Abstract
Worldwide, hepatocellular carcinoma (HCC) is one of the most common cancers. It is thought that 80% of hepatocellular carcinomas are linked to chronic infections with the hepatitis B (HBV) or hepatitis C (HCV) viruses. Chronic HBV and HCV infections can alter hepatocyte physiology in similar ways and may utilize similar mechanisms to influence the development of HCC. There has been significant progress towards understanding the molecular biology of HBV and HCV and identifying the cellular signal transduction pathways that are altered by HBV and HCV infections. Although the precise molecular mechanisms that link HBV and HCV infections to the development of HCC are not entirely understood, there is considerable evidence that both inflammatory responses to infections with these viruses, and associated destruction and regeneration of hepatocytes, as well as activities of HBV- or HCV-encoded proteins, contribute to hepatocyte transformation. In this review, we summarize progress in defining mechanisms that may link HBV and HCV infections to the development of HCC, discuss the challenges of directly defining the processes that underlie HBV- and HCV-associated HCC, and describe areas that remain to be explored.
Keywords: Hepatitis B virus, Hepatitis C virus, Hepatocellular carcinoma, Hepatocarcinogenesis, Liver
1. Introduction
Worldwide, primary liver cancer, hepatocellular carcinoma (HCC), is the third leading cause of cancer-associated death [1]. There is compelling evidence that hepatocarcinogenesis, regardless of the etiological agent, is a multistep process. HCC related to hepatitis B (HBV) and hepatitis C (HCV) virus infections is thought to account for more than 80% of primary liver cancers [1]. Despite the fact that HBV and HCV have distinct life cycles, and can have different pathogenic and clinical features, chronic HBV and HCV infections can alter hepatocyte physiology in similar ways and may utilize similar mechanisms to impact HCC development (reviewed in [1; 2]). Since the discovery of HBV and HCV, significant progress has been made towards understanding the molecular biology of these viruses as well as the molecular events and cellular signal transduction pathways that are altered by HBV and HCV infections. Classically, it has been proposed that HBV- and HCV-induced HCC develops in a microenvironment of chronic liver injury, inflammation, and regeneration (reviewed in [1; 3]). In this context, malignant transformation of hepatocytes in the infected liver could be driven by chronic inflammation and oxidative DNA damage, leading to genetic and epigenetic changes. There is also evidence that HBV- and HCV-encoded proteins may have a direct role in hepatocarcinogenesis, and proteins expressed from the genome of each of these viruses have been linked to alterations in hepatocyte physiology and hepatocellular signal transduction pathways (reviewed in [4; 5; 6]). Therefore, it is likely that indirect, inflammation-mediated, and direct, HBV or HCV protein-induced, mechanisms play a role in HBV- and HCV-associated hepatocyte transformation. Herein, we summarize progress in understanding mechanisms that may link HBV and HCV infections to HCC development, discuss potential links between HBV and HCV infections and the development of HCC that remain to be fully explored, and describe some current challenges of attempting to directly define the molecular mechanisms that underlie HBV- and HCV-associated HCC. Due to space and reference limitations, we have focused on major themes and cannot review or reference all the reported consequences of HBV and HCV infections that could potentially contribute to HCC development; we apologize to those whose work we could not include.
2. General overview of hepatocellular carcinoma
Primary liver cancer, hepatocellular carcinoma (HCC) is one of the most common cancers worldwide. Due to the paucity of available treatments options, HCC is associated with a poor prognosis and remains the third most common death-associated cancer (reviewed in [1]). While the incidence of HCC has historically been high in some Asian and African countries, there has been a more recent increase in the incidence of HCC in countries such as the United States (reviewed in [1]). HCC development has been linked to exposure to environment toxins, alcohol and drug abuse, autoimmune disorders, genetic factors, elevated hepatic iron levels, obesity, and infections with hepatotropic viruses (reviewed in ([7; 8]). In the context of many of these liver insults, chronic inflammation, liver regeneration, fibrosis, and cirrhosis are thought to contribute to the development of HCC (reviewed in [8]). Epidemiological studies suggest that infections with hepatotropic viruses are linked to the majority of cases of HCC. In fact, it has been estimated that chronic infections with HBV and HCV account for up to 80% of HCCs [1]. The mechanisms that underlie transformation of hepatocytes, the major cell of the liver, and the development of HCC have remained elusive and are likely to involve alterations of multiple cellular signal transduction pathways as well as alterations in the intracellular and extracellular profiles of signaling proteins, micro (mi)RNAs, and cytokines (reviewed in [7; 8]). It is also apparent that HBV- and HCV-encoded proteins and immune-mediated destruction of infected hepatocytes and concomitant liver regeneration can impact hepatocyte transformation and are likely to influence the development of HCC in individuals with persistent HCV or HBV infections (reviewed in [3; 4; 5; 6; 9]). While the exact molecular mechanisms that underlie HBV- or HCV-associated HCC are not entirely understood, the results of various studies have suggested putative mechanisms that could link infections with these viruses to alterations in hepatocyte physiology.
3. Hepatitis B virus and hepatocellular carcinoma
3.1. Natural history and epidemiology of HBV
Hepatitis, a consequence of inflammation of the liver, is often associated with malaise, fatigue, and jaundice, a yellowing of the skin and sclera that is caused by liver dysfunction and increased levels of bilirubin in the blood (reviewed [10]). While there appears to have been a long-standing recognition that individuals with persistent hepatitis have an increased risk for the development of liver cancer, Baruch Blumberg's discovery of the Australian antigen, the recognition that this antigen was the S antigen or envelope protein of HBV, and subsequent retrospective studies provided a direct correlation between the incidence of chronic HBV infections and the incidence of HCC (reviewed in [6]). Blumberg's discovery also facilitated isolation and characterization of HBV as well as the eventual development of a HBV vaccine. Although it is possible to experimentally infect some non-human primates with HBV, characterization of the virus demonstrated that humans are the only natural host for HBV and that hepatocytes are the primary target of the virus (reviewed in [6]). In addition, analyses of the global geographic distribution of the virus has led to the estimation that approximately 2 billion people have been infected with HBV and that about 350 million people worldwide remain chronically infected; high numbers of chronic infections have been observed in many Asian and African countries [1; 11]. Eight HBV genotypes have been identified, and these genotypes have distinct geographic distributions (reviewed in [6]). There is evidence that there may be HBV genotype-specific clinical outcomes of infections, including disease progression, persistence of infection, and risk for development of HCC. Chronic HBV infection, defined as the continual expression of the HBV S antigen for at least 6 months following the initial infection, is linked to the development of liver pathologies. These liver pathologies can include fibrosis and cirrhosis, which are thought to be major contributors to the eventual development of HCC; HCC can also develop in the absence of cirrhosis and fibrosis in HBV-infected individuals (reviewed in ([12]). More recently, it has become apparent that many individuals who were thought to have cleared their HBV infection remain infected with low levels of the virus; in these individuals, standard assays for HBV S antigen are negative but more sensitive polymerase chain reaction (PCR) assays can detect the presence of low levels of the HBV genome (reviewed in [13]). There is now mounting evidence that these individuals are also at increased risk for developing HCC, and this may significantly increase the number of HCC cases that can be directly associated with HBV infections.
3.2. Overview of HBV replication
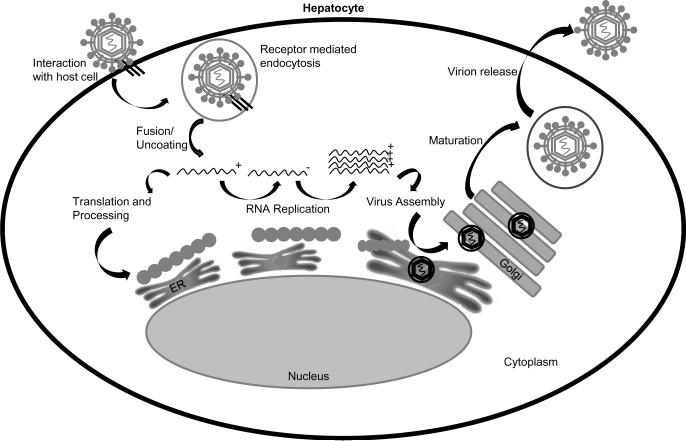
HBV is the human viral member of the Hepadnaviridae, a family of viruses that target the liver where they can establish chronic infections. Each member of the hepadnavirus family has a narrow host range that is thought to be defined by a cell-surface receptor as well as intracellular signaling factors (reviewed in [6]). Although numerous HBV candidate receptors have been proposed, a receptor or receptor complex has not been unequivocally identified, and the process for HBV entry into hepatocytes remains unclear (reviewed in [6]). Mechanisms that regulate the subsequent release of the viral genome and delivery of the genome to the nucleus are also unclear, and both direct transport of the encapsidated genome through nuclear pores as well as release of the viral genome from the capsid prior to its transport into the nucleus have been proposed [14; 15]. Following transport of the partially double-stranded, circular DNA HBV genome into the nucleus, the single-stranded DNA gaps are repaired, and the genome is converted to a covalently closed circular, double-stranded DNA (cccDNA) (Fig. 1) (reviewed in [6]). The cccDNA does not replicate but is the template for transcription and production of viral mRNAs. The HBV genome is compact and contains four overlapping open-reading-frames that encode the viral capsid (core), envelope (S antigen), reverse-transcriptase/polymerase (Pol) and HBx proteins (Fig. 2) (reviewed in [6]). The largest HBV transcript, the pre-genomic (pg)RNA, is terminally redundant and an intermediary in viral replication. Replication occurs in the cytoplasm of infected hepatocytes where the pgRNA is encapsidated by the HBV core protein and converted to the DNA genome by the viral reverse-transcriptase/polymerase (reviewed in [6]). The replicating, encapsidated viral genome is either directed back to the nucleus to amplify the nuclear pool of cccDNA or enveloped by the HBsAg proteins as capsids buds into the endoplasmic reticulum (Fig. 1) (reviewed in [6]). The enveloped virion is then secreted from the cell; various groups have reported that HBV secretion from cells involves multivesicular body factors [16; 17; 18; 19].
Fig. 1.
Hepatitis B virus life cycle: See text for detailed description.
Fig. 2.
Hepatitis B virus genome: See text for detailed description. Inner arrows represent open reading frames. Outer arrows represent mRNAs. EN (Enhancer); DR (Direct Repeat); PC (Precore); TP (terminal protein); 1/3182 (nucleotide 1 and 3182 of circular genome).
3.3. Possible mechanisms of HBV-associated HCC
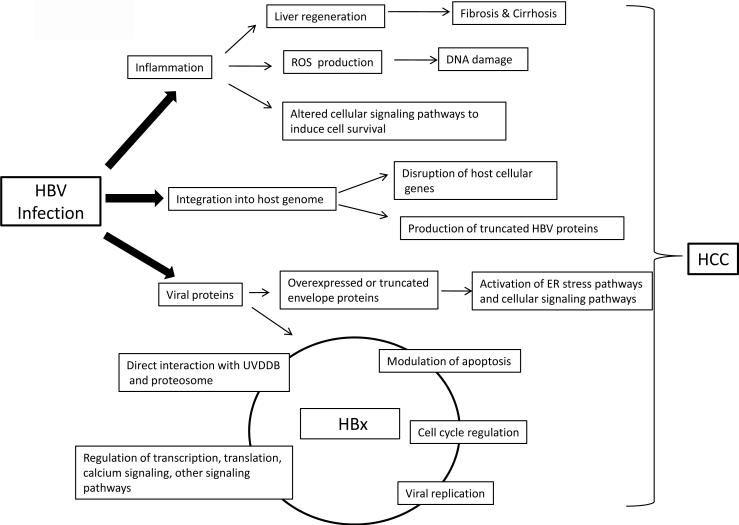
Numerous mechanisms are likely to contribute to progression and development of HCC in people persistently infected with HBV. Three mechanisms that have been the subject of intense investigation will be discussed here. These mechanisms are: 1.) inflammation-mediated destruction of HBV-infected hepatocytes and concomitant liver regeneration, which may contribute to fibrosis, cirrhosis, and the generation of a mutagenic environment, 2.) the potential consequence of HBV genome integration into host chromosomal DNA, and 3.) activities associated with expression of some HBV proteins (Fig. 3).
Fig. 3.
Hepatitis B virus hepatocarcinogenesis: Summary of putative mechanisms that could link chronic HBV infections to the development of HCC. Circle connecting HBx activities suggests that more than one HBx activity likely contributes to HCC development. See text for details.
3.3.1. HBV-associated liver inflammation and HCC
HBV-associated HCC predominately occurs in the context of liver injury caused by immune-mediated destruction of HBV-infected hepatocytes and resultant liver regeneration (reviewed in [3]). HBV is noncytopathic, and liver injury is associated with influx of immune cells into the liver and destruction of HBV-infected hepatocytes (reviewed in [20; 21; 22]). In chimpanzees that are experimentally infected with HBV, the initial influx of immune cells does not occur immediately but may take as long as two to three months. Prior to the influx of immune cells, most hepatocytes within the liver can be infected with HBV, and significant viral clearance is mediated by innate immune responses in the absence of cell damage [20]. Depletion of CD8+ lymphocytes delays the biochemical and histological consequences of HBV-associated liver damage and the clinical onset of liver disease, demonstrating the central role of the immune response in the pathological consequences of an HBV infection (reviewed in [20; 21; 22]).
While studies in chimpanzees that were directed towards understanding the inflammatory response that link HBV infections to liver disease have greatly contributed to our understanding of mechanisms involved in clearing an HBV infection and processes that contribute to the development of a chronic HBV infection, both cost and ethical reasons have limited the number and scope of studies in this animal model. Moreover, liver cirrhosis and the development of HCC are not usually observed in chimpanzees that are chronically infected with HBV [23]. An important, experimentally tractable model system has been HBV-transgenic mice; these mice have been developed and studied by various research groups [24; 25]. HBV-transgenic mice contain a copy of the HBV genome incorporated into the endogenous DNA of all hepatocytes, and presumably can produce HBV from all hepatocytes; these mice have served as a model for studying in vivo HBV replication and inflammation-mediated viral clearance (reviewed in [3]). Because the HBV genome is present as an endogenous part of the developing mouse, an anti-HBV immune response is not mounted in these mice, and an important caveat of this system is that immune responses must be studied in the context of transferred, primed immune cells. Although extremely rare, HCC has been observed in HBV transgenic mice, supporting the concept that HBV might be weakly carcinogenic in the absence of inflammation [25]. Transfer of HBV-primed cytotoxic T lymphocytes (CTLs) into HBV-transgenic mice produces an inflammatory liver disease that is similar to acute viral hepatitis in humans [26; 27]. While it is clear that specific CD4 + and CD8+ T-cells have an important role in viral clearance, these studies also provided evidence that clearance is aided by neutrophils, bystander lymphocytes, and natural killer cells; both antigen-specific and -non-specific cells are recruited and contribute to inflammation (reviewed in [3]). The mechanisms through which antigen-nonspecific inflammatory cells induce liver damage are not well understood but likely include production of proinflammatory and cytotoxic signals. Importantly, when HBV-infected hepatocytes are destroyed, hepatocyte regeneration is activated, and damaged or destroyed hepatocytes can be replaced by replication of mature hepatocytes [28]. Processes similar to wound healing are also activated, and deposition of extracellular matrix components can cause fibrosis, and if unchecked, cirrhosis [29]. While hepatocyte regeneration and liver fibrosis processes usually subside once the liver injury is repaired and the causative agent is removed, in individuals with chronic HBV infection, repeated cycles of immune-mediated clearance of HBV-infected hepatocytes causes continual proliferation of hepatocytes and constant liver regeneration (reviewed in [3; 6]). In the context of continual inflammation, the recruitment of immune cells, and resultant local increase in inflammatory cytokines, can generate a locally mutagenic microenvironment; local elevation in reactive oxygen species (ROS) levels can directly mediated mutagenic processes by damaging DNA. In addition, the presence of inflammatory cytokines may activate cellular signaling cascades that can regulate transcription, proliferation, or cell survival (reviewed in [28]). Inefficient immune responses in chronically HBV-infected individuals are thought to maintain a continuous cycle of low-level liver cell destruction that over long periods of time can lead to fibrosis, cirrhosis, and HCC [3]. In the context of a cirrhotic liver, continual hepatocyte proliferation is a major risk factor for HCC development. Cumulatively, it is likely that the continual stimulation of liver regeneration in the potentially mutagenic environment of HBV-associated liver inflammation may eventually select for transformed hepatocytes and cause the development of HCC.
