doi: 10.1073/pnas.0904739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, Zhou JM. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol. 2008;18:74–80. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, Mengiste T, Zhang Y, Zhou JM. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.