Although it is widely accepted that HBV-associated liver inflammation is an important factor in the development of HCC, the exact molecular mechanisms that link HBV-associated liver inflammation to hepatocyte transformation have not been precisely defined. Recent observations that have linked liver inflammation to cancer development provide intriguing possibilities of mechanisms that could link HBV infections to HCC. Among these, altered activation of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), stimulation of various signal transduction pathways, and the disruption of the normal balance of cytokines such as tumor necrosis factor alpha (TNFα) and interleukin 6 (IL6), have been linked to the development of HCC; similar types of alteration of the liver microenvironment occur in the context of an inflammatory response to an HBV infection and may be important contributors to the development of HBV-associated liver diseases and HCC (reviewed in [28]). With the development of mouse models in which the association between inflammatory responses in the liver and hepatocyte transformation can be studied, direct confirmation of the impact of specific inflammatory mechanisms on the development of HBV-associated HCC may now be open for direct investigation.
3.3.2. HBV Genome Integration and HCC
HBV-associated liver tumors frequently contain integrated HBV DNA sequences; it is thought that the double-stranded linear form of replicating HBV, an aberrant replication dead end product, is the preferred substrate for DNA integration [30; 31]. HBV does not encode an enzymatic activity that facilitates integration of its genome into host chromosomes, and integration is not a normal or required step in HBV replication (reviewed in [6]). Although the integrated HBV genome cannot support HBV replication because integration disrupts the circular nature of the viral genome (Fig. 2), the integrated genome could contribute to HCC development by activating or disrupting the expression or activity of cellular proteins. The woodchuck hepatitis virus (WHV) is highly related to HBV, and chronic infections of woodchucks with WHV frequently cause HCC (reviewed in [32]). Integration of the WHV genome into the host chromosome of chronically infected woodchucks is frequently located in the vicinity of the N-MYC2 gene, a functional N-MYC-related retrotransposon; N-myc2 is normally not expressed in hepatocytes [33; 34; 35; 36]. Disruption of the normal expression of members of the myc gene family can directly contribute to processes involved in cellular transformation, and this is thought to be a common mechanism associated with WHV-associated HCC in woodchucks [37]. In contrast, HBV integration appears to be mostly random [31]. While HBV-genome integration may contribute to HCC development in humans, the lack of consistent integration events has made it difficult to specifically show a direct correlation between HBV-genome integration and the development of HCC, and whether integration contributes to HCC development remains controversial. Integration of the HBV genome has been observed in the vicinity of genes that control cell immortalization, cell survival, or cellular signaling pathways that can impact cell proliferation, and these may have contributed to HCC development (reviewed in [31; 38]). Because integrated HBV genomes have been identified in individuals with chronic or acute HBV infections, and in the absence of tumors, integration can occur prior to tumor development. One complication in determining the role of HBV integration for the development of HCC is that transformed cells in tumors often contain multiple integrants, and it is difficult to determine when integration occurred and whether the integration initiated transformation, facilitated tumor progression, or had no role in the development of HCC. The presence of HBV genome integrants in the vicinity of genes such as hTERT provide putative pathways for contributing to hepatocyte immortalization and transformation; but in the absence of direct evidence for the role of a specific integration in the process of HCC development, it remains unknown whether HBV integration initiates or is a consequence of transformation [39]. In addition to potentially disrupting cellular genes, HBV genome integration into host chromosomes may also cause the production of altered or truncated HBV protein, and it has recently been reported that truncated forms of the HBx gene, a potential contributor to HCC development, is frequently observed in HBV-associated HCC [40; 41]. In addition, integration of the HBV genome has been reported to cause the production of truncated HBV envelope proteins that may also impact carcinogenic processes [42]. The potential role of specific HBV proteins in HCC development will be discussed below.
3.3.3. HBV-encoded proteins and HCC
3.3.3.1 HBV envelope proteins and HCC
There are three HBV envelope proteins; the small, middle, and large envelope proteins, also referred to as the small, middle, or large S antigens (reviewed in [6]). Excess production of the HBV envelope proteins, or truncated forms of the middle and large envelope protein, can activate cellular signal transduction pathways or endoplasmic reticulum stress pathways (reviewed in [6]). The truncated large envelope protein can induce cell proliferation by upregulating expression of cyclin A, and truncated M protein can activate c-Raf-1 and extracellular regulated kinase (ERK) signaling to stimulate cell proliferation. Alternatively, accumulation of envelope proteins in the ER can activate the unfolded protein response and cause oxidative stress, which can be mutagenic. In human hepatocytes, cytosolic accumulation of envelope protein causes the formation of “ground glass” hepatocytes containing altered ER structures [43]. Importantly, transgenic mice that over express the large envelope protein in hepatocytes undergo liver dysplasia and damage accompanied by liver regeneration and the eventual development of HCC; development of HCC in these model systems also involved continual liver inflammation resulting from over expressed envelope proteins and associated hepatocyte stress [43]. Cumulatively, these studies suggest that when over-expressed or truncated, the envelope proteins may have oncogenic potential, predominantly by activating hepatocyte stress-response pathways that alter hepatocyte physiology and may eventually stimulate transformation processes. Over expressed or truncated envelope proteins may arise from integration of the HBV genome into the host chromosomal DNA (reviewed in [31]).
3.3.3.2 HBx and HCC
The HBV protein that is thought to make the most significant contribution to the development of HBV-associated liver cancer is HBx. HBx is a 154 amino acid, multifunctional protein that is encoded by the smallest open reading frame of mammalian hepadnaviruses. HBx is predominately a cytoplasmic protein, but low levels of HBx are also present in the nucleus of HBV-infected hepatocytes (reviewed in [5]). Most of HBx, from amino acids 52–148, is essential for its various activities, and deletion of the amino-terminal 1–50 amino acid fragment up-regulates HBx transcriptional functions, suggesting that it is a negative regulatory element (reviewed in [44; 45]). The results of numerous studies have demonstrated that HBx can modulate cellular signal transduction, transcription, proliferation, and apoptotic pathways; HBx regulation of these signal transduction pathways may alter hepatocyte physiology and impact the development of HBV-associated HCC (reviewed in [5]). Unfortunately, there has been considerable confusion regarding the consequences of HBx expression, and the precise impact of HBx expression has varied depending on the cell type and conditions used in a particular assay. Although studies in various experimental systems have been invaluable for understanding the impact of HBx expression in different cellular environments, because most of these studies were conducted in immortalized or transformed cells, when HBx was over-expressed from a transfected plasmid, and when HBx was expressed in the absence of other HBV proteins, the consequence of HBx expression in normal hepatocytes or in the context of HBV replication is not completely understood. Because HBV-associated HCC usually requires years of a chronic HBV infection and associated inflammation, it is unlikely that HBx that is expressed in the context of HBV replication, is strongly oncogenic, and it is more likely that subtle HBx effects alter hepatocytes to generate an intracellular environment that may be more susceptible to other oncogenic signals [5; 6]. While many HBx activities have been reported, only major themes are discussed below with particular attention to how these HBx activities might contribute to oncogenic processes.
HBx and replication of mammalian hepadnaviruses
HBV-associated HCC occurs in the context of a chronic HBV infection and persistent HBV replication, and there is accumulating evidence that the X proteins of mammalian hepadnaviruses strongly impact viral replication. Studies from two groups demonstrated that WHx, the X protein of WHV is essential for WHV infection of woodchucks [46; 47] while another group found that a mutant WHV that lacked WHx expression produced a low level of viremia, suggesting that WHx was not absolutely required for WHV replication [48]. In transgenic mice that express a mutant HBV genome that lacks HBx expression, HBV replication was still observed; however, trans-complementation experiments carried out by crossing HBx-expressing mice with the mutant HBV-expressing mice demonstrated that HBx augmented HBV replication in the hybrid animals [49; 50]. Whether HBV replication in HBV-transgenic mice completely mimics all aspects of HBV replication in natural infection is unclear, suggesting that HBV-transgenic mice, while clearly important for many HBV studies, may not be the best system for studying all signals that are absolutely required for HBV replication. More recently, the use of two new model systems has provided additional support for an important role for HBx during HBV replication. The first model used in vivo hydrodynamic injection of the HBV genome into mouse livers to address the importance of HBx for HBV replication [51]. In these experiments, a hydrodynamically injected plasmid that contained the genome of wildtype HBV, or an HBx-deficient HBV mutant, revealed a significant impact of HBx on HBV replication, although there was not an absolute requirement. In the second model, a human hepatocyte chimeric mouse liver model was used [52]. In this model, human hepatocytes were used to reconstitute the mouse liver with a high percentage of human hepatocytes. When these chimeric mice were infected with a wildtype or HBx-deficient HBV, replication was only observed in human hepatocytes in the livers of mice infected with wildtype HBV [53; 54]. HBx also regulates HBV replication in HepG2 cells, a human hepatoblastoma cell line; however, replication of HBV in Huh7 cells, a human hepatoma cell line, is not influenced by HBx (reviewed in [5]). Collectively, these reports suggest that the X protein of mammalian hepadnaviruses performs an important but perhaps not absolute function in viral replication. In addition to a possible role for HBx regulation of cellular signaling pathways for HBx stimulation of HBV replication (described below), HBx interaction with the ultraviolet-damaged DNA-binding complex and HBx regulation of proteasome activity have been proposed to be essential for HBx stimulation of HBV replication; however, precisely how regulation of these HBx activities impacts HBV replication awaits further investigation [55; 56; 57; 58].
HBx regulates various cellular signal transduction and transcription pathways
HBx activates various transcription factors; activation of these factors by HBx can involve direct interactions as well as activation of cell signal transduction pathways. HBx can associate with components of the basal transcriptional machinery, including transcription factor (TF)IIB, TFIIH, the RPB5 subunit of RNA polymerases, and the TATA-binding protein (TBP). HBx also directly interacts with cAMP response element-binding protein (CREB) and CCAAT-enhancer binding protein α (c/EBPα) (reviewed in [5]). HBx indirectly activates transcription factors such as NF-κB, activator protein-1 (AP-1), and nuclear factor of activated T cells (NFAT) by stimulating cellular signal transduction pathways (reviewed in [5]). Significantly, HBx stimulates transcription in vivo in hepatocytes. When mice that express a reporter gene controlled by the Human Immunodeficiency Virus (HIV) long terminal repeat (LTR) were crossed with HBx-transgenic mice that express HBx in hepatocytes, transcription of the reporter was increased [59]. Another study reported an increase in HBV core promoter activity by HBx when HBV- and HBx-transgenic mice were crossed [60].
One pathway that HBx activates to stimulate transcription involves Pyk2 and Src kinases, Ras, Raf, and the mitogen activated protein kinases (MAPK) (reviewed in [5; 6]) and [61; 62; 63]. HBx activation of Pyk2 and Src kinases is also essential for its stimulation of cellular proliferation pathways and HBV replication in HepG2 cells [61; 63; 64]. HBx activates Pyk2 and Src kinases in HepG2 cells by stimulating cytosolic calcium signaling. We and others have demonstrated that HBx modulates cytosolic calcium levels and that many HBx activities, including some of its critical functions in HBV replication, can be explained by its regulation of calcium signaling pathways [63; 65; 66; 67; 68]. HBx regulation of calcium signaling in HepG2 cells and in cultured primary rat hepatocytes is dependent on the activity of the mitochondrial permeability transition pore (MPTP), a regulator of cytosolic calcium levels; specific inhibitors of the MPTP block the HBx-induced elevation of cytosolic calcium levels and HBV replication [63; 69; 70]. These observations are consistent with the reported interaction of HBx and the voltage-dependent anion channel, a component of the MPTP, and our recent observation that a fraction of HBx localizes to mitochondria in HepG2 cells and cultured primary rat hepatocytes [71; 72]. HBx activation of signal transducer and activator of transcription 3 (STAT3) is also dependent on calcium signaling, and HBx-induced changes in cytosolic calcium levels have been directly measured in HepG2 cells and Chang cells, a human liver cell line [62; 65; 66]. Mitochondrial localized HBx also elevates the levels of cellular ROS, and cellular ROS and calcium each regulate the other, generating a positive feedback mechanism that can ultimately elevate both ROS and calcium and activate numerous cellular signal transduction pathways. HBx alteration of cellular calcium, ROS, and cellular signal transduction and transcription pathways, especially in the context of inflammation, could alter cell physiology and contribute to cellular transformation.
HBx and Apoptosis
HBx has been reported to induce apoptosis, sensitize cells to pro-apoptotic stimuli, or prevent apoptosis. Some studies found enhanced hepatocyte apoptosis in mice expressing HBx, whereas others did not (reviewed in [5; 6]). In some cell culture systems, HBx blocked cell death mediated by TNFα, Fas, p53 or transforming growth factor β (TGFβ) [73; 74; 75]. In contrast, other studies indicate that HBx promotes apoptosis in cultured cells [76; 77; 78; 79]. The recognition that HBx interacts with the MPTP suggests a possible mechanism of HBx-induced modulation of apoptosis, although this association does not imply either a pro- or anti-apoptotic effect [71; 79]. HBx can induce mitochondrial aggregation and cytochrome c release, which is indicative of induction of apoptosis through mitochondrial dysfunction [79; 80]. Aggregation and hampered mobility of mitochondria was also observed during HBV replication in cultured cells; however, whether this was directly linked to apoptosis is not known [81]. HBx expressed during HBV replication in some cultured cells did not directly promote apoptosis, but caused hypersensitivity to TNFα-mediated cell death at levels below those at which cells are normally sensitive to TNFα [76; 82; 83]. In one study, the pro-apoptotic activity of HBx was reported to act through its inactivation of cellular fas-associated protein with death domain (FADD)-like interleukin-1 beta-converting enzyme (FLICE) inhibitor protein (c-FLIP), a protein that inhibits TNFα-mediated apoptosis [83]. However, over-expression of cFLIP did not fully block apoptosis stimulated by HBx, and it is unlikely that cFLIP is the only means through which HBx can modulate apoptotic pathways. Unfortunately, most studies that directly analyzed HBx regulation of apoptosis were performed in immortalized or transformed cells. Apoptotic pathways are often altered in immortalized or transformed cells, and this may account for the conflicting reports regarding HBx regulation of apoptosis. In cultured primary rat hepatocytes, HBx was both pro- and anti-apoptotic, depending on the status of NF-κB [67]. HBx activated NF-κB to inhibit apoptosis, but when activation of NF-κB was blocked, HBx expression induced apoptosis. HBx regulation of cytosolic calcium levels was linked to its pro-apoptotic activity when NF-κB activity was blocked; inhibitors of cellular calcium signaling pathways and the MPTP blocked HBx activation of apoptosis. A similar NF-κB dependence for HBx anti-apoptotic activity was also observed in various established cells lines [73; 82]. Collectively, these studies suggest that HBx can be either pro- or anti-apoptotic depending on the presence of other cellular signaling factors; the apoptotic activity of HBx might also change as hepatocyte physiology is altered during the process of transformation. Importantly, both pro- and anti-apoptotic activities of HBx could contribute to the development of HCC. Elevation of NF-κB and inhibition of apoptosis has been associated with the development of HCC. Similarly, inhibition of NF-κB activity and subsequent activation of apoptosis, followed by compensatory liver regeneration, has also been linked to the development of HCC (reviewed in [84]).
HBx and cell cycle progression
The results of various studies suggest that HBx can modulate cell cycle progression, although the precise impact of HBx on cell cycle progression has varied in different experimental systems [85]. In HepG2 and Chang cells, HBx activation of Src kinases, Ras, and MAPKs induced arrested cells to re-enter the cell cycle [86]. In other studies, HBx expression caused cells to enter the cell cycle and stall at the G1/S border, progress through the cell cycle more rapidly, or had no effect on cell cycle progression [5; 85]. HBx differentially regulated cell cycle progression in differentiated versus de-differentiated hepatocytic cells that were derived from the same parental liver cell line [87]. The differentiated hepatocytes displayed HBx-dependent G1, S, and G2/M progression, induction of cyclin D1, A and B1, and activation of cyclin-dependent kinase 1 (CDK1). The de-differentiated cells displayed HBx-dependent G1 and S phase entry but paused early in S phase. Three groups have used HBx-transgenic mouse models to analyze the effect of HBx expression on liver regeneration [88; 89; 90]; however, because each group used different experimental protocols and mice, and analyzed hepatocyte proliferation at different times after a partial hepatectomy, it is difficult to directly compare the reported observations. Two of the reports suggested that HBx expression inhibits liver regeneration while the results of one study suggested that HBx induces early cell cycle entry in a subpopulation of HBx-expressing hepatocytes. We recently demonstrated that HBx induces quiescent, cultured primary rat hepatocytes to exit quiescence and enter and stall in G1 phase of the cell cycle [68]. In this model system, HBx upregulated cyclin D1 levels and CDK4 activity while also increasing the levels of p21 and p27 to inhibit progression beyond G1. Similar observations were reported in normal mouse hepatocytes that expressed HBx [91]. In cultured primary rat hepatocytes, HBx modulation of cell cycle progression stimulated the activity of the HBV polymerase [92]. Additionally, HBx upregulated expression of active ribonucleotide reductase in cultured primary rat hepatocytes similar to observations in some established cell lines [92; 93]. Cumulatively, these observations suggest that HBx can modulate cell cycle pathways to stimulate HBV replication. An unfortunate consequence of HBx regulation of cell cycle pathways is that this activity also disrupts normal hepatocyte proliferation pathways, possibly contributing to hepatocyte transformation processes and the development of HCC.
The oncogenic potential of HBx
While all hepadnaviruses establish chronic infections in their hosts, only infection by the mammalian viruses are linked to HCC. As avian viruses either lack HBx or encode a divergent form, the potential role of HBx in the development of HCC in mammals is an area of intense interest (reviewed in [94]). There is evidence that HBx expression in different cell types, including hepatocytes, can enhance transformation in conjunction with other oncogenic signals (reviewed in [94]). Transgenic HBx mouse models also support its oncogenic potential, although whether it is directly oncogenic or functions more as a cofactor in cancer progression is not clear. Two reports in which HBx-transgenic mice were generated with an HBx-expression vector that included the HBV X gene sequence, as well as the HBV enhancer I region, the X gene promoter and the HBV poly-adenylation signal, demonstrated that HBx expression could directly cause HCC development [95; 96]. In contrast, several other groups derived HBx-transgenic mice in which the expression of HBx was controlled by the human alpha-1-antitrypsin or human anti-thrombin gene regulatory sequences [97; 98]. None of these studies supported a direct connection between HBx and tumorigenesis. The reasons for these discrepant results are unclear and may not simply reflect the use of different expression vectors. A strong role for HBx in liver carcinoma does not reflect the biology of HCC development, which involves decades of chronic HBV infection; however, subtle HBx-induced changes in cellular physiology may eventually have detrimental consequences for hepatocytes, sensitizing them to other oncogenic signals [12]. Several groups have described such an effect in HBx-transgenic mice treated with low levels of carcinogenic agents or in HBx-transgenic mice co-expressing activated myc. These HBx mice display higher levels of tumor development than control groups of mice [97; 98; 99]. Exactly what HBx-associated activities are involved in oncogenesis is unknown and may include modulation of cellular signal transduction, transcription, proliferation, and/or apoptotic pathways described above [5; 94]. In addition, a putative role for its direct interaction with ultraviolet-damaged DNA binding protein (UVDDB) has also been invoked, and some studies have suggested that HBx can inhibit DNA repair pathways [100]. In the context of mutagenic environment created during inflammation or by HBx-induced disruption of various signal transduction pathways, HBx inhibition of DNA repair pathways could also contribute to hepatocyte transformation and the development of HCC. Finally, there have been reports that HBx can interact with the tumor suppressor p53, which is involved in various cellular processes, including DNA repair, that protect cells from oncogenic damage (reviewed in [94]). Unfortunately, the potential impact of p53-regulated pathways on HBx activities, and whether HBx alters p53 functions, has varied so greatly in different systems, and in similar types of studies that were conducted in different laboratories, that it remains unclear whether p53 influences the contribution of HBx activities to HBV-associated liver diseases (reviewed in [94]).
4. Hepatitis C virus and hepatocellular carcinoma
4.1. Overview
Hepatitis C virus is a positive-stranded, enveloped, RNA virus belonging to the Flaviviridae family and is the only member of the genus hepacivirus [101]. HCV infection is a major cause of chronic liver diseases, with an estimated prevalence of HCV infection worldwide of 3% (> 170 million people) [102]. In contrast to HBV infections, most acute HCV infections are asymptomatic, complicating early diagnosis and treatment [102]. Although acute HCV infection resolves spontaneously in some patients [103], persistent infection with chronic liver disease develops in more than 70% of patients, of whom approximately 20% develop cirrhosis [104]. Soon after HCV was discovered in 1990 [101], Saito and coworkers [105] reported a correlation between HCV infection and the development of HCC. Despite the progress made over the past 20 years in elucidating the molecular biology of HCV and identifying the pathways involved in HCV-host interactions, the mechanisms driving disease progression to HCC in patients with chronic HCV infection remain poorly understood. Evidence shows that both indirect and virus-directed mechanisms may play a key role in HCV-related HCC [106]. In the following sections, we discuss the contribution of chronic inflammation, the interplay of HCV infection with various cellular pathways, and the potential role of HCV proteins in hepatocarcinogenesis.
4.2. HCV genome and life cycle
The lack of tissue culture systems that fully support HCV replication has hampered our understanding of the HCV life cycle. Recently, a tissue culture system that fully recapitulates HCV entry, replication, and complete virus production of various genotypes in hepatoma cell lines has been developed [107]. Although this breakthrough has generated a new model that should contribute to our understanding of HCV, most of the current knowledge of HCV cell biology has been generated using partial model systems such as tissue culture bi-cistronic replicons, HCV-like particles produced in insect cells, and various viral particles pseudotyped with HCV envelope E1 and E2 glycoproteins.
In common with other flaviviruses, HCV entry is pH dependent, and the virus is endocytosed in clathrin-coated pits [108]. HCV entry remains ill defined, and several molecules have been identified as receptors or cellular attachment factors. A striking property of HCV, when compared with other enveloped viruses, is its distinct lipid composition when produced in patients, infected chimpanzees, or in recently developed tissue culture systems. HCV can be found in low-density (1.06–1.15 g/ml) and high-density (1.17–1.25 g/ml) fractions; the low-density virus is more infectious than the high-density virus [109].
The HCV genome is approximately 9.6 kb in length and encodes a single polyprotein 3000 amino acids in length that is co- and posttranslationally processed by viral and cellular proteases into four structural proteins (core, E1, E2, and p7) and six nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) viral proteins [110] (Fig.4). The molecular biology of HCV has been recently reviewed [110]. The genome is flanked by 5' and 3' untranslated regions (UTRs) with extensive secondary structures that are essential for viral replication, translation, and likely for encapsidation through interaction of both UTRs with the core protein [110]. In contrast to cellular mRNAs, HCV translation is cap-independent and mediated by an internal ribosome entry site located within the 5'UTR [110]. HCV translation occurs on the rough ER [110]. A host signal peptidase processes the structural proteins [110]. Core is a basic protein that forms the viral nucleocapsid by interactions with the HCV RNA, and is further processed by host signal peptidase to yield a mature form that associates with lipids [110]. The two envelope glycoproteins E1 and E2 are glycosylated and form heterodimers in the ER but lack further modification by the Golgi [111]. E1 and E2 contain fusogenic regions and may cooperate to mediate entry into cells [110]. Viroporin (p7) is a small transmembrane protein with ion channel activity. p7 is dispensable for replication but required for the assembly and release of virus from cells; however, the exact role of p7 in the HCV life cycle is poorly understood [112]. A summary of the HCV life cycle is depicted in Fig. 5.
Fig. 4.
Hepatitis C virus genome organization and protein function: See text for detailed description. UTR (untranslated region).
Fig. 5.
Hepatitis C virus life cycle: See text for detailed description.
The nonstructural proteins are required for RNA replication and are cleaved from the HCV polyprotein by NS2 and NS3 viral proteases. NS2 is a cysteine protease that in addition to cleaving the NS2/NS3 junction in cis has a role in viral assembly [113]. A specific role for NS2 in HCV replication has not been defined. NS3 is a serine protease, helicase, and adenosine triphosphatase with complex activities. The amino terminal domain of NS3 and its cofactor NS4A are responsible for the cis or trans cleavage at four sites within the HCV polyprotein, generating the amino termini of NS4A, NS4B, NS5A, and NS5B [110]. The C-terminal portion of NS3 contains a helicase domain that colocalizes with NS5A and NS5B in the replication complex. Some of the activities of NS3 require a stable complex with NS4A, a 54-amino-acid protein that seems to act as an NS3 cofactor by directing the NS3/NS4A complex to the ER and the mitochondrial outer membrane, and by promoting adenosine triphosphate hydrolysis by the NS3 helicase [114]. Remarkably, the NS3A/4A complex plays a key role in the ability of the virus to counteract the innate immune response, leading to inactivation of the interferon regulatory factor 3 (IRF-3) [115], and cleavage of the Toll-like receptor 3 (TLR3) adaptor protein TRIF [116]. NS4B is an integral membrane protein found on the cytoplasmic side of the ER membrane, and is required for the formation of the membranous web that supports HCV replication [110]. NS5A is a phosphoprotein that is involved in HCV replication and virion assembly [110]. Mutations in NS5A are associated with increased replication efficiency of HCV replicons in hepatoma cells. NS5B is the RNA-dependent RNA polymerase [117]. Due to its error-prone RNA polymerase, HCV exhibits a high rate of variability. Genetic heterogeneity has been reported among different HCV isolates which have been classified into six genotypes and 52 subtypes, as well as quasispecies within infected individuals [118].
4.3. Natural history and risk factors of HCV-related HCC
The link between chronic HCV infection, cirrhosis, and HCC development has been demonstrated in prospective and retrospective cohort studies. Based on these studies, HCV-related HCC develops after many years of initial HCV exposure as a sequential occurrence of chronic infection, development of fibrosis and cirrhosis [102]. Most of the patients (approximately 75%) who are acutely infected with HCV remain asymptomatic, with no clinical manifestations of hepatitis [102]. It is estimated that approximately 80% of infected individuals will develop chronic infection with positive HCV RNA [102]. The age of the patient at the time of infection seems to be an important factor in estimating the time to development of cirrhosis. Among persons who acquire HCV infection before 40 years of age, 5% will develop cirrhosis within 20 years, in contrast to 20% of the individuals infected after 40 years of age [102]. Overall, it is estimated that in 75% of chronically HCV-infected patients, the disease will remain slowly progressive, whereas 25% of patients will develop cirrhosis that ultimately may lead to HCC. Although viral load and HCV genotype have not been clearly correlated with HCC development, chronic HCV-infected patients who have consistently normal aminotransferase levels show slower disease progression [119]. The story of disease progression is not well documented beyond 20 years of chronic infection.
The major factors known to be associated with fibrosis progression in patients with a chronic HCV infection are older age at the time of infection [120], steatosis [121], intake of more than 50 grams/day of alcohol [122], and male gender [123]. Patients coinfected with HBV have a higher risk of developing HCC, although the mechanism is ill defined, while the influence of HIV coinfection on HCC development depends on CD4 counts [124][125]. Finally, host genetic factors have been associated with different outcomes of HCV infection. The HLA class II DQ02 allele has been associated with HCC development, whereas the HLA-B27 allele seems to have a protective role [126].
4.4. Role of chronic inflammation in HCV-related HCC
The leading hypothesis for HCV-induced hepatocarcinogenesis is that HCC develops in the context of chronic liver injury followed by regeneration and cirrhosis [102]. This concept is supported at different levels: 1.) HCV is an RNA virus that completes its life cycle in the cytoplasm and does not have the ability to integrate in the host genome, 2.) Based on the natural course of infection and on epidemiologic studies, cirrhosis seems to be a prerequisite in the development of HCV-induced HCC, and 3.) The development of HCC in HCV-infected patients is an extremely long process (30–40 years), indirectly suggesting the role of the microenvironment in triggering HCC [102]. In recent years, oxidative stress, steatosis and insulin resistance have been recognized as major procarcinogenic cofactors in chronic HCV infection [127].
Since the identification of HCV in 1990, many laboratories have attempted to define the complex interplay between the host response and HCV infection in a variety of experimental settings. Various intracellular signaling pathways that may have a role in HCV hepatocarcinogenesis have been identified. Oxidative stress, endoplasmic reticulum stress, Ca2+ signaling, TGF-β, p53, retinoblastoma-susceptibility protein (RB), Raf/MAPK, and Wnt/β-catenin pathways have been shown to be modulated either during HCV infection or by some HCV proteins in overexpression experiments (as discussed in the following sections) [127; 128; 129; 130; 131; 132; 133; 134; 135]. Curiously, these pathways may represent common mechanisms in viral hepatocarcinogenesis as their modulation also occurs during HBV infection [136]. In contrast to HBV infection, steatosis is more frequently observed in HCV-infected patients and is significantly associated with increased fibrosis and progression rate in HCV infection [102; 137]. DNA methylation [138] and iron deposition are also common in patients with HCV chronic infection. Iron overload induces mitochondrial injury and increases the risk of HCC development in transgenic mice expressing the HCV polyprotein [139]. HCV-related HCC chromosomal aberrations are uncommon, and genomic instability and oncogene activation are not driven by insertional mutagenesis [127].
4.5. HCV interference with molecular pathways: role in HCC
ER stress is a homeostatic mechanism that regulates cellular metabolism and protein synthesis in response to perturbations in protein folding and biosynthesis, which is also linked to oxidative stress [140]. Markers of intracellular oxidative stress such as reactive aldehydes produced by lipid peroxidation and DNA base-modified products generated by ROS are increased in patients with chronic HCV infection [141] and in human hepatocyte chimeric severe combined immunodeficiency (SCID)/Alb-uPA mice that are experimentally infected with HCV [129]. Antioxidant cellular enzymes, such as glutathione peroxidase (GSH) that function to maintain the redox balance of the cell are also modulated in patients with HCV infection and in hepatoma cell lines [130]. The complexity of oxidative stress during HCV infection is being elucidated, and synergy between chronic inflammation and oxidative stress directly triggered by HCV may explain its unique role in the progression of HCV-associated liver diseases. The fact that transgenic mice expressing the HCV core protein show an increased accumulation of ROS that correlates with HCC development in the absence of inflammation [131] suggests that at least some HCV proteins may have a direct role in triggering oxidative stress. In this regard, the core, NS3, and NS5A proteins have been shown to increase oxidative stress, although different mechanisms may be involved. HCV core protein is associated with the outer membrane of the mitochondria and may facilitate the uptake of Ca2+ into the mitochondria by increased oxidation of mitochondrial GSH [132]. NS3 triggers ROS production via activation of NADPH oxidase 2 (Nox2) [142]. NS5A localizes to the outer membrane of the ER and induces oxidative stress by inducing release of Ca2+ from the ER [133; 143]. Cellular responses to oxidative stress are regulated by MAPKs and other signaling proteins and may lead to double-stranded DNA breaks that affect regulation of cell growth and may promote cell transformation.
A unique property of HCV is its interference with the lipid metabolism of the cell by affecting lipid synthesis, oxidative stress, lipid peroxidation, insulin resistance, and the assembly and secretion of very-low-density lipoproteins (VLDLs). Clinical and experimental data suggest that HCV directly induces intrahepatic lipid accumulation (hepatosteatosis). Steatosis disappears in patients who respond to antiviral treatment, and relapse at the end of therapy results in steatosis reappearance [144]. In a recent meta-analysis of individual data from 3068 patients with histologically confirmed chronic HCV infection, the association between steatosis and fibrosis invariably was dependent on a simultaneous association between steatosis and hepatic inflammation [145]. Steatosis also develops in transgenic mice expressing the core protein [146]. Although steatotegenesis in HCV infection may be multifactorial, at least four pathways are directly affected by the virus, likely through the core protein: 1.) impaired secretion and/or assembly of VLDLs through inhibition of the activity of microsomal triglyceride transfer protein (MTP) [147], 2.) increased de novo synthesis of free fatty acids by upregulation of the sterol regulatory element binding protein (SREBP)-1c signaling pathway (core, NS2 and NS4 have been shown to up-regulate (SREBP)-1c at the transcription level [148]), 3.) impaired fatty acid degradation through core-induced inhibition of nuclear receptors regulating genes responsible for fatty acid degradation such as the peroxisome proliferators-activated receptor (PPAR)-α [149], and 4.) inhibition of tyrosine phosphorylation of insulin receptor substrate (IRS)-1 [150]. It has been also suggested that hepatic steatosis in HCV infection may act as a fuel for oxidative stress overproduction [151]. The production of ROS, as discussed above, may also result in the peroxidation of membrane lipids and structural proteins involved in lipid trafficking and secretion, ultimately leading to VLDL secretion, and to the development of steatosis.
Hepatitis C virus hepatocarcinogenesis could also be promoted through viral modulation of cellular tumor suppressors and regulatory proteins that control cell proliferation and apoptosis. HCV core, NS3 and NS5A proteins have been found to interfere with the p53 pathway (reviewed in [127]). Interestingly, HCV NS5B contains a conserved L-x-C/N-x-D amino acid motif that is homologous to Rb-binding domains in the oncoproteins of DNA viruses, and NS5B-dependent ubiquitination of Rb leads to proteosomal degradation of Rb in HCV-infected Huh7 cells [152]. Recently, some HCV proteins have been found to modulate cellular DExD/H helicases, such as DEX3 (which seems to be required for HCV replication) and DEX5 (a transcriptional coactivator of p53) [153]. The Wnt/β-catenin pathway controls cell proliferation, DNA synthesis and cell-cycle progression. This pathway has also been found to be modulated by some HCV proteins [134; 135] (discussed in sections below). A summary of putative mechanism in HCV hepatocarcinogenesis is depicted in Fig.6.
Fig. 6.
Hepatitis C virus hepatocarcinogenesis. Summary of putative mechanisms that could link chronic HCV infections to the development of HCC. See text for details
4.6. Virus triggers: HCV as a direct oncogenic virus
Although the leading hypothesis holds that HCV-related HCC results from chronic inflammation and factors in the microenvironment, other epidemiologic and experimental data underscore the potential direct oncogenic properties of HCV: 1.) Although a rare event, chronic HCV infection can also progress to HCC in the absence of cirrhosis, but the incidence and prevalence of this condition are very difficult to ascertain [154]; 2.) The combination of necrosis and inflammation alone, as is seen in autoimmune hepatitis, does not cause HCC; 3.) Some HCV proteins drive cell transformation in cell culture experiments. 4.) Data using transient and constitutive HCV protein expression systems in mice (a species that is not susceptible to HCV infection) suggest a possible direct oncogenic role of HCV; however, the genetic background of the mice is also an important factor in the development of HCV-induced HCC [155; 156]. It is important to note that in patients with HCV-related HCC, HCC develops 30 to 40 years after initial exposure to the virus, and extrapolating in vitro data to the understanding of HCV-related HCC is difficult [102]. Moreover, another major caveat is that HCV-infected chimpanzees, the only animal species other than humans permissive for HCV infection, liver disease is minimal, and there is no evidence of HCC [157].
4.6.1. Role of HCV structural proteins in hepatocyte transformation
The HCV structural proteins, core (C), E1 and E2 have been studied in both in vitro culture systems and transgenic mice for their potential oncogenic properties. An overlapping reading frame (frameshift protein, F) in the core has recently been identified [158]. The function of F remains to be elucidated, and there are no data in the literature on its potential contribution to hepatocarcinogenesis. Studies in transgenic mice expressing the complete HCV polyprotein suggest that structural HCV proteins contribute to the development of steatosis rather than HCC in the absence of anti-viral immune response [159]. Nevertheless a considerable number of studies in which HCV structural proteins were expressed in cell culture and in mice lead to the hypothesis that some HCV structural proteins may have a direct oncogenic role.
HCV core
In addition to its role in viral assembly, the core protein is a key player in HCV-cell interaction by modulating cellular gene products and cell regulatory pathways. Among these, core protein has been involved in cellular transformation, transcription regulation, apoptosis, lipid metabolism, and oxidative stress (reviewed in [160]).
How does HCV core protein drive cellular transformation? Evidence from cell-culture systems shows that core protein indirectly regulates cellular proliferation and cell cycle control by different mechanisms, such as binding and modulation of various tumor suppressor proteins or their targets, and activation of some intracellular pathways. With respect to tumor suppressor proteins, the HCV core protein has been shown to bind to p53 [161], p73 [162], and Rb [163]. A transcriptional target of p53 that regulates the activity of cyclin/CDK complexes, p21/Waf, is also modulated by the HCV core protein, suggesting a role for HCV core in modulating cell-cycle control [164]. Core has also been shown to affect NF-κB transcription, but data in the literature are controversial [165; 166]. There is also evidence that core protein modulates at least three intracellular pathways known to have a direct role in cell proliferation: 1.) Core activates the Raf1/MAPK pathway, thereby promoting cell proliferation [167]; 2.) core activates the Wnt/β-catenin pathway, which can control DNA synthesis and cell-cycle progression [168]; and 3.) core modulates TGFβ signaling, which can control cell proliferation, apoptosis, and differentiation [169]. Interestingly, both cell culture-based studies and data from patients with chronic HCV infection support the notion that core protein modulates the TGFβ pathway. Several studies have demonstrated that the degree of fibrosis also correlates with high levels of TGFβ in chronically HCV-infected patients [170]. Pavio and coworkers have recently isolated core variants from liver tumor but not from adjacent nontumor tissue [171]. Interestingly, only the core variants isolated from the tumor liver tissue inhibited the TGFβ pathway and interacted with Smad3. Conversely, the expression of TGFβ was transcriptionally upregulated by wild-type core [172], and its expression was also induced in hepatic stellate cells, a direct mechanism for promoting fibrogenesis [173]. To reconcile these apparently conflicting data, it seems that the phase of the disease is a critical factor in understanding the effects of core on the TGFβ pathway. Therefore, a dual action of core on the TGFβ pathway has been suggested. First, during the early phase of HCV infection, core might trigger TGFβ synthesis and promote fibrogenesis. Second, in the cirrhotic liver (resulting from long-term inflammation and fibrosis), core variants would contribute to cellular transformation by inhibiting TGFβ–dependent anti-proliferative pathways, driving clonal cell expansion [171].
Perhaps the most direct evidence of the oncogenic activity of HCV core comes from studies in the core transgenic mouse [131; 146]. In this model, mice expressing the core protein develop histologic characteristics of chronic hepatitis infection, similar to those observed in patients with HCV infection (steatosis, bile duct damage, and lymphoid follicle formation) [174], and late in life, the mice develop HCC [131]. A striking finding in core-transgenic mice is the absence of prominent inflammation in the liver with increased production of oxidative stress. When inflammation is induced in this mouse model, oxidative stress is significantly enhanced. Interestingly, only the Jun N-terminal kinase (JNK) signaling pathway has been shown to be activated in the core transgenic mouse model prior to HCC development. Downstream of the JNK pathway, the activation of the transcription factor AP-1 was significantly enhanced, with transcriptional and translational up-regulation of cyclin D1 and CDK4, indicating a causative effect on the development of cellular transformation [175].
Envelope glycoproteins E1 and E2
In contrast to core protein, E1 and E2 expression has not been linked to the development of HCC. Although cell culture data demonstrate that E2 can interfere with dsRNA-activated protein kinase (PKR) [176] and ER-stress signaling kinase (PERK) signaling pathways [177], and promote cellular proliferation through the MAPK/ERK via the cellular receptor CD81 and low-density lipoprotein receptor (LDLR) [178], there is no evidence of HCC development in transgenic mice expressing E1 and/or E2 glycoproteins [179].
4.6.2. Role of HCV non-structural proteins in hepatocyte transformation
Recently, several groups have reported interesting findings indicating that some of the HCV non-structural proteins, NS2, NS3, NS4A, NS4B, NS5A, and NS5B, play a key role in virus-host interactions and in the evasion of the immune response (reviewed in [180]). However, their potential contribution to hepatocarcinogenesis is considered more indirect, and in general, it is based on the overexpression of HCV NS proteins in cell culture systems.
NS2
Although less information is available in the literature regarding the potential contribution of NS2 to HCV pathogenesis, it has been reported that NS2 may inhibit gene expression via downregulation of transcription [181]. NS2-mediated inhibition of a cellular proapoptotic molecule, CIDE-B (cell death-inducing DFFA-like effector b) has been shown [182]. To date, there is no evidence that NS2 has a direct oncogenic activity.
NS3
HCV NS3 is a multifunctional protein with RNA helicase, NTPase, and protease activity. NS3 expression has been linked to cellular transformation in cell culture experiments using murine fibroblast NIH 3T3 cells [183], and non-tumorigenic rat fibroblast RT cells [184]. Interestingly, cell transformation seems to require the NS3 serine protease activity, and tumor development is observed in athymic nude mice in which transformed NS3-expressing RT cells are injected [184]. Others have suggested that NS3 could promote cellular transformation via its interaction with several cellular proteins, such as tumor suppressor p53, which leads to transcriptional repression of p21 (wafI) [185]. Although there is no direct evidence for NS3 oncogenic activity, recent investigations have demonstrated that NS3, together with its cofactor NS4A, plays a remarkable role in HCV evasion of the innate immune response by interfering with the retinoic acid-inducible gene 1 (RIG-1), melanoma differentiation associated gene-5 (MDA-5), and toll-like receptor 3 (TLR3) signaling pathways [186].
NS4B
Much remains to be learned about the role of NS4B protein in HCV life cycle and pathogenesis (recently reviewed in [187]). Although, there are no in vivo data suggesting a role of NS4B in HCC, both direct and indirect contributions of NS4B in cellular transformation have been suggested. Transgenic mice expressing NS4B in the liver did not display any tumor development [188]. However, overexpression of NS4B has been found to transform NIH 3T3 cells in cooperation with the HA-ras oncogene [189]. Recently, the nucleotide binding motif of NS4B has been shown to mediate cellular transformation activity [190]. Moreover, an indirect role of NS4B in carcinogenesis has been speculated based on its ability to induce ER stress and modulate lipid metabolism. In a cell culture system, expression of NS4B induces activation of the sterol regulatory element binding proteins (SREBPs) signaling pathway, and lipid accumulation is dependent on the phosphoinositide-3 kinase (PI3K) pathway [148]. Whether this interaction has a role in steatosis in chronic HCV infection is unknown.
NS5A
HCV NS5A [191] is a membrane-bound cytoplasmic protein that is part of the HCV RNA replication complex; however, a truncated form of NS5A is localized to the nucleus and acts as a transcriptional activator. In addition to a potential role for NS5A in the modulation of several intracellular pathways and in evasion of the host immune response, studies have shown that C57BL/6J transgenic mice harboring the HCV NS5A gene (genotype 1b) under the control of HBV enhancer developed steatosis over a 6-month period, and in some mice HCC was induced [155]. In this model, high levels of ROS, NF-κB, and STAT3 were detected in hepatocytes of NS5A transgenic mice, and NS5A colocalized with apolipoprotein A-I in fatty hepatocytes. However, whether NS5A has a direct oncogenic activity remains controversial. In this regard, previous studies using HCV genotype 1a and FBV mice did not find any abnormality in the liver of NS5A transgenic mice [156]. Overall, it seems that the HCV genotype and the mouse strain used in these studies may account for the observed differences. In contrast, there is agreement regarding the antiapoptotic activity of NS5A [192]. Mankouri et al. [193] have recently shown that in hepatoma cells infected with HCV, oxidative stress failed to initiate apoptosis via Kv2.1, a host cell K+ channel that mediates the amplification of an outward K+ current in response to oxidative stress that precedes apoptosis. Interestingly, NS5A inhibited oxidative stress-induced p38 MAPK phosphorylation of Kv2.1. Therefore, NS5A may play a key role in maintaining infected cells refractory to proapoptotic stimuli, contributing to the establishment of chronic HCV infection. NS5A also interacts with the proto-oncogene β-catenin, stimulating its transcriptional activity and activating the p85 regulatory subunit of PI3K [135]. Therefore, core and NS5A may contribute synergistically to HCV-induced hepatocarcinogenesis through the β-catenin pathway.
5. Emerging areas of investigation
5.1. Role of microRNAs in HBV and HCV-related HCC
MicroRNAs (miRNAs) are a novel class of small non-coding single-stranded RNAs that regulate gene expression at the posttranscriptional level [194]. Although all their functions have not been fully identified, evidence increasingly indicates that miRNAs play a major role in fundamental cellular processes, in the regulation of the immune system, and in the onset and progression of many diseases, such as cancer [195]. Interestingly, evidence is also emerging regarding the contribution of miRNAs in viral pathogenesis.
Recent data support the notion that miRNAs may be key players in the regulation of liver functions, and in hepatocarcinogenesis, regardless of HCC etiology. Using global miRNA profiling of HCC cell lines or liver tissue, the expression of several miRNAs has been found to be either up-regulated (miR-18, miR-21, miR-221, miR-222, miR-224, miR-373, and miR-301) [196; 197], or down-regulated (miR-122, miR-223, miR-125, miR-130a, miR-150, miR-199, miR-200, and let-7 family members) [198; 199; 200] in HCC. These studies have identified miRNA targets in genes potentially involved in cellular transformation, such as cyclin-dependent inhibitors p27/CDKN1B and p57/CDKN1C, PI3K, cyclins D2 and E2, or stathmin1 (STMN1), a key microtubule-regulatory protein that controls the S phase of the cell cycle, and cellular proliferation. A central question is whether the alteration of the miRNAome is a consequence of, or a driving force in hepatocarcinogenesis. Based on data currently available, and the emerging complexity of the miRNA/mRNA regulatory networks, it is likely that some miRNA play a triggering role in HCC. Conversely, deregulation of the expression of some miRNAs may be indirectly induced in a context in which other cellular transformation processes have been activated.
Alterations of miRNA profiles in the context of HBV or HCV-related hepatocarcinogenesis has emerged as an area of active investigation, but how miRNAs may impact viral replication and associated liver disease progression remains incompletely understood. Interestingly, differential miRNAs expression patterns have been found in the livers of HBV- and HCV- infected individuals with HCC [201]. Moreover, using pathway analysis of infection-associated miRNA-targeted genes, it has been possible to identify pathways related to cell death, DNA damage, recombination, and signal transduction as potential targets in HBV-infected livers, whereas miRNA-targeted pathways were often related to immune response, antigen presentation, cell cycle, proteasome, and lipid metabolism in HCV-infected livers [201]. However, because most HBV and HCV miRNA profiling studies have been conducted in established cell lines or by comparing tumor tissues to non-tumor tissues, precisely how HBV- or HCV-induced alterations of normal miRNA profiles impacts HCC development or progression is not entirely understood and remains an area of active investigation. While informative, the miRNA profiles conducted in established cell lines or liver tumors are currently unable to answer whether HBV- or HCV-induced changes in miRNA profiles are a cause or consequence of transformation processes.
The first miRNA shown to be related to HCV infection, miRNA-122, is a liver-specific miRNA that exhibits a remarkable role in the HCV life cycle. Two functions of miRNA-122 that impact HCV replication have been identified: 1.) miRNA-122 enhances the replication of HCV by targeting the viral 5' non-coding region [202], and 2.) miR-122 stimulates HCV translation by enhancing the association of ribosomes with the viral RNA [203]. Interestingly, a recent study demonstrated that treatment of chronically HCV-infected chimpanzees with a locked nucleic acid (LNA)-modified oligonucleotide complementary to miR-122 caused long-lasting suppression of HCV viremia, with no evidence of viral resistance or side effects in the treated animals; diminished HCV-induced liver abnormalities were also observed [204]. This study suggests that therapeutic silencing of miRNA-122 may be a promising novel antiviral treatment against HCV infection. Although miR-122 expression has been shown to be down-regulated in some HCCs in humans and rodents, its expression can be up-regulated in HCV-related HCC [205; 206; 207], and the role of miRNA-122 in HCV-related hepatocarcinogenesis is controversial. Interesting, there is evidence that HCV core and NS5B may modulate expression of host cell miRNAs. Core protein can suppress the activity of Dicer, an enzyme involved in the production of miRNAs [208], and NS5B has been shown to use miRNAs as primers to synthesize dsRNA and amplify the process of RNA interference [209]. Although HCV core and NS5A proteins can alter the levels of various cellular miRNAs, whether these changes in miRNA levels directly impact HCC development remains to be determined.
Regulation of miRNAs by HBV and HBx has been studied in established cell lines and HBV-associated tumors. Similar to studies with HCV, our understanding of HBV-induced alteration in cellular miRNA levels is still incomplete. Lethal-7a (Let7a) miRNAs were shown to be down-regulated in HBx-expressing HepG2 cells [210]. Let7a miRNAs negatively regulated cell proliferation, and downregulation of Let 7a by HBx, and associated up-regulation of STAT 3, induced cell proliferation. HBx expression and HBV replication in HepG2 cells also caused overexpression of miRNA-602, which can inhibit the expression of the tumor suppressor, RASSF1A, providing another mechanisms that might link HBV infections to the development of HCC [211]. The results of another study demonstrated that miRNA-152 was frequently down-regulated in HBV-related HCC [212]. HBV-induced downregulation of miRNA-152 caused global DNA hypermethylation by facilitating upregulation of DNA methyltransferase 1, providing an epigenetic signal that silenced two tumor suppressor genes, glutathione S-transferase pi 1 and E-cadherin and suggesting that miRNA152 may have a tumor suppressor role that is blocked in HBV-associated HCC. Finally, there is evidence that miR-122 may inhibit HBV replication; however, considering the high level of miR-122 that is expressed in hepatocytes, it is unclear precisely how miR-122 impacts HBV replication in the liver where high levels of HBV replication occur in the presence of miR-122 [213]. Overall, although it is clear that HBV proteins can alter the levels of various cellular miRNAs, whether these changes in miRNA levels directly impact HCC development remains to be determined.
6. Perspective
While much is known regarding the impact of HBV and HCV infections on hepatocyte and liver physiology, and there is clear evidence that infections with either HBV or HCV are linked to the development of HCC, there are still many unanswered question regarding the molecular mechanisms that link HBV or HCV infections to liver disease progression and HCC development. How the combination of viral proteins and inflammatory responses specifically alter the liver intra- and intercellular environment remains incompletely understood, and a complete evaluation of these processes is paramount to developing effective strategies for treating HBV or HCV infections and preventing HCC. To develop a complete understanding of HCV liver disease and hepatocarcinogenesis, it will be important to define the basis of spontaneous clearance of HCV infection and its long-term consequences, and to understand why HCV infections sometimes causes cirrhosis and sometimes do not. Similarly, HBV-associated HCC can also occur in the presence or absence of cirrhosis, and understanding mechanisms that impact HCC development in the presence or absence of cirrhosis will be necessary to fully elucidate carcinogenic pathways that are stimulated during an HBV infection. Current knowledge of the proteomics of HBV- and HCV-induced liver disease is in its infancy, and another challenge for the HBV and HCV fields in the coming years will be the study of potential proteomic signatures associated with these infections and associated disease progression. Improved technologies for studying the proteome in diagnosis and treatment must be developed. Finally, the role of epigenetics in HBV and HCV liver disease is being elucidated but also requires additional focus; this endeavor may develop into a major research field during the coming years.
A major impediment to understanding the connection between HBV and/or HCV infections and the development of HCC remains the absence of experimentally tractable animal models that mimic HBV or HCV infections in humans and can be used to study the entire carcinogenic process. Consequently, the biological relevance of the laboratory observations that we have discussed awaits the development of animal models in which natural HBV or HCV infections lead to HCC. Most studies in cell lines that have addressed the potential direct oncogenic role of HCV proteins have used the Huh7 cell line, which is of malignant origin and expresses a mutant p53 [214]. Because Huh7 and it derivatives are the only cell lines that fully support the HCV replication cycle using the newly developed full-length HCV chimeric genomes, additional permissive cell culture systems must be developed to fully elucidate the impact of HCV infection on cell physiology. Studies in the HBV field have clearly demonstrated cell-specific affects of HBV proteins. Oncogenic processes associated with HBV replication or HBx expression have predominately been studied in transformed or immortalized cell lines or in systems where viral proteins are over expressed. Cell culture systems in which transformation processes more closely recapitulate in vivo hepatocyte transformation processes are needed to fully understand the oncogenic capacity of HBV as are animal models, such as transgenic mice, where viral protein levels more closely mimic those observed in an authentic infection. Similarly, animal models where anti-HBV or HCV immune responses can be studied will be necessary to fully understand the role of the inflammatory response to disease progression. The narrow host range of HBV and HCV may make it difficult to develop these types of experimentally tractable animal models, but it is possible that the full characterization of host specific receptors for these viruses may facilitate generation of transgenic animals that could serve as novel model systems.
Finally, ongoing research in many laboratories is aimed at identifying the miRNAs that are differentially expressed during HBV and HCV infection. Our understanding of the role of miRNAs in HBV and HCV pathogenesis is incomplete, but data accumulated to date warrant further in-depth investigations. miRNA profiling systems are needed that not only compare miRNA profiles in tumors relative to non-tumor tissues but also HCV or HBV-induced alterations in miRNA profiles that may occur early in an infection, and in the context of inflammation. These types of studies may identify changes in miRNAs that initiate or contribute to processes that alter hepatocyte physiology and contribute to transformation processes. Overall, the precise identification of molecular processes that link HBV or HCV infections to HCC development should provide a means to understand replication mechanisms of these viruses as well as potential targets that could help decrease the global incidence of HCC.
Acknowledgments
We thank members of the Bouchard and Navas-Martin laboratories for critical reading of the manuscript. We thank Connie Lin, Siddhartha Rawat, and Tricia Gearhart for figure designs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors have no conflicting interests.
References
- [1].El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- [2].Wang XW, Hussain SP, Huo T, Wu CG, Forgues M, Hofseth LJ, Brechot C, Harris CC. Molecular pathogenesis of human hepatocellular carcinoma. Toxicology. 2002;181–182:43–47. doi: 10.1016/s0300-483x(02)00253-6. [DOI] [PubMed] [Google Scholar]
- [3].Guidotti LG, Chisari FV. Immunobiology and pathogenesis of viral hepatitis. Annu Rev Pathol. 2006;1:23–61. doi: 10.1146/annurev.pathol.1.110304.100230. [DOI] [PubMed] [Google Scholar]
- [4].Bartosch B, Thimme R, Blum HE, Zoulim F. Hepatitis C virus-induced hepatocarcinogenesis. J Hepatol. 2009;51:810–820. doi: 10.1016/j.jhep.2009.05.008. [DOI] [PubMed] [Google Scholar]
- [5].Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Seeger C, Zoulim F, Mason W. Hepadnaviridae. In: Knipe D, Howley P, editors. Fields Virology, Wolters Kluwer Health. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2977–3030. [Google Scholar]
- [7].Sherman M. Hepatocellular carcinoma: epidemiology, risk factors, and screening. Semin Liver Dis. 2005;25:143–154. doi: 10.1055/s-2005-871194. [DOI] [PubMed] [Google Scholar]
- [8].Roberts LR, Gores GJ. Hepatocellular carcinoma: molecular pathways and new therapeutic targets. Semin Liver Dis. 2005;25:212–225. doi: 10.1055/s-2005-871200. [DOI] [PubMed] [Google Scholar]
- [9].Anzola M. Hepatocellular carcinoma: role of hepatitis B and hepatitis C viruses proteins in hepatocarcinogenesis. J Viral Hepat. 2004;11:383–393. doi: 10.1111/j.1365-2893.2004.00521.x. [DOI] [PubMed] [Google Scholar]
- [10].Greenberger N. History Taking and Physical Examination for the Patient with Liver Diseases. In: Schiff ER, Sorell MF, Maddrey WC, editors. Schiff's Diseases of The Liver. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 3–19. [Google Scholar]
- [11].Beasley R, Lin C-C, Hwang L-Y, Chien C-S. Hepatocellular carcinoma and hepatitis B virus: a prospective study of 22 707 men in Taiwan. Lancet. 1981:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- [12].Hollinger F, Liang T. Viral Hepatitis. Lippincott Williams and Wilkins; Philadelphia: 2002. Hepatitis B Virus; pp. 103–168. [Google Scholar]
- [13].Besisik F, Karaca C, Akyuz F, Horosanh S, Onel D, Badur S, Sever M, Danahoglu A, Demir K, Kaymakoglu S, Cakaloglu Y, Okten A. Occult HBV infection--both hidden and mysterious. Gastroenterology. 2003;125:1903–1905. doi: 10.1053/j.gastro.2003.05.011. [DOI] [PubMed] [Google Scholar]
- [14].Guidotti LG, Martinez V, Loh YT, Rogler CE, Chisari FV. Hepatitis B virus nucleocapsid particles do not cross the hepatocyte nuclear membrane in transgenic mice. J Virol. 1994;68:5469–5475. doi: 10.1128/jvi.68.9.5469-5475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rabe B, Delaleau M, Bischof A, Foss M, Sominskaya I, Pumpens P, Cazenave C, Castroviejo M, Kann M. Nuclear entry of hepatitis B virus capsids involves disintegration to protein dimers followed by nuclear reassociation to capsids. PLoS Pathog. 2009;5:e1000563. doi: 10.1371/journal.ppat.1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hartmann-Stuhler C, Prange R. Hepatitis B virus large envelope protein interacts with gamma2-adaptin, a clathrin adaptor-related protein. J Virol. 2001;75:5343–5351. doi: 10.1128/JVI.75.11.5343-5351.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kian Chua P, Lin MH, Shih C. Potent inhibition of human Hepatitis B virus replication by a host factor Vps4. Virology. 2006;354:1–6. doi: 10.1016/j.virol.2006.07.018. [DOI] [PubMed] [Google Scholar]
- [18].Rost M, Mann S, Lambert C, Doring T, Thome N, Prange R. Gamma-adaptin, a novel ubiquitin-interacting adaptor, and Nedd4 ubiquitin ligase control hepatitis B virus maturation. J Biol Chem. 2006;281:29297–29308. doi: 10.1074/jbc.M603517200. [DOI] [PubMed] [Google Scholar]
- [19].Watanabe T, Sorensen EM, Naito A, Schott M, Kim S, Ahlquist P. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc Natl Acad Sci U S A. 2007;104:10205–10210. doi: 10.1073/pnas.0704000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Guidotti L, Rochford R, Chung J, Shapiro M, Purcell R, Chisari F. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- [21].Guidotti LG. The role of cytotoxic T cells and cytokines in the control of hepatitis B virus infection. Vaccine. 2002;20(Suppl 4):A80–82. doi: 10.1016/s0264-410x(02)00392-4. [DOI] [PubMed] [Google Scholar]
- [22].Guidotti LG, Chisari FV. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu Rev Immunol. 2001;19:65–91. doi: 10.1146/annurev.immunol.19.1.65. [DOI] [PubMed] [Google Scholar]
- [23].Prince AM, Brotman B. Perspectives on hepatitis B studies with chimpanzees. ILAR J. 2001;42:85–88. doi: 10.1093/ilar.42.2.85. [DOI] [PubMed] [Google Scholar]
- [24].Guidotti L, Matzke B, Schaller H, Chisari F. High-level hepatitis B virus replication in transgenic mice. J. Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zheng Y, Chen WL, Louie SG, Yen TS, Ou JH. Hepatitis B virus promotes hepatocarcinogenesis in transgenic mice. Hepatology. 2007;45:16–21. doi: 10.1002/hep.21445. [DOI] [PubMed] [Google Scholar]
- [26].Ando K, Guidotti LG, Wirth S, Ishikawa T, Missale G, Moriyama T, Schreiber RD, Schlicht HJ, Huang SN, Chisari FV. Class I-restricted cytotoxic T lymphocytes are directly cytopathic for their target cells in vivo. J Immunol. 1994;152:3245–3253. [PubMed] [Google Scholar]
- [27].Guidotti LG, Ishikawa T, Hobbs MV, Matzke B, Schreiber R, Chisari FV. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity. 1996;4:25–36. doi: 10.1016/s1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- [28].Berasain C, Castillo J, Perugorria MJ, Latasa MU, Prieto J, Avila MA. Inflammation and liver cancer: new molecular links. Ann N Y Acad Sci. 2009;1155:206–221. doi: 10.1111/j.1749-6632.2009.03704.x. [DOI] [PubMed] [Google Scholar]
- [29].Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bill CA, Summers J. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc Natl Acad Sci U S A. 2004;101:11135–11140. doi: 10.1073/pnas.0403925101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bonilla Guerrero R, Roberts LR. The role of hepatitis B virus integrations in the pathogenesis of human hepatocellular carcinoma. J Hepatol. 2005;42:760–777. doi: 10.1016/j.jhep.2005.02.005. [DOI] [PubMed] [Google Scholar]
- [32].Menne S, Cote PJ. The woodchuck as an animal model for pathogenesis and therapy of chronic hepatitis B virus infection. World J Gastroenterol. 2007;13:104–124. doi: 10.3748/wjg.v13.i1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wei Y, Fourel G, Ponzetto A, Silvestro M, Tiolais P, Buendia M-A. Hepadnavirus integration: mechanisms of activation of the N-myc2 retrotransposon in woodchuck liver tumors. J. Virol. 1992:5265–5276. doi: 10.1128/jvi.66.9.5265-5276.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ogston CW, Jonak GJ, Rogler CE, Astrin SM, Summers J. Cloning and structural analysis of integrated woodchuck hepatitis virus sequences from hepatocellular carcinomas of woodchucks. Cell. 1982;29:385–394. doi: 10.1016/0092-8674(82)90155-6. [DOI] [PubMed] [Google Scholar]
- [35].Moroy T, Marchio A, Etiemble J, Trepo C, Tiollais P, Buendia MA. Rearrangement and enhanced expression of c-myc in hepatocellular carcinoma of hepatitis virus infected woodchucks. Nature. 1986;324:276–279. doi: 10.1038/324276a0. [DOI] [PubMed] [Google Scholar]
- [36].Fourel G, Trepo C, Bougueleret L, Henglein B, Ponzetto A, Tiollais P, Buendia MA. Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumours. Nature. 1990;347:294–298. doi: 10.1038/347294a0. [DOI] [PubMed] [Google Scholar]
- [37].Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- [38].Feitelson MA, Lee J. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 2007;252:157–170. doi: 10.1016/j.canlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- [39].Ferber MJ, Montoya DP, Yu C, Aderca I, McGee A, Thorland EC, Nagorney DM, Gostout BS, Burgart LJ, Boix L, Bruix J, McMahon BJ, Cheung TH, Chung TK, Wong YF, Smith DI, Roberts LR. Integrations of the hepatitis B virus (HBV) and human papillomavirus (HPV) into the human telomerase reverse transcriptase (hTERT) gene in liver and cervical cancers. Oncogene. 2003;22:3813–3820. doi: 10.1038/sj.onc.1206528. [DOI] [PubMed] [Google Scholar]
- [40].Ma NF, Lau SH, Hu L, Xie D, Wu J, Yang J, Wang Y, Wu MC, Fung J, Bai X, Tzang CH, Fu L, Yang M, Su YA, Guan XY. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin Cancer Res. 2008;14:5061–5068. doi: 10.1158/1078-0432.CCR-07-5082. [DOI] [PubMed] [Google Scholar]
- [41].Wang Y, Lau SH, Sham JS, Wu MC, Wang T, Guan XY. Characterization of HBV integrants in 14 hepatocellular carcinomas: association of truncated X gene and hepatocellular carcinogenesis. Oncogene. 2004;23:142–148. doi: 10.1038/sj.onc.1206889. [DOI] [PubMed] [Google Scholar]
- [42].Kekule AS, Lauer U, Meyer M, Caselmann WH, Hofschneider PH, Koshy R. The preS2/S region of integrated hepatitis B virus DNA encodes a transcriptional transactivator. Nature. 1990;343:457–461. doi: 10.1038/343457a0. [DOI] [PubMed] [Google Scholar]
- [43].Chisari F, Pinkert C, Milich D, Filippi P, McLachlan A, Palmiter R, Brinster R. A transgenic mouse model of the chronic hepatitis B surface antigen carrier state. Science. 1985;230:1157–1160. doi: 10.1126/science.3865369. [DOI] [PubMed] [Google Scholar]
- [44].Yen TS. Hepadnaviral X protein: review of recent progress. J. Biomed. Sci. 1996;3:20–30. doi: 10.1007/BF02253575. [DOI] [PubMed] [Google Scholar]
- [45].Murakami S, Cheong J, Kaneko S. Human hepatitis virus X gene encodes a regulatory domain that represses transactivation of X protein. J. Biol. Chem. 1994;269:15118–15123. [PubMed] [Google Scholar]
- [46].Chen H, Kaneko S, Girones R, Anderson R, Hornbuckle W, Tennant B, Cote P, Gerin J, Purcell R, Miller R. The woodchuck helpatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhang Z, Torii N, Hu Z, Jacob J, Liang T. X-deficient woodchuck hepatitis virus mutants behave like attenuated viruses andinduce protective immunity in vivo. J. Clin. Invest. 2001;108:1523–1531. doi: 10.1172/JCI13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xu Z, Yen T, Wu L, Madden C, Tan W, Slagle B, Ou J.-h. Enhancement of hepatitis B virus replication by its X protein in transgenic mice. J. Virol. 2002;76:2579–2584. doi: 10.1128/jvi.76.5.2579-2584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Reifenberg K, Nusser P, Lohler J, Spindler G, Kuhn C, von Weizsacker F, Kock J. Virus replication and virion export in X-deficient hepatitis B virus transgenic mice. J Gen Virol. 2002;83:991–996. doi: 10.1099/0022-1317-83-5-991. [DOI] [PubMed] [Google Scholar]
- [51].Keasler VV, Hodgson AJ, Madden CR, Slagle BL. Enhancement of hepatitis B virus replication by the regulatory X protein in vitro and in vivo. J Virol. 2007;81:2656–2662. doi: 10.1128/JVI.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tateno C, Yoshizane Y, Saito N, Kataoka M, Utoh R, Yamasaki C, Tachibana A, Soeno Y, Asahina K, Hino H, Asahara T, Yokoi T, Furukawa T, Yoshizato K. Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol. 2004;165:901–912. doi: 10.1016/S0002-9440(10)63352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tsuge M, Hiraga N, Akiyama R, Tanaka S, Matsushita M, Mitsui F, Abe H, Kitamura S, Hatakeyama T, Kimura T, Miki D, Mori N, Imamura M, Takahashi S, Hayes CN, Chayama K. HBx protein is indispensable for development of viraemia in human hepatocyte chimeric mice. J Gen Virol. 2010;91:1854–1864. doi: 10.1099/vir.0.019224-0. [DOI] [PubMed] [Google Scholar]
- [54].Tsuge M, Hiraga N, Takaishi H, Noguchi C, Oga H, Imamura M, Takahashi S, Iwao E, Fujimoto Y, Ochi H, Chayama K, Tateno C, Yoshizato K. Infection of human hepatocyte chimeric mouse with genetically engineered hepatitis B virus. Hepatology. 2005;42:1046–1054. doi: 10.1002/hep.20892. [DOI] [PubMed] [Google Scholar]
- [55].Leupin O, Bontron S, Schaeffer C, Strubin M. Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death. J Virol. 2005;79:4238–4245. doi: 10.1128/JVI.79.7.4238-4245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sitterlin D, Bergametti F, Tiollais P, Tennant BC, Transy C. Correct binding of viral X protein to UVDDB-p127 cellular protein is critical for efficient infection by hepatitis B viruses. Oncogene. 2000;19:4427–4431. doi: 10.1038/sj.onc.1203770. [DOI] [PubMed] [Google Scholar]
- [57].Zhang Z, Sun E, Ou JH, Liang TJ. Inhibition of Cellular Proteasome Activities Mediates HBX-independent Hepatitis B Virus Replication In Vivo. J Virol. 2010 doi: 10.1128/JVI.00579-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhang Z, Protzer U, Hu Z, Jacob J, Liang TJ. Inhibition of cellular proteasome activities enhances hepadnavirus replication in an HBX-dependent manner. J Virol. 2004;78:4566–4572. doi: 10.1128/JVI.78.9.4566-4572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Balsano C, Billet O, Bennoun M, Cavard C, Zider A, Grimber G, Natoli G, Briand P, Levrero M. Hepatitis B virus X gene product acts as a transactivator in vivo. J. of Hepat. 1994;21:103–109. doi: 10.1016/s0168-8278(94)80144-4. [DOI] [PubMed] [Google Scholar]
- [60].Reifenberg K, Wilts H, Lohler J, Nusser P, Hanano R, Guidotti LG, Chisari FV, Schlicht HJ. The hepatitis B virus X protein transactivates viral core gene expression in vivo. J Virol. 1999;73:10399–10405. doi: 10.1128/jvi.73.12.10399-10405.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Klein N, Bouchard M, Wang L-H, Kobarg C, Schneider R. Src kinases involved in hepatitis B virus replication. EMBO J. 1999;18:5019–5027. doi: 10.1093/emboj/18.18.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tarn C, Zou L, Hullinger R, Andrisani O. Hepatitis B virus X protein activates the p38 mitogen-activated protein kinase pathway in dedifferentiated hepatocytes. J. Virol. 2002;76:9763–9772. doi: 10.1128/JVI.76.19.9763-9772.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Bouchard M, Wang L-H, Schneider R. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- [64].Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- [65].Chami M, Ferrari D, Nicotera P, Paterlini-Brechot P, Rizzuto R. Caspase-dependent alterations of Ca2+ signaling in the induction of apoptosis by hepatitis B virus X protein. J Biol Chem. 2003;278:31745–31755. doi: 10.1074/jbc.M304202200. [DOI] [PubMed] [Google Scholar]
- [66].Oh JC, Jeong DL, Kim IK, Oh SH. Activation of calcium signaling by hepatitis B virus-X protein in liver cells. Exp Mol Med. 2003;35:301–309. doi: 10.1038/emm.2003.41. [DOI] [PubMed] [Google Scholar]
- [67].Clippinger AJ, Gearhart TL, Bouchard MJ. Hepatitis B virus X protein modulates apoptosis in primary rat hepatocytes by regulating both NF-kappaB and the mitochondrial permeability transition pore. J Virol. 2009;83:4718–4731. doi: 10.1128/JVI.02590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gearhart TL, Bouchard MJ. The Hepatitis B Virus X Protein Modulates Hepatocyte Proliferation Pathways to Stimulate Viral Replication. J Virol. 2010 doi: 10.1128/JVI.02196-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Bouchard MJ, Puro RJ, Wang L, Schneider RJ. Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication. J Virol. 2003;77:7713–7719. doi: 10.1128/JVI.77.14.7713-7719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].McClain SL, Clippinger AJ, Lizzano R, Bouchard MJ. HBV replication is associated with an HBx-dependent mitochondrial regulated increase in cytosolic calcium levels. J Virol. 2007 doi: 10.1128/JVI.00740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Rahmani Z, Huh K-W, Lasher R, Siddiqui A. Hepatitis B virus X protein colocalizes to mitochondria with human voltage -dependent anion channel, HVDAC3, and alters its transmembrane potential. J. Virol. 2000;74:2840–2846. doi: 10.1128/jvi.74.6.2840-2846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Clippinger AJ, Bouchard MJ. The Hepatitis B Virus Hbx Protein Localizes to Mitochondria in Primary Rat Hepatocytes and Modulates Mitochondrial Membrane Potential. J Virol. 2008 doi: 10.1128/JVI.00154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Pan J, Duan LX, Sun BS, Feitelson MA. Hepatitis B virus X protein protects against anti-Fas-mediated apoptosis in human liver cells by inducing NF-kappa B. J Gen Virol. 2001;82:171–182. doi: 10.1099/0022-1317-82-1-171. [DOI] [PubMed] [Google Scholar]
- [74].Elmore J, Hancock A, Chang S-F, Wang X, Chang S, Callahan C, Geller D, Will H, Harris C. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc. Natl. Acad. Sci. USA. 1997;94:14707–14712. doi: 10.1073/pnas.94.26.14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Shih W-L, Kuo M-L, Chuang S-E, Cheng A-L, Doong S-L. Hepatitis B virus X protein inhibits transforming growth factor-β-induced apoptosis through the activation of phosphatidylinositol 3-kinase pathway. J. Biol. Chem. 2000;275:25858–25864. doi: 10.1074/jbc.M003578200. [DOI] [PubMed] [Google Scholar]
- [76].Su F, Schneider R. Hepatits B virus HBx protein sensitizes cells to apoptotic killing by tumor necrosis factor α. Proc. Natl. Acad. Sci. USA. 1997;94:8744–8749. doi: 10.1073/pnas.94.16.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Terradillos O, Pollicino T, Lecoeur H, Tripodi M, Gougeon ML, Tiollais P, Buendia MA. p53-independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene. 1998;17:2115–2123. doi: 10.1038/sj.onc.1202432. [DOI] [PubMed] [Google Scholar]
- [78].Shintani Y, Yotsuyanagi H, Moriya K, Fujie H, Tsutsumi T, Kanegae Y, Kimura S, Saito I, Koike K. Induction of apoptosis after switch-on of the hepatitis B virus X gene mediated by the Cre/loxP recombination system. J. Gen. Virol. 1999;80:3257–3265. doi: 10.1099/0022-1317-80-12-3257. [DOI] [PubMed] [Google Scholar]
- [79].Shirakata Y, Koike K. Hepatitis B virus X protein induces cell death by causing loss of mitochondrial membrane potential. J Biol Chem. 2003;278:22071–22078. doi: 10.1074/jbc.M301606200. [DOI] [PubMed] [Google Scholar]
- [80].Takada S, Shirakata Y, Kaneniwa N, Koike K. Association of hepatitis B virus X protein with mitochondria causes mitochondrial aggregation at the nuclear periphery, leading to cell death. Oncogene. 1999;18:6965–6973. doi: 10.1038/sj.onc.1203188. [DOI] [PubMed] [Google Scholar]
- [81].Kim S, Kim HY, Lee S, Kim SW, Sohn S, Kim K, Cho H. Hepatitis B virus x protein induces perinuclear mitochondrial clustering in microtubule- and Dynein-dependent manners. J Virol. 2007;81:1714–1726. doi: 10.1128/JVI.01863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Su F, Theodosis C, Schneider R. Role of NF-κB and myc proteins in apoptosis induced by hepatitis B virus HBx protein. J. Virol. 2001;75:215–225. doi: 10.1128/JVI.75.1.215-225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kim KH, Seong BL. Pro-apoptotic function of HBV X protein is mediated by interaction with c-FLIP and enhancement of death-inducing signal. Embo J. 2003;22:2104–2116. doi: 10.1093/emboj/cdg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Karin M, Lawrence T, Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- [85].Madden CR, Slagle BL. Stimulation of cellular proliferation by hepatitis B virus X protein. Dis Markers. 2001;17:153–157. doi: 10.1155/2001/571254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Bouchard M, Giannakopoulos S, Wang EH, Tanese N, Schneider RJ. Hepatitis B virus HBx protein activation of cyclin A-cyclin-dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J Virol. 2001;75:4247–4257. doi: 10.1128/JVI.75.9.4247-4257.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lee S, Tarn C, Wang WH, Chen S, Hullinger RL, Andrisani OM. Hepatitis B virus X protein differentially regulates cell cycle progression in X-transforming versus nontransforming hepatocyte (AML12) cell lines. J Biol Chem. 2002;277:8730–8740. doi: 10.1074/jbc.M108025200. [DOI] [PubMed] [Google Scholar]
- [88].Hodgson AJ, Keasler VV, Slagle BL. Premature cell cycle entry induced by hepatitis B virus regulatory HBx protein during compensatory liver regeneration. Cancer Res. 2008;68:10341–10348. doi: 10.1158/0008-5472.CAN-08-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Tralhao J, Roudier J, Morosan S, Giannini C, Tu H, Goulenok C, Carnot F, Zavala F, Joulin V, Kremsdorf D, Brechot C. Paracrine in vivo inhibitory effects of hepatitis B virus X protein (HBx) on liver proliferation: an alternative mechanism of HBx-related pathogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:6991–6996. doi: 10.1073/pnas.092657699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wu BK, Li CC, Chen HJ, Chang JL, Jeng KS, Chou CK, Hsu MT, Tsai TF. Blocking of G1/S transition and cell death in the regenerating liver of Hepatitis B virus X protein transgenic mice. Biochem Biophys Res Commun. 2006;340:916–928. doi: 10.1016/j.bbrc.2005.12.089. [DOI] [PubMed] [Google Scholar]
- [91].Qiao L, Leach K, McKinstry R, Gilfor D, Yacoub A, Park J-S, Grant S, Hylemon P, Fisher P, Dent P. Hepatitis B virus X protein increases expression of p21Cip-1/WAF1/MDA6 and p27Kip-1 in primary mouse hepatocytes, leading to a reduced cell cycle progression. Hepatology. 2001;34:906–917. doi: 10.1053/jhep.2001.28886. [DOI] [PubMed] [Google Scholar]
- [92].Gearhart TL, Bouchard MJ. Replication of the hepatitis B virus requires a calcium-dependent HBx-induced G1 phase arrest of hepatocytes. Virology. 2010;407:14–25. doi: 10.1016/j.virol.2010.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Cohen D, Adamovich Y, Reuven N, Shaul Y. Hepatitis B virus activates deoxynucleotide synthesis in nondividing hepatocytes by targeting the R2 gene. Hepatology. 2010;51:1538–1546. doi: 10.1002/hep.23519. [DOI] [PubMed] [Google Scholar]
- [94].Koike K. Hepatitis B virus X gene is implicated in liver carcinogenesis. Cancer Lett. 2009;286:60–68. doi: 10.1016/j.canlet.2009.04.010. [DOI] [PubMed] [Google Scholar]
- [95].Kim C, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;353:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- [96].Yu DY, Moon HB, Son JK, Jeong S, Yu SL, Yoon H, Han YM, Lee CS, Park JS, Lee CH, Hyun BH, Murakami S, Lee KK. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J Hepatol. 1999;31:123–132. doi: 10.1016/s0168-8278(99)80172-x. [DOI] [PubMed] [Google Scholar]
- [97].Slagle BL, Lee TH, Medina D, Finegold MJ, Butel JS. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol Carcinog. 1996;15:261–269. doi: 10.1002/(SICI)1098-2744(199604)15:4<261::AID-MC3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- [98].Terradillos O, Billet O, Renard CA, R. L, Molina T, Briand P, Buendia M-A. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- [99].Madden C, Finegold M, Slagle B. Hepatitis B virus X protein acts a a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J. Virol. 2001;75:3851–3858. doi: 10.1128/JVI.75.8.3851-3858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Becker S, Lee T-H, Butel J, Slagle B. Hepatitis B virus X protein interferes with cellular DNA repair. J. Virol. 1998;72:266–272. doi: 10.1128/jvi.72.1.266-272.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- [102].Recommendations from the National Institutes of Health consensus development conference statement: management of hepatitis C: 2002. Hepatology. 2002;36:1039. doi: 10.1002/hep.510360502. [DOI] [PubMed] [Google Scholar]
- [103].Micallef JM, Kaldor JM, Dore GJ. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J Viral Hepat. 2006;13:34–41. doi: 10.1111/j.1365-2893.2005.00651.x. [DOI] [PubMed] [Google Scholar]
- [104].Alter MJ. Epidemiology of hepatitis C in the West. Semin Liver Dis. 1995;15:5–14. doi: 10.1055/s-2007-1007259. [DOI] [PubMed] [Google Scholar]
- [105].Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y, et al. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Chisari FV. Unscrambling hepatitis C virus-host interactions. Nature. 2005;436:930–932. doi: 10.1038/nature04076. [DOI] [PubMed] [Google Scholar]
- [107].Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, Rouille Y. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J Virol. 2006;80:6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Helle F, Dubuisson J. Hepatitis C virus entry into host cells. Cell Mol Life Sci. 2008;65:100–112. doi: 10.1007/s00018-007-7291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Suzuki T, Aizaki H, Murakami K, Shoji I, Wakita T. Molecular biology of hepatitis C virus. J Gastroenterol. 2007;42:411–423. doi: 10.1007/s00535-007-2030-3. [DOI] [PubMed] [Google Scholar]
- [111].Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, Hahn YS, Rice CM, Dubuisson J. Formation of native hepatitis C virus glycoprotein complexes. J Virol. 1997;71:697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Brohm C, Steinmann E, Friesland M, Lorenz IC, Patel A, Penin F, Bartenschlager R, Pietschmann T. Characterization of determinants important for hepatitis C virus p7 function in morphogenesis by using trans-complementation. J Virol. 2009;83:11682–11693. doi: 10.1128/JVI.00691-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Yi M, Ma Y, Yates J, Lemon SM. Trans-complementation of an NS2 defect in a late step in hepatitis C virus (HCV) particle assembly and maturation. PLoS Pathog. 2009;5:e1000403. doi: 10.1371/journal.ppat.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Beran RK, Lindenbach BD, Pyle AM. The NS4A protein of hepatitis C virus promotes RNA-coupled ATP hydrolysis by the NS3 helicase. J Virol. 2009;83:3268–3275. doi: 10.1128/JVI.01849-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Foy E, Li K, Wang C, Sumpter R, Jr., Ikeda M, Lemon SM, Gale M., Jr. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- [116].Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- [117].Behrens SE, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- [118].Timm J, Roggendorf M. Sequence diversity of hepatitis C virus: implications for immune control and therapy. World J Gastroenterol. 2007;13:4808–4817. doi: 10.3748/wjg.v13.i36.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Lawson A. Hepatitis C virus-infected patients with a persistently normal alanine aminotransferase: do they exist and is this really a group with mild disease? J Viral Hepat. 2010;17:51–58. doi: 10.1111/j.1365-2893.2009.01148.x. [DOI] [PubMed] [Google Scholar]
- [120].Pradat P, Voirin N, Tillmann HL, Chevallier M, Trepo C. Progression to cirrhosis in hepatitis C patients: an age-dependent process. Liver Int. 2007;27:335–339. doi: 10.1111/j.1478-3231.2006.01430.x. [DOI] [PubMed] [Google Scholar]
- [121].Kumar D, Farrell GC, Kench J, George J. Hepatic steatosis and the risk of hepatocellular carcinoma in chronic hepatitis C. J Gastroenterol Hepatol. 2005;20:1395–1400. doi: 10.1111/j.1440-1746.2005.04007.x. [DOI] [PubMed] [Google Scholar]
- [122].Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127:S87–96. doi: 10.1053/j.gastro.2004.09.020. [DOI] [PubMed] [Google Scholar]
- [123].Williams MJ, Lang-Lenton M. Progression of initially mild hepatic fibrosis in patients with chronic hepatitis C infection. J Viral Hepat. 2010 doi: 10.1111/j.1365-2893.2009.01262.x. [DOI] [PubMed] [Google Scholar]
- [124].Potthoff A, Manns MP, Wedemeyer H. Treatment of HBV/HCV coinfection. Expert Opin Pharmacother. 2010;11:919–928. doi: 10.1517/14656561003637659. [DOI] [PubMed] [Google Scholar]
- [125].Clifford GM, Rickenbach M, Polesel J, Dal Maso L, Steffen I, Ledergerber B, Rauch A, Probst-Hensch NM, Bouchardy C, Levi F, Franceschi S. Influence of HIV-related immunodeficiency on the risk of hepatocellular carcinoma. AIDS. 2008;22:2135–2141. doi: 10.1097/QAD.0b013e32831103ad. [DOI] [PubMed] [Google Scholar]
- [126].Bengsch B, Thimme R, Blum HE. Role of host genetic factors in the outcome of hepatitis C virus infection. Viruses. 2009;1:104–105. doi: 10.3390/v1020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].McGivern DR, Lemon SM. Tumor suppressors, chromosomal instability, and hepatitis C virus-associated liver cancer. Annu Rev Pathol. 2009;4:399–415. doi: 10.1146/annurev.pathol.4.110807.092202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Fujita N, Sugimoto R, Ma N, Tanaka H, Iwasa M, Kobayashi Y, Kawanishi S, Watanabe S, Kaito M, Takei Y. Comparison of hepatic oxidative DNA damage in patients with chronic hepatitis B and C. J Viral Hepat. 2008;15:498–507. doi: 10.1111/j.1365-2893.2008.00972.x. [DOI] [PubMed] [Google Scholar]
- [129].Joyce MA, Walters KA, Lamb SE, Yeh MM, Zhu LF, Kneteman N, Doyle JS, Katze MG, Tyrrell DL. HCV induces oxidative and ER stress, and sensitizes infected cells to apoptosis in SCID/Alb-uPA mice. PLoS Pathog. 2009;5:e1000291. doi: 10.1371/journal.ppat.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Swietek K, Juszczyk J. Reduced glutathione concentration in erythrocytes of patients with acute and chronic viral hepatitis. J Viral Hepat. 1997;4:139–141. doi: 10.1111/j.1365-2893.1997.tb00217.x. [DOI] [PubMed] [Google Scholar]
- [131].Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065–1067. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- [132].Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366–375. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- [133].Robinson LC, Marchant JS. Enhanced Ca2+ leak from ER Ca2+ stores induced by hepatitis C NS5A protein. Biochem Biophys Res Commun. 2008;368:593–599. doi: 10.1016/j.bbrc.2008.01.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Park CY, Choi SH, Kang SM, Kang JI, Ahn BY, Kim H, Jung G, Choi KY, Hwang SB. Nonstructural 5A protein activates beta-catenin signaling cascades: implication of hepatitis C virus-induced liver pathogenesis. J Hepatol. 2009;51:853–864. doi: 10.1016/j.jhep.2009.06.026. [DOI] [PubMed] [Google Scholar]
- [135].Milward A, Mankouri J, Harris M. Hepatitis C virus NS5A protein interacts with beta-catenin and stimulates its transcriptional activity in a phosphoinositide-3 kinase-dependent fashion. J Gen Virol. 2010;91:373–381. doi: 10.1099/vir.0.015305-0. [DOI] [PubMed] [Google Scholar]
- [136].Tsai WL, Chung RT. Viral hepatocarcinogenesis. Oncogene. 2010;29:2309–2324. doi: 10.1038/onc.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Lonardo A, Adinolfi LE, Loria P, Carulli N, Ruggiero G, Day CP. Steatosis and hepatitis C virus: mechanisms and significance for hepatic and extrahepatic disease. Gastroenterology. 2004;126:586–597. doi: 10.1053/j.gastro.2003.11.020. [DOI] [PubMed] [Google Scholar]
- [138].Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908–918. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Kew MC. Hepatic iron overload and hepatocellular carcinoma. Cancer Lett. 2009;286:38–43. doi: 10.1016/j.canlet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- [140].Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Fujita N, Sugimoto R, Motonishi S, Tomosugi N, Tanaka H, Takeo M, Iwasa M, Kobayashi Y, Hayashi H, Kaito M, Takei Y. Patients with chronic hepatitis C achieving a sustained virological response to peginterferon and ribavirin therapy recover from impaired hepcidin secretion. J Hepatol. 2008;49:702–710. doi: 10.1016/j.jhep.2008.05.014. [DOI] [PubMed] [Google Scholar]
- [142].Bureau C, Bernad J, Chaouche N, Orfila C, Beraud M, Gonindard C, Alric L, Vinel JP, Pipy B. Nonstructural 3 protein of hepatitis C virus triggers an oxidative burst in human monocytes via activation of NADPH oxidase. J Biol Chem. 2001;276:23077–23083. doi: 10.1074/jbc.M100698200. [DOI] [PubMed] [Google Scholar]
- [143].Gong G, Waris G, Tanveer R, Siddiqui A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc Natl Acad Sci U S A. 2001;98:9599–9604. doi: 10.1073/pnas.171311298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Poynard T, Ratziu V, McHutchison J, Manns M, Goodman Z, Zeuzem S, Younossi Z, Albrecht J. Effect of treatment with peginterferon or interferon alfa-2b and ribavirin on steatosis in patients infected with hepatitis C. Hepatology. 2003;38:75–85. doi: 10.1053/jhep.2003.50267. [DOI] [PubMed] [Google Scholar]
- [145].Leandro G, Mangia A, Hui J, Fabris P, Rubbia-Brandt L, Colloredo G, Adinolfi LE, Asselah T, Jonsson JR, Smedile A, Terrault N, Pazienza V, Giordani MT, Giostra E, Sonzogni A, Ruggiero G, Marcellin P, Powell EE, George J, Negro F. Relationship between steatosis, inflammation, and fibrosis in chronic hepatitis C: a meta-analysis of individual patient data. Gastroenterology. 2006;130:1636–1642. doi: 10.1053/j.gastro.2006.03.014. [DOI] [PubMed] [Google Scholar]
- [146].Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Ishibashi K, Matsuura Y, Miyamura T, Koike K. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J Gen Virol. 1997;78(Pt 7):1527–1531. doi: 10.1099/0022-1317-78-7-1527. [DOI] [PubMed] [Google Scholar]
- [147].Mirandola S, Bowman D, Hussain MM, Alberti A. Hepatic steatosis in hepatitis C is a storage disease due to HCV interaction with microsomal triglyceride transfer protein (MTP) Nutr Metab (Lond) 2010;7:13. doi: 10.1186/1743-7075-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Park CY, Jun HJ, Wakita T, Cheong JH, Hwang SB. Hepatitis C virus nonstructural 4B protein modulates sterol regulatory element-binding protein signaling via the AKT pathway. J Biol Chem. 2009;284:9237–9246. doi: 10.1074/jbc.M808773200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Dharancy S, Malapel M, Perlemuter G, Roskams T, Cheng Y, Dubuquoy L, Podevin P, Conti F, Canva V, Philippe D, Gambiez L, Mathurin P, Paris JC, Schoonjans K, Calmus Y, Pol S, Auwerx J, Desreumaux P. Impaired expression of the peroxisome proliferator-activated receptor alpha during hepatitis C virus infection. Gastroenterology. 2005;128:334–342. doi: 10.1053/j.gastro.2004.11.016. [DOI] [PubMed] [Google Scholar]
- [150].Shintani Y, Fujie H, Miyoshi H, Tsutsumi T, Tsukamoto K, Kimura S, Moriya K, Koike K. Hepatitis C virus infection and diabetes: direct involvement of the virus in the development of insulin resistance. Gastroenterology. 2004;126:840–848. doi: 10.1053/j.gastro.2003.11.056. [DOI] [PubMed] [Google Scholar]
- [151].Koike K. Hepatitis C as a metabolic disease: Implication for the pathogenesis of NASH. Hepatol Res. 2005;33:145–150. doi: 10.1016/j.hepres.2005.09.023. [DOI] [PubMed] [Google Scholar]
- [152].Munakata T, Liang Y, Kim S, McGivern DR, Huibregtse J, Nomoto A, Lemon SM. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 2007;3:1335–1347. doi: 10.1371/journal.ppat.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].McGivern DR, Villanueva RA, Chinnaswamy S, Kao CC, Lemon SM. Impaired replication of hepatitis C virus containing mutations in a conserved NS5B retinoblastoma protein-binding motif. J Virol. 2009;83:7422–7433. doi: 10.1128/JVI.00262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Madhoun MF, Fazili J, Bright BC, Bader T, Roberts DN, Bronze MS. Hepatitis C prevalence in patients with hepatocellular carcinoma without cirrhosis. Am J Med Sci. 2010;339:169–173. doi: 10.1097/MAJ.0b013e3181c4af27. [DOI] [PubMed] [Google Scholar]
- [155].Wang AG, Lee DS, Moon HB, Kim JM, Cho KH, Choi SH, Ha HL, Han YH, Kim DG, Hwang SB, Yu DY. Non-structural 5A protein of hepatitis C virus induces a range of liver pathology in transgenic mice. J Pathol. 2009;219:253–262. doi: 10.1002/path.2592. [DOI] [PubMed] [Google Scholar]
- [156].Majumder M, Steele R, Ghosh AK, Zhou XY, Thornburg L, Ray R, Phillips NJ, Ray RB. Expression of hepatitis C virus non-structural 5A protein in the liver of transgenic mice. FEBS Lett. 2003;555:528–532. doi: 10.1016/s0014-5793(03)01337-1. [DOI] [PubMed] [Google Scholar]
- [157].Thomson M, Nascimbeni M, Havert MB, Major M, Gonzales S, Alter H, Feinstone SM, Murthy KK, Rehermann B, Liang TJ. The clearance of hepatitis C virus infection in chimpanzees may not necessarily correlate with the appearance of acquired immunity. J Virol. 2003;77:862–870. doi: 10.1128/JVI.77.2.862-870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Branch AD, Stump DD, Gutierrez JA, Eng F, Walewski JL. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin Liver Dis. 2005;25:105–117. doi: 10.1055/s-2005-864786. [DOI] [PubMed] [Google Scholar]
- [159].Lerat H, Honda M, Beard MR, Loesch K, Sun J, Yang Y, Okuda M, Gosert R, Xiao SY, Weinman SA, Lemon SM. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122:352–365. doi: 10.1053/gast.2002.31001. [DOI] [PubMed] [Google Scholar]
- [160].Roingeard P, Hourioux C. Hepatitis C virus core protein, lipid droplets and steatosis. J Viral Hepat. 2008;15:157–164. doi: 10.1111/j.1365-2893.2007.00953.x. [DOI] [PubMed] [Google Scholar]
- [161].Ray RB, Steele R. Separate domains of MBP-1 involved in c-myc promoter binding and growth suppressive activity. Gene. 1997;186:175–180. doi: 10.1016/s0378-1119(96)00693-2. [DOI] [PubMed] [Google Scholar]
- [162].Alisi A, Giambartolomei S, Cupelli F, Merlo P, Fontemaggi G, Spaziani A, Balsano C. Physical and functional interaction between HCV core protein and the different p73 isoforms. Oncogene. 2003;22:2573–2580. doi: 10.1038/sj.onc.1206333. [DOI] [PubMed] [Google Scholar]
- [163].Cho J, Baek W, Yang S, Chang J, Sung YC, Suh M. HCV core protein modulates Rb pathway through pRb down-regulation and E2F-1 up-regulation. Biochim Biophys Acta. 2001;1538:59–66. doi: 10.1016/s0167-4889(00)00137-3. [DOI] [PubMed] [Google Scholar]
- [164].Uchida M, Hino N, Yamanaka T, Fukushima H, Imanishi T, Uchiyama Y, Kodama T, Doi T. Hepatitis C virus core protein binds to a C-terminal region of NS5B RNA polymerase. Hepatol Res. 2002;22:297–306. doi: 10.1016/s1386-6346(02)00005-0. [DOI] [PubMed] [Google Scholar]
- [165].Joo M, Hahn YS, Kwon M, Sadikot RT, Blackwell TS, Christman JW. Hepatitis C virus core protein suppresses NF-kappaB activation and cyclooxygenase-2 expression by direct interaction with IkappaB kinase beta. J Virol. 2005;79:7648–7657. doi: 10.1128/JVI.79.12.7648-7657.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Tai DI, Tsai SL, Chang YH, Huang SN, Chen TC, Chang KS, Liaw YF. Constitutive activation of nuclear factor kappaB in hepatocellular carcinoma. Cancer. 2000;89:2274–2281. [PubMed] [Google Scholar]
- [167].Hayashi J, Aoki H, Kajino K, Moriyama M, Arakawa Y, Hino O. Hepatitis C virus core protein activates the MAPK/ERK cascade synergistically with tumor promoter TPA, but not with epidermal growth factor or transforming growth factor α. Hepatology. 2000;32:958–961. doi: 10.1053/jhep.2000.19343. [DOI] [PubMed] [Google Scholar]
- [168].Fukutomi T, Zhou Y, Kawai S, Eguchi H, Wands JR, Li J. Hepatitis C virus core protein stimulates hepatocyte growth: correlation with upregulation of wnt-1 expression. Hepatology. 2005;41:1096–1105. doi: 10.1002/hep.20668. [DOI] [PubMed] [Google Scholar]
- [169].Shin JY, Hur W, Wang JS, Jang JW, Kim CW, Bae SH, Jang SK, Yang SH, Sung YC, Kwon OJ, Yoon SK. HCV core protein promotes liver fibrogenesis via up-regulation of CTGF with TGF-beta1. Exp Mol Med. 2005;37:138–145. doi: 10.1038/emm.2005.19. [DOI] [PubMed] [Google Scholar]
- [170].Neuman MG, Benhamou JP, Bourliere M, Ibrahim A, Malkiewicz I, Asselah T, Martinot-Peignoux M, Shear NH, Katz GG, Akremi R, Benali S, Boyer N, Lecomte L, Le Breton V, Le Guludec G, Marcellin P. Serum tumour necrosis factor-alpha and transforming growth factor-beta levels in chronic hepatitis C patients are immunomodulated by therapy. Cytokine. 2002;17:108–117. doi: 10.1006/cyto.2001.0997. [DOI] [PubMed] [Google Scholar]
- [171].Pavio N, Battaglia S, Boucreux D, Arnulf B, Sobesky R, Hermine O, Brechot C. Hepatitis C virus core variants isolated from liver tumor but not from adjacent non-tumor tissue interact with Smad3 and inhibit the TGF-beta pathway. Oncogene. 2005;24:6119–6132. doi: 10.1038/sj.onc.1208749. [DOI] [PubMed] [Google Scholar]
- [172].Taniguchi H, Kato N, Otsuka M, Goto T, Yoshida H, Shiratori Y, Omata M. Hepatitis C virus core protein upregulates transforming growth factor-beta 1 transcription. J Med Virol. 2004;72:52–59. doi: 10.1002/jmv.10545. [DOI] [PubMed] [Google Scholar]
- [173].Bataller R, Paik YH, Lindquist JN, Lemasters JJ, Brenner DA. Hepatitis C virus core and nonstructural proteins induce fibrogenic effects in hepatic stellate cells. Gastroenterology. 2004;126:529–540. doi: 10.1053/j.gastro.2003.11.018. [DOI] [PubMed] [Google Scholar]
- [174].Bach N, Thung SN, Schaffner F. The histological features of chronic hepatitis C and autoimmune chronic hepatitis: a comparative analysis. Hepatology. 1992;15:572–577. doi: 10.1002/hep.1840150403. [DOI] [PubMed] [Google Scholar]
- [175].Tsutsumi T, Suzuki T, Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Matsuura Y, Kimura S, Koike K, Miyamura T. Alteration of intrahepatic cytokine expression and AP-1 activation in transgenic mice expressing hepatitis C virus core protein. Virology. 2002;304:415–424. doi: 10.1006/viro.2002.1702. [DOI] [PubMed] [Google Scholar]
- [176].Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- [177].Pavio N, Romano PR, Graczyk TM, Feinstone SM, Taylor DR. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2alpha kinase PERK. J Virol. 2003;77:3578–3585. doi: 10.1128/JVI.77.6.3578-3585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Zhao LJ, Wang L, Ren H, Cao J, Li L, Ke JS, Qi ZT. Hepatitis C virus E2 protein promotes human hepatoma cell proliferation through the MAPK/ERK signaling pathway via cellular receptors. Exp Cell Res. 2005;305:23–32. doi: 10.1016/j.yexcr.2004.12.024. [DOI] [PubMed] [Google Scholar]
- [179].Koike K, Moriya K, Ishibashi K, Matsuura Y, Suzuki T, Saito I, Iino S, Kurokawa K, Miyamura T. Expression of hepatitis C virus envelope proteins in transgenic mice. J Gen Virol. 1995;76(Pt 12):3031–3038. doi: 10.1099/0022-1317-76-12-3031. [DOI] [PubMed] [Google Scholar]
- [180].Dustin LB, Rice CM. Flying under the radar: the immunobiology of hepatitis C. Annu Rev Immunol. 2007;25:71–99. doi: 10.1146/annurev.immunol.25.022106.141602. [DOI] [PubMed] [Google Scholar]
- [181].Dumoulin FL, von dem Bussche A, Li J, Khamzina L, Wands JR, Sauerbruch T, Spengler U. Hepatitis C virus NS2 protein inhibits gene expression from different cellular and viral promoters in hepatic and nonhepatic cell lines. Virology. 2003;305:260–266. doi: 10.1006/viro.2002.1701. [DOI] [PubMed] [Google Scholar]
- [182].Erdtmann L, Franck N, Lerat H, Le Seyec J, Gilot D, Cannie I, Gripon P, Hibner U, Guguen-Guillouzo C. The hepatitis C virus NS2 protein is an inhibitor of CIDE-B-induced apoptosis. J Biol Chem. 2003;278:18256–18264. doi: 10.1074/jbc.M209732200. [DOI] [PubMed] [Google Scholar]
- [183].Smirnova IS, Aksenov ND, Vonsky MS, Isaguliants MG. Different transformation pathways of murine fibroblast NIH 3T3 cells by hepatitis C virus core and NS3 proteins. Cell Biol Int. 2006;30:915–919. doi: 10.1016/j.cellbi.2006.06.020. [DOI] [PubMed] [Google Scholar]
- [184].Zemel R, Gerechet S, Greif H, Bachmatove L, Birk Y, Golan-Goldhirsh A, Kunin M, Berdichevsky Y, Benhar I, Tur-Kaspa R. Cell transformation induced by hepatitis C virus NS3 serine protease. J Viral Hepat. 2001;8:96–102. doi: 10.1046/j.1365-2893.2001.00283.x. [DOI] [PubMed] [Google Scholar]
- [185].Kwun HJ, Jung EY, Ahn JY, Lee MN, Jang KL. p53-dependent transcriptional repression of p21(waf1) by hepatitis C virus NS3. J Gen Virol. 2001;82:2235–2241. doi: 10.1099/0022-1317-82-9-2235. [DOI] [PubMed] [Google Scholar]
- [186].Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Gouttenoire J, Penin F, Moradpour D. Hepatitis C virus nonstructural protein 4B: a journey into unexplored territory. Rev Med Virol. 2010;20:117–129. doi: 10.1002/rmv.640. [DOI] [PubMed] [Google Scholar]
- [188].Wang AG, Moon HB, Kim JM, Hwang SB, Yu DY, Lee DS. Expression of hepatitis C virus nonstructural 4B in transgenic mice. Exp Mol Med. 2006;38:241–246. doi: 10.1038/emm.2006.29. [DOI] [PubMed] [Google Scholar]
- [189].Park JS, Yang JM, Min MK. Hepatitis C virus nonstructural protein NS4B transforms NIH3T3 cells in cooperation with the Ha-ras oncogene. Biochem Biophys Res Commun. 2000;267:581–587. doi: 10.1006/bbrc.1999.1999. [DOI] [PubMed] [Google Scholar]
- [190].Einav S, Sklan EH, Moon HM, Gehrig E, Liu P, Hao Y, Lowe AW, Glenn JS. The nucleotide binding motif of hepatitis C virus NS4B can mediate cellular transformation and tumor formation without Ha-ras co-transfection. Hepatology. 2008;47:827–835. doi: 10.1002/hep.22108. [DOI] [PubMed] [Google Scholar]
- [191].Gimenez-Barcons M, Wang C, Chen M, Sanchez-Tapias JM, Saiz JC, Gale M., Jr. The oncogenic potential of hepatitis C virus NS5A sequence variants is associated with PKR regulation. J Interferon Cytokine Res. 2005;25:152–164. doi: 10.1089/jir.2005.25.152. [DOI] [PubMed] [Google Scholar]
- [192].Chung YL, Sheu ML, Yen SH. Hepatitis C virus NS5A as a potential viral Bcl-2 homologue interacts with Bax and inhibits apoptosis in hepatocellular carcinoma. Int J Cancer. 2003;107:65–73. doi: 10.1002/ijc.11303. [DOI] [PubMed] [Google Scholar]
- [193].Mankouri J, Dallas ML, Hughes ME, Griffin SD, Macdonald A, Peers C, Harris M. Suppression of a pro-apoptotic K+ channel as a mechanism for hepatitis C virus persistence. Proc Natl Acad Sci U S A. 2009;106:15903–15908. doi: 10.1073/pnas.0906798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- [195].Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: tiny players in a big field. Immunity. 2007;26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- [196].Wang WX, Wilfred BR, Baldwin DA, Isett RB, Ren N, Stromberg A, Nelson PT. Focus on RNA isolation: obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim Biophys Acta. 2008;1779:749–757. doi: 10.1016/j.bbagrm.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- [199].Fornari F, Gramantieri L, Giovannini C, Veronese A, Ferracin M, Sabbioni S, Calin GA, Grazi GL, Croce CM, Tavolari S, Chieco P, Negrini M, Bolondi L. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761–5767. doi: 10.1158/0008-5472.CAN-08-4797. [DOI] [PubMed] [Google Scholar]
- [200].Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- [201].Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- [202].Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- [203].Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [206].Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, Croce CM, Bolondi L, Negrini M. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- [207].Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- [208].Wang Y, Kato N, Jazag A, Dharel N, Otsuka M, Taniguchi H, Kawabe T, Omata M. Hepatitis C virus core protein is a potent inhibitor of RNA silencing-based antiviral response. Gastroenterology. 2006;130:883–892. doi: 10.1053/j.gastro.2005.12.028. [DOI] [PubMed] [Google Scholar]
- [209].Lipardi C, Wei Q, Paterson BM. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell. 2001;107:297–307. doi: 10.1016/s0092-8674(01)00537-2. [DOI] [PubMed] [Google Scholar]
- [210].Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow P, Chung AY, Jooi LL, Lee CG. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol. 2010;53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- [211].Yang L, Ma Z, Wang D, Zhao W, Chen L, Wang G. MicroRNA-602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer Biol Ther. 2010;9 doi: 10.4161/cbt.9.10.11440. [DOI] [PubMed] [Google Scholar]
- [212].Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- [213].Qiu L, Fan H, Jin W, Zhao B, Wang Y, Ju Y, Chen L, Chen Y, Duan Z, Meng S. miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem Biophys Res Commun. 2010;398:771–777. doi: 10.1016/j.bbrc.2010.07.021. [DOI] [PubMed] [Google Scholar]
- [214].Bressac B, Galvin K, Liang T, Isselbacher K, J. W, Ozturk M. Abnormal structure and expression of p53 gene in human hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA. 1990;87:1973–1977. doi: 10.1073/pnas.87.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]